ABSTRACT
MicroRNA (miRNA)-mediated repression controls expression of more than half of protein-coding genes in metazoan animals. Translation repression is associated with target mRNA degradation initiated by decapping and deadenylation of the repressed mRNAs. Earlier evidence suggests the endoplasmic reticulum (ER) as the site where microRNPs (miRNPs) interact with their targets before translation repression sets in, but the subcellular location of subsequent degradation of miRNA-repressed messages is largely unidentified. Here, we explore the subcellular distribution of essential components of degradation machineries of miRNA-targeted mRNAs. We have noted that interaction of target mRNAs with AGO2 protein on the ER precedes the relocalization of repressed messages to multivesicular bodies (MVBs). The repressed messages subsequently get deadenylated, lose their interaction with AGO2, and become decapped. Blocking maturation of endosomes to late endosome and MVBs by targeting the endosomal protein HRS uncouples miRNA-mediated translation repression from target RNA degradation. HRS is also targeted by the intracellular parasite Leishmania donovani, which curtails the HRS level in infected cells to prevent uncoupling of mRNA-AGO2 interaction, preventing degradation of translationally repressed messages, and thus stops recycling of miRNPs preengaged in repression.
KEYWORDS: RNA binding proteins, RNA stability, mRNA, mRNA degradation, translational control
INTRODUCTION
MicroRNAs (miRNAs) are 20- to 22-nucleotide (nt)-long noncoding RNAs that regulate gene expression by imperfectly base pairing with the 3′ untranslated region (3′UTR) of target mRNAs, resulting in translation repression and degradation of the target messages. AGO2, miRNAs, and target mRNAs have all been found to localize to processing bodies (P bodies), the cytoplasmic sites for mRNA degradation (1, 2). However, it has been observed that visible P bodies are not required for RNA interference (RNAi), and their formation is a consequence rather than a cause of silencing (3). Hence, the exact subcellular sites where miRNA interaction and translational repression of target mRNA happen have remained obscure despite ample conceptual advancement in understanding the mechanisms of miRNA-mediated gene regulation. Recently, the rough endoplasmic reticulum (rER) membrane has been proposed to be the central nucleation site of miRNA/small interfering RNA (siRNA) assembly and to encounter target mRNA (4). The role of ER in miRNA function was strengthened further when the requirement of Argonaute1 interaction with AMP1, an ER integral membrane protein, in miRNA-mediated translation inhibition was reported in Arabidopsis (5).
Do the other organelles have any role to play in miRNA-mediated gene regulatory processes? Internalization and sorting of protein cargos are ensured by regulated endocytosis, and the endosomal pathway sorts the proteins to their final destinations. Maturation of endosomes is a multistep process. In early endosomes, molecules targeted for lysosomal degradation are sorted in a ubiquitin-dependent manner and segregated from those that are to be recycled to the cell surface. Normally, cargo from early endosomes arrives at the late endosome on their way to the lysosome (6). Assisted by proteins of the ESCRT complex, the endosomal limiting membrane invaginates into the interior of endosome to form structures replete with intralumenal vesicles called multivesicular endosomes (MVEs) or multivesicular bodies (MVBs). These MVBs and late endosomes fuse with lysosomes, resulting in degradation of the cargo, or they can fuse with the cell membrane (exocytosis) to deliver the luminal vesicles as exosomes in the extracellular medium (7). MVBs therefore act as a transport intermediate between early and late endosomes.
Interestingly, perturbing the endosomal trafficking pathway affects silencing by small RNAs. Lee et al. reported that blocking MVB formation from early endosomes inhibits silencing, whereas blocking MVB turnover or fusion with lysosomes stimulates silencing (8). Another group also reported endosomes and MVBs as sites of accumulation of miRNA-induced silencing complexes (miRISCs) (9). This suggests a possible interplay between these vesicular structures and small RNA-mediated gene regulation.
Vacuolar protein sorting (VPS) proteins that were initially identified in yeast serve as the major components of vesicular sorting. The VPS class E proteins form three endosomal sorting complexes required for transport (ESCRT-I, -II, and -III) that assemble sequentially to facilitate sorting and MVB formation. One of the VPS proteins that have not been assigned to any of the ESCRT complexes is HRS (hepatocyte growth factor-regulated tyrosine kinase substrate), whose yeast homolog is Vps27p. HRS contains an N-terminal VHS domain, a FYVE domain, two coiled-coil domains (CC1 and CC2), and a C-terminal proline- and glutamine-rich domain (10, 11). Notably, HRS interacts with ubiquitin and thereby localizes ubiquitylated cargo on HRS-containing microdomains on the endosomes. Recruitment of HRS on endosomal membranes initiates membrane targeting of ESCRT-I via recruitment of Tsg101 (a component of ESCRT-I). Assembly of ESCRT-I triggers sequential recruitment of ESCRT-II and -III. They promote inward vesiculation of the endosomal membrane to generate the MVBs. Interaction of HRS with ESCRT-I complex (more specifically, Tsg101) is crucial for inward vesiculation, as loss of ESCRT-I recruitment impairs the successive assembly of ESCRT-II and -III. Hence, knockdown of HRS leads to reduced intralumenal vesiculation and consequently impairs formation of MVBs (10, 12).
Compartmentalization of cellular processes provides a mechanism of regulation of these processes with exquisite spatial and temporal control. Currently, a model of compartmentalized protein synthesis on the ER has evolved where all mRNAs are targeted to ER membranes before translation (13, 14). However, not much is known regarding the compartmentalization of the mRNAs postrepression. Here, we report that degradation of miRNA-targeted messages is compartmentalized and is functionally coupled with the formation of MVBs. miRNA-repressed target mRNAs are enriched in MVBs/late endosomes. Interestingly, in late endosomes these target mRNAs show reduced association with AGO2, suggesting an uncoupling of repressed mRNAs from microRNPs (miRNPs) before their degradation. Furthermore, we observed that hampering MVB formation by knocking down HRS results in impaired degradation of target mRNAs, thereby substantiating the role of these vesicular structures in controlling cellular mRNA homeostasis. HRS depletion also hampers miRNP recycling from one target mRNA to a new target. The protozoan parasite Leishmania donovani, the causative agent of visceral leishmaniasis, infects macrophages and upregulates anti-inflammatory responses to establish infection. We documented a reduction in HRS protein level upon infection, which contributes to impaired miRNP recycling and enhanced expression of infection promoting interleukin-6 (IL-6).
RESULTS
miRNP components and mRNA degradation machinery show differential cellular distribution in human cells.
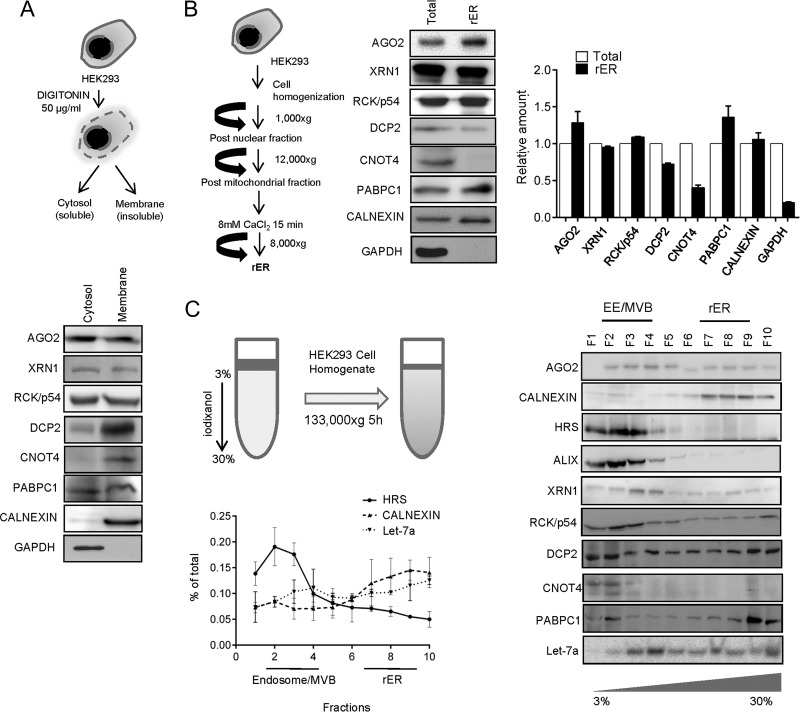
To gain insight into the subcellular localization of factors involved in miRNA-mediated repression and associated degradation of target mRNAs, we treated human HEK293 cells with the detergent digitonin to selectively release the cytosolic components and to separate them from the detergent-insoluble membranous structures. Although both AGO2 and RCK/p54, the key components required for miRNA-mediated gene repression, were found to be evenly distributed in both digitonin-soluble cytosolic and insoluble membrane fractions, DCP2 and CNOT4, required for decapping and degradation of miRNA-repressed messages, were primarily associated with the membrane fraction (Fig. 1A). XRN1 was found to be present in both fractions. PABPC1, the protein that binds the poly(A) tail of mRNAs in mammalian cells, was found to be equally distributed between two fractions (Fig. 1A).
FIG 1.
Differential distribution of miRNPs and RNA degradation components in human cells. (A) Schematic representation of digitonin fractionation in HEK293 cells. AGO2, XRN1, RCK/p54, DCP2, CNOT4, and PABPC1 were immunoblotted using cell equivalent amounts of cytosolic and membrane fractions. Calnexin and GAPDH served as membrane and cytosolic markers, respectively. (B) Isolation of microsome/rER from HEK293 cells. Western blotting was done for AGO2 (miRNP component) and degradation factors XRN1, RCK/p54, DCP2, CNOT4, and PABPC1 for protein equivalent amounts of total cell lysates and microsomal/rER fractions. Calnexin served as an rER marker. The absence of GAPDH (cytosolic marker) confirmed the purity of the isolated rER/microsome fraction. Shown are the relative amounts of proteins present in total cellular extracts and rER. (C) A cartoon representation of organelle separation by 3 to 30% iodixanol gradient (OptiPrep). Distribution of miRNP components (AGO2 and let-7a) and RNA degradation factors (XRN1, RCK/p54, ALIX, DCP2, CNOT4, and PABPC1) were shown in different fractions of iodixanol gradient (OptiPrep) by Western blotting and Northern blotting. Differential distribution of calnexin (rER marker) and HRS (MVB marker) confirmed the separation of individual organelles on the gradient. Quantification of blots have been calculated from two or more data sets and plotted.
rER has been described previously as the site where the target RNA meets the miRNPs (4). To assess the importance of rER structures in miRNA-mediated degradation of target RNAs, we analyzed the attachment of miRNP components and target RNA degradation-associated factors with isolated rER from HEK293 cells. As also reported earlier, AGO2 was found to be enriched with isolated rER (15, 16). Although PABPC1, RCK/p54, and XRN1 all showed association with the rER, the level of the decapping enzyme DCP2 was found to be relatively low on the rER, while CNOT4, a component of the mRNA deadenylation complex, was almost nondetectable in the rER-enriched fraction (Fig. 1B). To get a better understanding of the subcellular distribution of these components within human cells, we resolved the HEK293 cell homogenate on an iodixanol gradient (3 to 30% OptiPrep gradient) and analyzed the distribution pattern of these proteins in different fractions enriched for specific subcellular organelles. As evident from the presence of HRS, an early endosomal marker, fractions 2 to 4 of the above-mentioned gradient are enriched for endosomes and MVBs, while fractions 7 to 9 are enriched for calnexin, the ER marker protein. RNA degradation and deadenylation components XRN1 and CNOT4 were found to get enriched predominantly in MVB/endosome dominant fractions (Fig. 1C). AGO2, as reported earlier, was present both in rER and endosome/MVB-enriched fractions; endogenous miRNA let-7a and the decapping enzyme DCP2 were also observed to be present in endosomal as well as ER compartments. These findings suggest differential distribution of the proteins involved in mRNA degradation and repression machineries and hints at a possible compartmentalization of the components of mRNA decay machinery in human cells.
miRNA-repressed mRNAs are targeted to late endosome/MVBs postrepression.
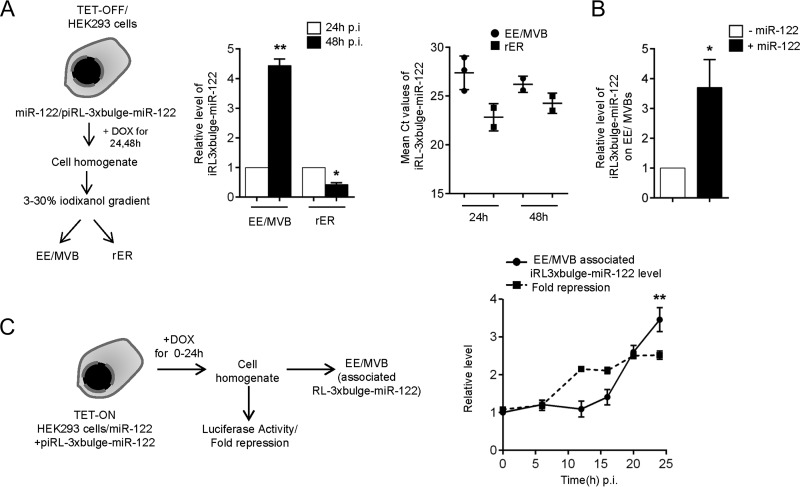
Since the key components of the mRNA degradation machinery were found to be predominantly localized to endosome/MVBs, it would be rational to monitor the distribution and association of an miRNA-targeted mRNA with these fractions to get an idea of the specific subcellular sites where miRNA-targeted mRNA degradation occurs. We used a doxycycline-inducible expression system in HEK293 cells either to shut down or express miR-122 target mRNA, RL-3xbulge-miR-122 (TET-OFF and TET-ON, respectively). Using the TET-OFF system, we tracked the residual mRNA levels in MVB and ER fractions after 24 h and 48 h of switching off the transcription of corresponding reporter mRNAs. This was done to monitor the relative distribution of an already-synthesized and repressed RL-3xbulge-miR-122 mRNA with different subcellular compartments against time. Our data indicated that the localization of RL-3xbulge-miR-122 mRNA was shifted from the rER to endosome/MVB fraction within 48 h of doxycycline treatment (Fig. 2A). However, reporter mRNA RL-3xbulge-miR-122 was found to be much less targeted to MVB in the absence of miR-122, confirming that the MVB targeting of the mRNA is dependent on the presence of its cognate miRNA (Fig. 2B).
FIG 2.
miRNA-repressed mRNAs are targeted to endosome/MVB postrepression. (A) Experimental setup. (Left) Relative level of RL-3xbulge-miR-122 was quantified in isolated MVB and ER fractions after 24 and 48 h of doxycycline treatment in TET-OFF stable HEK293 cells. (Right) Mean threshold cycle (CT) values of RL-3xbulge-miR-122 quantification have also been plotted to show absolute distribution of mRNA between the two fractions. (B) MVB targeting of an mRNA is dependent on the presence of its cognate miRNA. Relative quantification of RL-3xbulge-miR-122 in the MVB fraction in the presence of miR-122-expressing plasmids is presented. Cells not expressing miR-122 served as a control. (C) MVB targeting of mRNA occurs postrepression. A model of the experiments has been outlined. Relative luciferase expression of RL-3xbulge-miR-122 was plotted after 0, 6, 12, 16, 20, and 24 h of induction. MVB association of RL-3xbulge-miR-122 was measured quantitatively and plotted. Experiments were done in TET-ON stable HEK293 cells. In both cases, 0 h of doxycycline treatment was taken as 1. In all reverse transcriptase qPCR (RT-qPCR) experiments, 18S rRNA served as the endogenous control. For estimation of RNA and proteins, we used cell equivalent amounts in individual reactions. RT-qPCR results from three independent experiments ± standard deviations (SD) are shown, and the values of the control are normalized to 1 (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Does miRNA-mediated repression occur before mRNA targeting to MVBs or otherwise? To answer this question, repression was scored for de novo-synthesized reporter mRNAs in the tetracycline-inducible expression system (TET-ON) after 0 to 24 h of induction. We observed that the onset of repression happens around 10 h, while MVB targeting of target RNA was documented only after 16 h of induction (Fig. 2C). According to a previous report, within 4 h of induction, the de novo-synthesized mRNA should get targeted to rER (17). Therefore, following the cellular distribution of RL-3xbulge-miR-122 mRNA in a TET-ON system, we observed an enrichment of the de novo-synthesized mRNA in MVB fractions only during the late hours of induced expression. This was in contrast to the rER association and repression kinetics of induced mRNA, as enrichment and translation repression of de novo mRNAs was detected within the first few hours of its induction (Fig. 2C) (17).
Degradation of miRNA-targeted messages begins on late endosomes/MVBs.
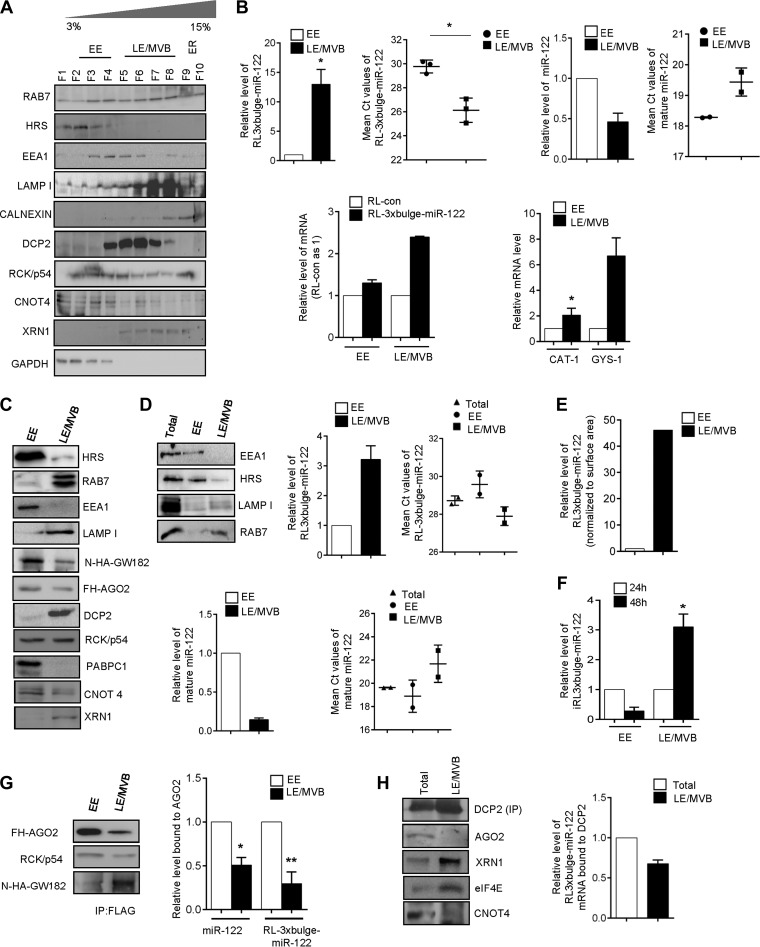
Using the 3 to 30% iodixanol gradient, the early endosomes (EE) cannot be separated from mature late endosomes (LE) and MVBs. To achieve separation, we fractionated cells using a 3 to 15% iodixanol gradient. The distribution of HRS, EEA1 (early endosome marker), RAB7, and LAMP1 (late endosome marker) in this gradient suggests fractions 2 to 4 are enriched for the early endosomes, while fractions 5 to 8 represent late endosomes and MVBs (Fig. 3A). Further confirmation was obtained from the Western blot data for respective markers done with pooled fractions 2, 3, and 4 and pooled fractions 5, 6, 7, and 8 (Fig. 3C). Fractions 2 to 4 were HRS positive, whereas fractions 5 to 8 are RAB7 positive. DCP2 and XRN1 were localized predominantly to MVBs/late endosomes, while CNOT4 localization was observed to be early endosomal (Fig. 3A).
FIG 3.
Degradation of miRNA-targeted messages occurs on late endosomes/MVBs. (A) Resolution of early and late endosomes/MVBs on 3 to 15% iodixanol gradient. Differential distribution of DCP2, RCK/p54, CNOT4, and XRN1 are shown. The distribution of HRS, EEA1, LAMP1, and RAB7 confirms the position of early and late endosomes on the 3 to 15% iodixanol gradient. GAPDH distribution confirms the absence of cytosolic contamination in LE/MVB fractions. (B) miRNA-targeted mRNA accumulates in MVB/LE. (Upper) Relative quantification of RL-3xbulge-miR-122 and miR-122 with RNA isolated from early (fractions 2, 3, and 4) and late (fractions 5, 6, 7, and 8) endosomal fractions. A cell equivalent amount of RNA was used for quantification. In each case, levels on early endosomes were taken as 1. Mean threshold cycle (CT) values have also been plotted to give an idea of the absolute distribution of the molecules. (Lower) Distribution of RL-con mRNA with no miR-122 binding site has been compared to that of RL-3xbulge-miR-122 on both early and late endosomes. In both cases, the level of RL-con was taken as 1. Distribution of endogenous miR-122 target mRNAs, CAT-1 and GYS-1, also has been shown. (C) Fractions 2, 3, and 4 and fractions 5, 6, 7, and 8 were pooled and Western blotted for miRNP and mRNA degradation components. (D) Distribution of target mRNA was reconfirmed by using protein equivalent amounts of EE and MVB/LE. Fractions 2, 3, and 4 and fractions 5, 6, 7, 8 were pooled, diluted, and ultracentrifuged at 285,000 × g to obtain pellets of EE and MVB/LE. Protein equivalent amounts of the isolated organellar pellets were used for Western blotting and RNA isolation. The distribution of EEA1, HRS, RAB7, and LAMP1 was checked to verify the purity of the fractions. RL-3xbulge-miR-122 showed enrichment in the LE/MVB fractions. (E) Surface areas of EE and LE were calculated using IMARIS software, and the RL-3xbulge-miR-122 level was normalized against organelle surface area. (F) miRNA-targeted reporter mRNA shifts from EE to LE/MVB. The relative level of RL-3xbulge-miR-122 was quantified in isolated EE and LE/MVB fractions after 24 and 48 h of doxycycline treatment in TET-OFF stable HEK293 cells. (G) FLAG-tagged AGO2 was immunoprecipitated (IP) from both early and late endosomes. Association of RCK/p54 and N-HA-tagged GW182 were measured by Western blotting. Association of miR-122 and RL-3xbulge-miR-122 was also measured and plotted. In each case, levels in early endosome were taken as 1. (H) Endogenous DCP2 was immunoprecipitated from late endosomal fractions, and association of AGO2, XRN1, eIF4E, and CNOT4 was monitored. RL-3xbulge-miR-122 association was also measured and plotted. In both cases total levels served as a control for comparison. In all RT-qPCR experiments, 18S rRNA served as the endogenous control. RT-qPCR results from three independent experiments ± SD are shown, and the values of the control were taken as 1 (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
A miRNA-repressed mRNA makes its way to endosome/MVBs postrepression (Fig. 2C). Therefore, it would be exciting to dissect which of these endosomal structures are the sites for degradation of miRNA-targeted messages. Interestingly, the distribution of target mRNA between early and late endosomal/MVB fractions was observed to be highly preferential. We observed a more than 10-fold enrichment of RL-3xbulge-miR-122 mRNAs in the LE/MVBs compared to the EE fractions, suggesting that repressed mRNAs accumulate in LE/MVBs before the degradation. Notably, the miR-122 level was lower in LE/MVBs than in early endosomes. Accumulation of mRNAs in LE/MVB fractions is also dependent on its repression by the miRNA, as a reporter mRNA without the miRNA binding sites was found to be localized less to LE/MVBs. Endogenous miR-122-targeted mRNAs, CAT-1 and GYS-1, also were enriched preferentially in LE/MVB fractions (Fig. 3B). The distribution of the protein factors involved in mRNA decay was also different between EE and LE/MVB fractions. AGO2 was less abundant in LE/MVBs, while PABPC1 was exclusively associated with EE. DCP2 and XRN1 were primarily late endosomal, while CNOT4 showed preferential localization to EE (Fig. 3C).
Notably, our inferences are based on relative enrichment of miRNA-targeted mRNA between early and late endosomal compartments. However, the surface area of early endosomes and late endosomes are different inside the cell. Taking this into consideration, the amount of target mRNA associated with individual organelles should be normalized against the surface area of EE and LE/MVBs. Using protein equivalent amounts of EE and LE/MVB fractions, we observed increased RL-3xbulge-miR-122 accumulation in LE (Fig. 3D). Additionally, we normalized the RL-3xbulge-miR-122 level against the surface area of the individual organelles and reconfirmed that target mRNAs accumulate more in LE/MVB fractions (Fig. 3E). Using the TET-OFF system, we also monitored the subcellular dynamics of the already synthesized RL-3xbulge-miR-122 with time and observed a net shift of the mRNA from EE to LE/MVB fractions against time, reconfirming the compartmentalization of miRNA-repressed mRNAs in mammalian cells (Fig. 3F).
We immunoprecipitated AGO2 from early endosomal and MVB fractions and measured the associated miRNA and target mRNA content. AGO2 content of LE/MVBs was lower than that of EE, although it showed higher association with GW182 but not with RCK/p54. Interestingly, we observed that both RL-3xbulge-miR-122 and miR-122 were preferentially bound to early endosomal AGO2 compared to AGO2 found in LE/MVBs (Fig. 3G). This indicates dissociation of target mRNA and miRNA from AGO2 happens before LE/MVB targeting. Therefore, we hypothesize that miRNPs and target mRNAs are uncoupled on MVBs, where the target mRNA gets associated with the components of degradation machinery that eventually leads to degradation of the miRNA-targeted mRNAs. In support of our hypothesis, we observed that almost two-thirds of DCP2-associated RL-3xbulge-miR-122 mRNA was present in the LE/MVB fractions. From immunoprecipitated DCP2, we found significant association with XRN1 but not CNOT4. DCP2 did not show any association with AGO2 in LE/MVB fractions, further supporting the uncoupling of target mRNA from AGO2 on LE/MVBs (Fig. 3H).
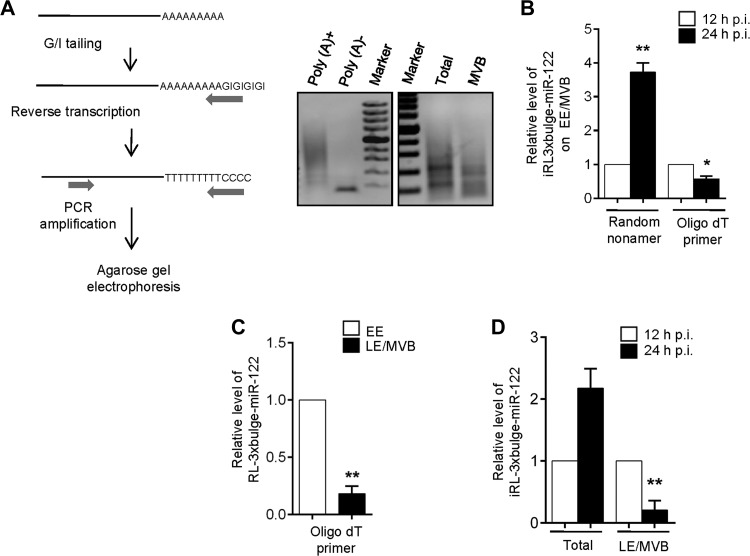
mRNA degradation occurs either by XRN1-mediated 5′ to 3′ exoribonuclease action on decapped mRNAs or by exosome-mediated 3′ to 5′ decay of deadenylated mRNAs. Both of these pathways are preceded by a common step of deadenylation which involves removal of the poly(A) tail by cellular deadenylases. In the 5′ pathway, deadenylation is followed by DCP2-catalyzed decapping and subsequent 5′ to 3′ degradation of the remaining 5′ monophosphorylated mRNA by exoribonuclease XRN1 (18–20). In the 3′ decay pathway, deadenylation is followed by exosome-mediated 3′ to 5′ decay; the remaining capped species is taken care of by DcpS scavenger decapping enzyme (21) (22). We observed that the deadenylated mRNA population was larger in endosome/MVB fractions (obtained from 3 to 30% OptiPrep gradient) than in the total sample (Fig. 4A). With inducible expression of RL-3xbulge-miR-122, we also found that the amount of deadenylated mRNA, associated with the endosome/MVB fraction, showed an increase with time (Fig. 4B). Probing the mechanism further, we observed that the deadenylated pool of RL-3xbulge-miR-122 was primarily enriched in LE/MVB fractions compared to EE in steady state (Fig. 4C). Using the inducible system, we observed an increase in the amount of the deadenylated species of the mRNA specifically in LE/MVBs (Fig. 4D).
FIG 4.
Deadenylation of target mRNA occurs on late endosomes/MVBs. (A) Pictorial representation of poly(A) tail length assay. The assay was verified using in vitro-transcribed m7G-capped RL-3xbulge-miR-122, either polyadenylated or nonpolyadenylated. Final PCR products of total cellular RNA and MVB-bound RNA were shown by agarose gel electrophoresis after poly(A) length assay. (B) De novo-synthesized RL-3xbulge-miR-122 accumulates on endosomes and gets deadenylated with time. The relative level of EE/MVB-bound RL-3xbulge-miR-122 was quantified after 12 and 24 h of induction in TET-ON stable HEK293 cells. A random nonamer has been used for cDNA synthesis to quantitate total mRNA and oligo(dT) to quantify the extent of deadenylation. (C) Relative deadenylation of RL-3xbulge-miR-122 was measured on both early and late endosomes. Oligo(dT) primers were used. (D) Extent of deadenylation of RL-3xbulge-miR-122 increases with time in LE/MVB fractions. Using oligo(dT) primers, RL-3xbulge-miR-122 was quantified in total samples and LE/MVB fractions in TET-ON stable miR-122-expressing HEK293 cells. In all RT-qPCR experiments 18S rRNA serves as the endogenous control. RT-qPCR results ± SD from three independent experiments are shown, and the values of the control have been taken as 1 (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Impaired MVB targeting of miRNA-targeted mRNAs increases their stability in cells.
To understand the significance of these multivesicular structures in degradation of miRNA-repressed messages, we intended to inhibit the endosomal maturation. Early endosomes are formed by several mechanisms, including the classical clathrin-dependent pathway or caveolin-dependent and -independent endocytotic pathways (23). Early endosomes then subsequently mature into late endosomes and MVBs. HRS initiates the step of LE/MVB formation from early endosomes. We performed siRNA-mediated knockdown of several proteins that function upstream or downstream of HRS in the endosomal pathways. ALIX works in concert with ESCRT-III components, facilitating budding of intralumenal vesicles within MVBs (24). RAB7 GTPase is involved in maturation of late endosomes, and RILP facilitates fusion of late endosomes with lysosomes (25). RAB5 is one of the best-studied Rab GTPases, is present on endosomes, and plays a well-characterized role in homotypic endosome fusion and clathrin-dependent endocytosis (26).
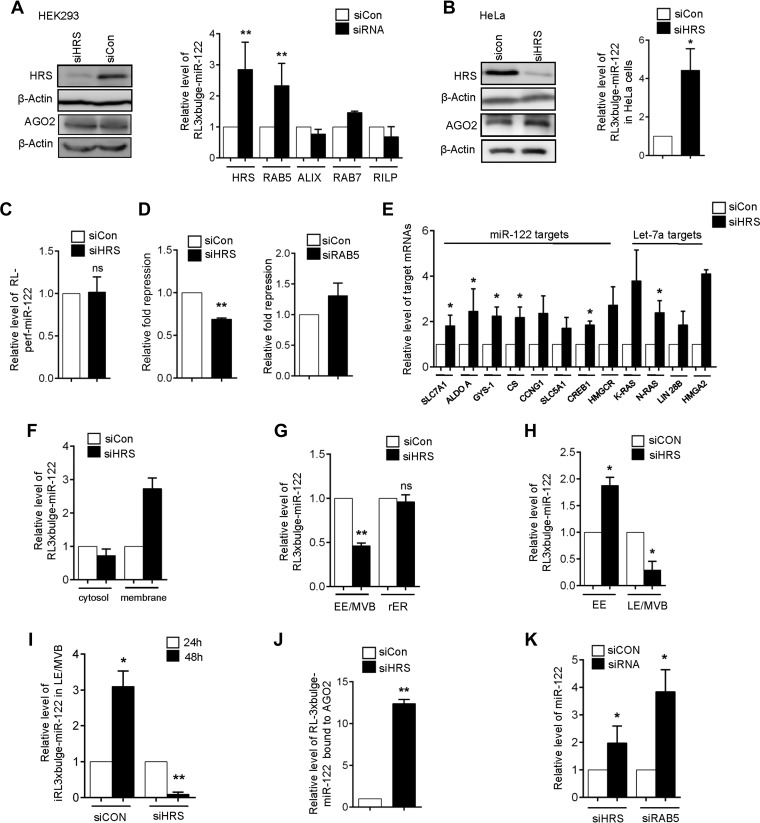
HRS, being the central player in triggering sequential ESCRT assembly on endosomes that eventually leads to formation of MVBs, was the key protein that we targeted to measure the effect of its depletion on miRNA repression and target RNA degradation. We documented an increase in target mRNA levels in HEK293 cells depleted of HRS. HRS depletion was carried out by siRNA treatment of the cells (Fig. 5A). In these cells, the RL-3xbulge-miR-122 mRNA level was more than 2.5-fold higher than that in control siRNA-treated cells. Apart from HRS, a significant increase in cellular target mRNA level was observed in RAB5-depleted cells. Since RAB7, ALIX, and RILP function downstream of HRS, blocking these factors did not affect mRNA degradation significantly (Fig. 5A). A similar observation was found in HeLa cells where siRNA-mediated knockdown of HRS was carried out (Fig. 5B). Target RNAs bearing a perfectly complementary site are cleaved endonucleolytically by the cognate miRNA-AGO2 (miRISC) complex. Stalder et al. have shown previously that siRNA-mediated target mRNA cleavage occurs on the rER membrane (4). Therefore, a message bearing perfectly complementary miRNA sites should not be targeted to MVBs. Consistent with that observation, no significant increase in mRNA levels of a reporter mRNA with a perfect site for miR-122 was detected (RLperf-miR-122) in HRS-depleted cells (Fig. 5C). This confirmed that target mRNA-miRNP interaction happening on rER is independent of HRS depletion. Supporting this observation, minimal effects on miRNA-mediated repression levels in cells depleted for RAB5 or HRS were noted (Fig. 5D). Since depletion of HRS interferes with the formation or maturation of MVBs, repressed mRNA targeting to MVBs necessary for their subsequent degradation probably is impaired, resulting in increased stability of miRNA-repressed mRNAs in HRS-compromised cells. Levels of a few endogenous target mRNAs of miR-122 and let-7a were quantified in HRS-depleted cells expressing these miRNAs. Consistent with our earlier observation, HRS depletion significantly enhanced the cellular levels of all target mRNAs checked (Fig. 5E).
FIG 5.
Impairment of MVB formation increases mRNA stability. (A) Effect of depletion of various components of endosome pathways on the level of miRNA-targeted mRNA. The relative level of RL-3xbulge-miR-122 was measured in HRS, RAB5, ALIX, RAB7, and RILP knockdown HEK293 cells. Knockdown of HRS has been confirmed by Western blotting in HEK293 cells. In each case values obtained from control siRNA-treated cells were taken as 1. (B) Western blot of HRS knockdown in HeLa cells. The AGO2 protein level was also quantified in HRS knockdown cells. Relative quantification of RL-3xbulge-miR-122 in HRS-depleted HeLa cells. The values of control siRNA-treated cells were taken as 1. (C) Effect of HRS depletion on the level of a target RNA with a perfect complementary miRNA binding site. The relative level of RL-perf-miR-122 reporter in HRS knockdown HEK293 cells was quantified. (D) Relative repression of RL-3xbulge-miR-122 was measured in HRS knockdown HEK293 cells. Relative repression of RL-3xbulge-miR-122 in RAB5 knockdown HEK293 cells also was plotted. The siCON value was taken as 1. (E) Depletion of HRS leads to accumulation of several miRNA-targeted mRNAs. Relative levels of endogenous targets of miR-122 and let-7a in siHRS- and siCON-treated HEK293 cells are shown. (F) Elevated RL-3xbulge-miR-122 mRNA accumulates in crude digitonin-insoluble membrane fractions in HRS-depleted cells. (G) Reduced association of target mRNA with endosomal fractions in HRS-depleted cells. The level of RL-3xbulge-miR-122 was monitored in MVB and rER fractions isolated from both control and HRS knockdown HEK293 cells. (H and I) Reduced transfer of target mRNA from early to late endosomes in HRS-depleted cells leads to increased accumulation of target in early endosomal fractions. (H) RL-3xbulge-miR-122 was quantified in both early and late endosomal fractions isolated from HRS knockdown cells. The values obtained from control cells were taken as 1. (I) Using TET-OFF stable HEK293 cells, RL-3xbulge-miR-122 was quantified in early and late endosomal fractions after 24 and 48 h of doxycycline treatment. (J) Increased association of AGO2 with target mRNA in HRS-depleted cells. HA-tagged Ago2 was immunoprecipitated from siRNA-treated HEK293 cells, and associated RL-3xbulge-miR-122 was measured. The level of RL-3xbulge-miR-122 in control siRNA-treated cells was taken as 1. (K) Mature miRNA levels are increased on HRS depletion. The relative level of mature miR-122 in HRS- and RAB5-depleted HEK293 cells is shown. In all RT-qPCR experiments, 18S rRNA serves as the endogenous control. RT-qPCR results ± SD from three independent experiments are shown, and the value of the control was normalized to 1 (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant).
To investigate the subcellular distribution of the increased mRNAs in HRS-depleted cells, we isolated crude digitonin-insoluble membranes. The increased RL-3xbulge-miR-122 was observed to accumulate exclusively in membrane fractions and not in the cytosol (Fig. 5F). We observed a reduced abundance of RL-3xbulge-miR-122 in endosome/MVBs in HRS-depleted cells (Fig. 5G). We have documented higher enrichment of target mRNAs in LE/MVB compared to early endosomes (Fig. 3B). However, with HRS depletion, repressed mRNAs accumulate more with EE, as they showed reduced association with LE/MVBs (Fig. 5H). Using the TET-OFF inducible system, we further showed that an already synthesized RL-3xbulge-miR-122 mRNA failed to localize to LE/MVBs with time in siHRS-treated cells compared to that of control siRNA-treated cells (Fig. 5I). From the data presented in Fig. 3G, it seems that the target mRNA may get uncoupled from miRNPs before their degradation happening on LE/MVBs. With depletion of HRS, which prevents targeting of miRNA-repressed message from EE to LE/MVBs, we observed an increased AGO2 association of target mRNAs (Fig. 5J), indicating a failure of miRNPs to get uncoupled from target mRNA in HRS-depleted cells.
Downregulation of HRS in Leishmania-infected macrophages results in elevated levels of miRNA targets.
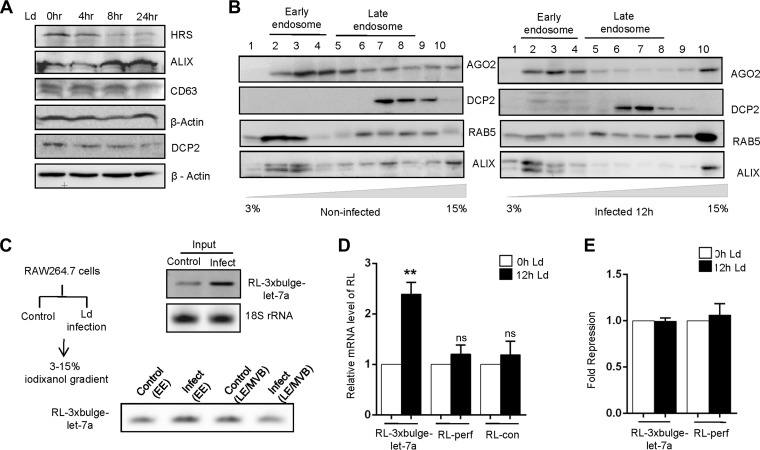
L. donovani is a pathogenic parasite that invades mammalian macrophage cells in the infected hosts and resides within. During the course of its infection, it affects several components of host cell signaling and miRNP machineries to alter the gene regulatory function of the host for its own benefit. Infection of mouse macrophage cells, RAW 264.7, with L. donovani promastigotes leads to depletion of HRS. RAW 264.7 cells were infected with the parasite, and levels of proteins involved in endosomal trafficking were measured against time. With infection, a marked decrease in HRS level against time was noted (Fig. 6A). The reduction in HRS level can be attributed to the activity of the parasite-derived proteases, which are reported to cleave several host proteins upon infection (27, 28). Interestingly, upon infection both ALIX and AGO2 also failed to be localized to LE/MVBs, although DCP2 showed no altered localization (Fig. 6B). The level of reporter mRNA, RL-3xbulge-let-7a, was increased significantly in L. donovani-infected cells and was consistent with the observation in HRS-depleted HEK293 cells (Fig. 6C). In analyzing the compartmentalization of a let-7a targeted reporter mRNA, it was observed that upon infection the reporter mRNA was found to be associated predominantly with EE, in contrast to that in noninfected cells, where it was enriched with late endosomes (Fig. 6C). In accordance with our observation in HRS-depleted HEK293 cells, an increased mRNA level was noted only for RL-3xbulge-let-7a mRNA but not for RL-perf or RL-con mRNAs, having one perfect or no let-7a binding site, respectively (Fig. 6D). These data further affirm the fact that the blocking of MVB formation could impair degradation of miRNA-targeted mRNAs without affecting the miRNA-mediated repression of target messages at the protein level (Fig. 6E).
FIG 6.
Leishmania donovani infection leads to HRS depletion in macrophage cells. (A) RAW 264.7 cells were infected with Leishmania donovani (Ld) at 1:10 (ratio of macrophage to parasite) for 4, 8, and 24 h. Western blotting for HRS, ALIX, CD63, and DCP2 was done. β-Actin was used as a loading control. (B) OptiPrep density gradient fractionation was performed, and the distribution of AGO2, DCP2, RAB5, and ALIX was confirmed by Western blotting. (C) Schematic representation of the Leishmania infection experiment. RL-3xbulge-let-7a levels were measured in total, early, and late endosomes of control and infected cells by semiquantitative PCR. (D) RAW 264.7 cells were transfected with reporter plasmids, and the mRNA levels were quantified at 0 h and 12 h postinfection by RT-qPCR. (E) Relative fold repression of RL-3xbulge-let-7a and RL-perf was measured at 0 h and 12 h postinfection. RT-qPCR results ± SD from three independent experiments are shown, and the values of the control are set to 1 (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant).
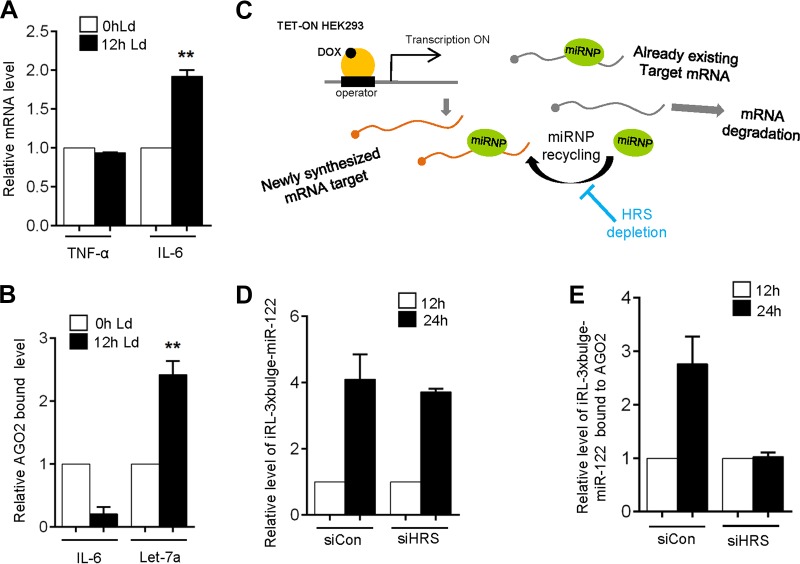
It has been reported that high levels of circulating IL-6 accompany progressive Leishmania infections (29). Considering the role of IL-6 as an anti-inflammatory cytokine in the context of Leishmania infection, we measured the IL-6 mRNA levels and observed a significant increase in IL-6 mRNA within 12 h postinfection (p.i.). Proinflammatory cytokine tumor necrosis factor alpha (TNF-α) mRNA levels were not increased (Fig. 7A). Transcriptional upregulation contributes to the enhanced IL-6 level in infected macrophage (30). However, IL-6 is a let-7a target and therefore should be repressed by let-7a miRNPs in infected macrophages. Interestingly, the increased IL-6 formed postinfection was not AGO2 associated in the L. donovani-infected macrophage (Fig. 7B). We have observed in HRS-depleted cells that AGO2 association with repressed messages gets enhanced (Fig. 5J). This led us to hypothesize that de novo-formed mRNAs fail to get associated with miRNPs that are preengaged in repression of other targets in HRS-depleted cells. To verify this, we ensured the induced expression of RL-3xbulge-miR-122 mRNA in HEK293 cells and measured its association with AGO2 in control and HRS-depleted cells against time (Fig. 7C). Induction of mRNA was confirmed in both control and HRS-depleted cells (Fig. 7D). However, the de novo-formed mRNAs failed to associate with AGO2 specifically in HRS-depleted cells (Fig. 7E). This suggests HRS depletion restricts recycling of miRNPs from the bound target mRNAs, and as the miRNPs remain retained to previously bound target mRNAs, it could not get freed upon degradation of its prebound targets on LE/MVBs in the absence of functional LE/MVB formation. This results in defective recycling of bound miRNPs (Fig. 7C).
FIG 7.
HRS depletion affects miRNP recycling from one target mRNA to another. (A) Relative mRNA level of TNF-α and IL-6 at 0 h and 12 h postinfection. Ld, L. donovani. (B) Ago2-bound IL-6 and let-7a levels after 0 h and 12 h of infection. (C to E) De novo-synthesized target mRNA fail to associate with AGO2 RISC in HRS-depleted cells. (C) Rationale of the experiment described. (D) RL-3xbulge-miR-122 level was quantified in total RNA after 12 and 24 h of induction in both control and HRS knockdown HEK293 cells. (E) HA-tagged AGO2 was immunoprecipitated from control and knockdown HEK293 cells, and associated RL-3xbulge-miR-122 was measured after 12 and 24 h of induction. In each case, values obtained from 12 h were taken as 1. RT-qPCR results ± SD from three independent experiments are shown, and the values of the control are normalized to 1 (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
DISCUSSION
Involvement of subcellular structures in miRNA-mediated gene regulatory processes is an underexplored research domain. Although the importance of endosomal trafficking for miRNA-mediated repression processes has been documented previously, the molecular details have never been investigated. In recent times, findings related to secretion of small RNAs via exosomes (7, 31) for intercellular cross talk or other regulatory purposes justify the possible link between the endocytic pathway and miRNA-mediated gene silencing. In this paper, we report that LE/MVB targeting is crucial for degradation of miRNA-targeted mRNAs. Our findings highlight how miRNA-mediated mRNA repression and the endocytic pathway interact. The work described here also reveals how the mRNA degradation process is compartmentalized in mammalian cells.
Cytoplasm of eukaryotic cells is subdivided into membrane-bound compartments called organelles. Compartmentalization of cellular processes to specific subcellular locales is a mechanism that ensures stringent regulation. Compartmentalization of transcription to specific nuclear territories or compartmentalized translation on rER membrane serves to regulate these processes with exquisite spatial and temporal control (32, 33). Assembly of the miRNA machinery and their interaction with target mRNA binding have been reported to be compartmentalized on rER membrane (4, 17). Therefore, it is rational that in the subsequent steps of repression, mRNA degradation also takes place in some specific subcellular domains. We found that components of mRNA degradation machinery are differentially distributed in human cells, and as opposed to rER membranes, where factors involved in miRNA assembly get enriched, most of the mRNA catabolic factors localize to endosome/MVB fractions. Interestingly, miRNA-repressed target mRNAs also get enriched on LE/MVBs in a time-dependent manner.
In an earlier report, Djuranovic et al. illustrated the exact order of the closely spaced events of mRNA repression, deadenylation, and decay (34). They showed that translation repression by miRNA is followed by mRNA deadenylation and decay, suggesting that these two events are separated temporally. In another report from our laboratory, we have shown that polysomal sequestration of miRNPs with target mRNAs in growth-retarded cells can uncouple mRNA repression from degradation (35). This implies that the two events can be separated by space too. In this work, we show that translation repression and deadenylation/decay of miRNA-targeted mRNA are separated spatially as well. This is achieved by compartmentalization of the two processes in distinct subcellular addresses. Translation repression occurs on rER, while degradation of the mRNAs takes place on endosome/MVBs. Our evidence also suggests that postrepression mRNAs are targeted to MVBs, where the subsequent steps of deadenylation and degradation occur. Dissecting further, we observed that the miRNA-targeted mRNAs are specifically associated with MVB/late endosomes as opposed to early endosomes. Interestingly, this pool of mRNAs was less bound to miRNPs and more associated with DCP2, the key decapping enzyme. DCP2 and XRN1 exonuclease enriched almost exclusively on LE/MVBs, further confirming the observation. A very recent finding has reported cotranslational mRNA degradation that is initiated by decapping and occurs 5′ to 3′ by XRN1 (36). Earlier we reported miRNA-mediated repression on translating polyribosomes (35). Therefore, it is plausible that postrepression, polyribosome-bound target mRNAs are trafficked from rER to endosomes, where they are subsequently degraded. HuR-mediated export of miRNAs in mammalian cells has been shown recently, and from our published data it seems that HuR replaces the miRNPs and unloads miRNAs from Ago2 for their export on rER. Therefore, it may be the rER compartment where decisions (mRNA translation or degradation) on the fate of miRNA-repressed messages are already made before the mRNAs are targeted to EE (37). More interestingly, we have also pointed out that the two closely knit processes can be functionally uncoupled from each other by reducing levels of a key player in MVB maturation. Reducing cellular HRS levels can therefore prevent degradation of the repressed mRNAs and also inhibit recycling of miRNPs to new targets.
It has been reported that RCK/p54 (DDX6) acts downstream of the CCR4-NOT complex to repress translation initiation and stimulate decapping in human cells (38–42). We have observed abundant amounts of RCK/p54 in endosomes (Fig. 1C). Furthermore, RCK/p54 was present in early as well as late endosomal fractions. Hence, it is plausible that RCK/p54 interaction occurs postrepression in endosomal compartments. P bodies, believed to be cytoplasmic foci required for mRNA degradation, also contain GW182 (GW bodies), which are required for miRNA-mediated mRNA deadenylation and degradation (43). P bodies also contain 5′ to 3′ exonuclease XRN1 (44, 45), but components of the exosome complex, involved in 3′ to 5′ degradation of mRNAs, are absent from P bodies (3, 44, 46). Therefore, it is likely that mRNA degradation in P bodies occurs predominantly via the 5′ to 3′ XRN1 pathway postdeadenylation (47).
Lee et al. reported that in Drosophila, blocking MVB formation inhibits mRNA silencing by miRISC (Ago1-miRNA complex), whereas blocking MVB turnover stimulates silencing (8). They have observed that GW bodies are associated with MVBs, and their formation or stability is also dependent on the formation of MVBs. Since GW bodies or P bodies are cytoplasmic granules replete with factors essential for mRNA catabolism (48), their association with MVBs implies cross talk between these two compartments. It is plausible that repressed mRNAs are shuttled from MVBs to P bodies for degradation. MVB formation is hampered by knockdown of HRS, and we observed an increase in cellular target mRNA level upon HRS depletion without any drastic change in miRNA-mediated translation repression level. The increased target RNAs were AGO2 bound, and their MVB association was reduced but without much drop in miRNA-mediated repression. Therefore, it is evident that owing to impaired degradation, the cellular level of target mRNAs shoots up when MVB formation is blocked. Interestingly, we could identify a physiological context where HRS depletion can lead to defective degradation of miRNA-repressed mRNAs. During Leishmania infection of mammalian macrophages, HRS gets reduced, contributing to enhanced reporter mRNA levels in infected macrophage cells.
It was tempting to analyze the fate of miR-122 in the HRS-depleted cells. We found that mature miR-122 levels also shoot up (Fig. 5K). However, the increased miR-122 was not functional in repressing target messages, as evident from the reduction of fold repression in HRS-depleted cells (Fig. 5D). A similar increase in mature miR-122 level was also observed in RAB5-depleted cells (Fig. 5K). Knockdown of the components of the endocytic pathway leads to increased cellular abundance of miRNAs due to their reduced exosomal export (49). This accounts for the rise in mature miR-122 level and opens up avenues for future studies.
Our observations also indicate that uncoupling of the miRNP from its target mRNA happens as the repressed RNA progresses from EE to LE/MVB compartments. This could act as a mechanism that ensures the recycling of released miRNPs after one round of repression to become engaged with new sets of target mRNAs for a fresh round of repression (8). Notably, IL-6, a cytokine known to have pleiotropic effects in Leishmania infection, showed reduced AGO2 association within 12 h of infection along with a concomitant increase in let-7a miRNPs. Since let-7a miRNPs fail to uncouple from prebound target mRNAs and repress newly formed IL-6 mRNA, enhanced translation of IL-6 in host cells helps Leishmania to suppress host macrophage activation and thereby promote infection. This further supports the notion of defective recycling of miRNPs in cells impaired for HRS.
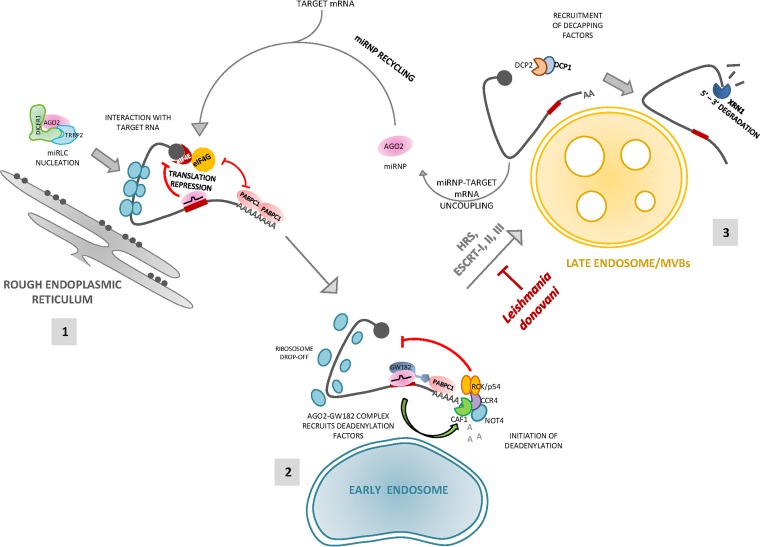
Overall, our observations not only revalidate the connection between RNA silencing and endosomal pathways but also identify the specific subcellular compartments where mRNAs are targeted for degradation postrepression by miRNAs. We also showed that degradation can be blocked by inhibiting biogenesis of these endosomal compartments, further confirming that miRNA-mediated mRNA decay is functionally coupled with MVB formation (Fig. 8).
FIG 8.
Schematic representation of the fate of an miRNA-targeted mRNA from rER membrane to late endosomal membranes. A diagrammatic summary of the model that was inferred from the data is presented. (1) miRNP nucleation and interaction with target mRNA on rER is followed by initiation of translation repression of the target mRNA. (2) Postrepression, the target mRNA-miRNP complex is trafficked to early endosomes, where deadenylation of the target begins by recruitment of cellular deadenylases. (3) As early endosomes mature to MVBs/late endosomes, uncoupling of miRNP occurs from target mRNA followed by decapping and degradation of the mRNA. The miRNP is recycled for another fresh round of repression or can be exported outside the cell via exosomes.
MATERIALS AND METHODS
Cell culture and transfection.
HEK293 and HeLa cells were grown in Dulbecco's modified Eagle's medium (DMEM) (Life Technologies) containing 2 mM l-glutamine and 10% heat-inactivated fetal bovine serum (FBS) (Life Technologies). All plasmids were transfected with Lipofectamine 2000 (Life Technologies), whereas all siRNAs were transfected using RNAiMAX (Life Technologies). For tetracycline-inducible gene expression, doxycycline (DOX; Sigma) was used at 300 ng/ml. RAW 264.7 cells were cultured in RPMI 1640 medium (Gibco) supplemented with 2 mM l-glutamine, 0.5% β-mercaptoethanol, 10% heat-inactivated fetal calf serum, and 1% penicillin-streptomycin (Pen-Strep) solution. Leishmania donovani strain AG83 (MAOM/IN/1083/AG83) was maintained in M199 medium (Gibco) supplemented with 10% heat-inactivated fetal calf serum and 1% Pen-Strep solution. Second- to fourth-passage cultures of L. donovani promastigotes were used to infect RAW 264.7 cells at a 10:1 ratio for all experiments.
Plasmid constructs.
Plasmids containing humanized renilla luciferase coding sequences (RL-con), three miR-122 binding sites downstream of the renilla luciferase (RL) coding region (RL-3xbulge-miR-122), three let-7a binding sites (RL-3xbulge-let-7a), and firefly luciferase (FL) under a simian virus 40 (SV40) promoter (pGL3FF) were kind gifts from Witold Filipowicz. The miR-122 expression plasmid, pmiR-122, was described by Chang et al. (50). FLAG- and hemagglutinin (HA)-tagged human AGO2 expression plasmid (FH-AGO2) was described by Meister et al. (51). The plasmid used for generating TET-inducible stable HEK293 cells, pTet-On-Advanced vector, was purchased from Clontech. The inducible mRNA expression construct iRL-3xbulge-miR-122 has been described by Barman and Bhattacharyya (17). SMARTpool ON-TARGETplus HGS siRNA (siHRS) and a nontargeting pool (siCON) were purchased from Dharmacon.
RNA isolation and Northern blotting.
Total cellular or organellar RNA was isolated using TRIzol reagent or TRIzol LS (Life Technologies). Northern blotting of total cellular RNA (5 to 10 μg) was performed as described elsewhere (2). For detection, γ-32P-labeled 22-nt miRCURY complementary LNA probes for let-7a (Exiqon) were used. Phosphorimaging of the blots was performed in a Cyclone Plus storage phosphor system (PerkinElmer).
Real-time quantification.
For real-time quantification of mRNAs, cDNA was prepared using random nonamers (Eurogentec reverse transcriptase core kit) and PCR was done with gene-specific primers using Mesa Green quantitative PCR (qPCR) master mix plus (Eurogentec) by following the manufacturer's protocol. For miRNA quantification, a TaqMan reverse transcriptase kit was used, followed by a TaqMan universal PCR mix to amplify cDNAs per the manufacturer's protocols (Applied Biosystems). The following primers were used for the amplification of miRNAs: human let-7a (assay 000377), human miR-122 (assay 000445), and U6 snRNA (assay 001973). All reactions were carried out in a 7500 Applied Biosystems real-time system or a Bio-Rad CFX96 real-time system. Cycles were set per the manufacturer's protocol.
Immunoprecipitation and Western blotting.
For immunoprecipitation, recombinant protein G-agarose beads (Life Technologies) or FLAG-M2 agarose beads (Sigma) were used. HA-tagged Ago2 or endogenous DCP2 was pulled from HEK293 cells as described previously (52). Proteins and RNAs were extracted from immunoprecipitated beads for further analysis. For Western blotting, after SDS-PAGE proteins were transferred to polyvinylidene difluoride (PVDF) membrane, followed by blocking and probing with the following primary antibodies at 4°C for 16 h: mouse anti-Ago2 (Abnova), 1:1,000; rabbit anticalnexin (Cell Signaling), 1:1,000; rabbit anti-XRN1 (Bethyl Laboratories), 1:5,000; rabbit anti-RCK/p54 (Bethyl Laboratories), 1:10,000; rabbit anti-DCP2 (Bethyl Laboratories), 1:5,000; rabbit anti-PABPC1 (Abnova), 1:1,000; mouse anti-CNOT4 (Abnova), 1:1,000; rabbit anti-HRS (Bethyl Laboratories), 1:1,000; mouse anti-ALIX (Santa Cruz), 1:200; rabbit anti-GRP78 (Sigma), 1:3,000; rabbit anti-RAB7 (Cell Signaling), 1:1,000; rabbit anti-RAB5 (Cell Signaling), 1:1,000; rabbit anti-GW182 (Bethyl Laboratories), 1:5,000; mouse anti-CD63 (BD Pharmingen), 1:500; rabbit anti-eukaryotic translation initiation factor 4E (anti-eIF4E; Bethyl Laboratories), 1:10,000; rat anti-HA (Roche), 1:1,000; and horseradish peroxidase (HRP)-conjugated β-actin (Sigma), 1:10,000. HRP-conjugated glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (1:50,000; Sigma) was used as a cytosolic marker.
Deadenylation and poly(A) length measurement.
To measure poly(A) tail length, 1 μg of total RNA was taken following G/I tailing, reverse transcription, and PCR amplification using a poly(A) tail length measurement kit (Affymetrix) according to the manufacturer's protocol. To quantify deadenylated mRNA levels, total RNAs were subjected to reverse transcription using oligo(dT) primers (reverse transcriptase core kit; Eurogentec) followed by PCR amplification with gene-specific primers using Mesa Green qPCR master mix plus (Eurogentec) per the manufacturer's protocols. Control polyadenylated and deadenylated mRNA transcripts were prepared using an mMESSAGE mMACHINE kit (Life Technologies) and poly(A) tailing kit (Life Technologies) according to the manufacturer's protocol.
Cell fractionation.
For digitonin fractionation, cells were treated with 50 μg/ml digitonin (Calbiochem) for 10 min at 4°C, followed by centrifugation at 2,500 × g as described previously (17). Cell fractionation using 3 to 30% OptiPrep (iodixanol) density gradient centrifugation was carried out as described previously (53). For cell fractionation in iodixanol gradient (OptiPrep gradient), roughly 1 × 107 cells were used. The cell pellet was incubated in a hypotonic buffer (50 mM HEPES, pH 7.8, 78 mM KCl, 4 mM MgCl2, 8.4 mM CaCl2, 10 mM EGTA, 250 mM sucrose, 100 μg/ml of cycloheximide, 5 mM vanadyl ribonucleoside complex, and 1× EDTA-free protease inhibitor cocktail) and homogenized by a glass Dounce homogenizer (Sartorius). The cell homogenate was cleared by centrifugation at 1,000 × g twice, and the supernatant was loaded on a 3 to 30% iodixanol gradient (OptiPrep) and ultracentrifuged for 5 h at 36,000 rpm in an SW60 rotor (Beckman Coulter). Fractions were collected manually; RNA and protein were isolated and analyzed. To resolve early and late endosome/MVB fractions, 3 to 15% OptiPrep gradient was used. To get microsomal fractions, postmitochondrial fractions were precipitated by 8 mM CaCl2 and centrifuged at 8,000 × g as described earlier (17).
Quantitative analysis of surface area of early and late endosome.
Z-stack images were acquired using an Andor spinning disc confocal microscope with an Ixon3 electron-multiplying charge-coupled-device (EMCCD) detection system. Images were then used to perform three-dimensional surface reconstruction using Imaris x64 7.6.3 (Bitplane AG). The surpass module was used to construct the surface, followed by manual threshold adjustment. All parameters were kept constant for independent studies. Surface area was calculated from three independent cells, and the mean value was calculated to obtain the overall surface area of early endosome and late endosome.
ACKNOWLEDGMENTS
We are thankful to Witold Filipowicz for different clones and constructs we used in this study.
This work has been supported by the Wellcome Trust Senior Research Fellowship fund and the Swarnajayanti Fellowship Fund of Department of Science and Technology, Government of India, to S.N.B. M.B., B.B., and A.G. received support from CSIR, Government of India.
REFERENCES
- 1.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. 2005. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol 7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. 2005. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science 309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 3.Eulalio A, Behm-Ansmant I, Izaurralde E. 2007. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol 8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- 4.Stalder L, Heusermann W, Sokol L, Trojer D, Wirz J, Hean J, Fritzsche A, Aeschimann F, Pfanzagl V, Basselet P, Weiler J, Hintersteiner M, Morrissey DV, Meisner-Kober NC. 2013. The rough endoplasmatic reticulum is a central nucleation site of siRNA-mediated RNA silencing. EMBO J 32:1115–1127. doi: 10.1038/emboj.2013.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li S, Liu L, Zhuang X, Yu Y, Liu X, Cui X, Ji L, Pan Z, Cao X, Mo B, Zhang F, Raikhel N, Jiang L, Chen X. 2013. MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell 153:562–574. doi: 10.1016/j.cell.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piper RC, Katzmann DJ. 2007. Biogenesis and function of multivesicular bodies. Annu Rev Cell Dev Biol 23:519–547. doi: 10.1146/annurev.cellbio.23.090506.123319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stoorvogel W, Kleijmeer MJ, Geuze HJ, Raposo G. 2002. The biogenesis and functions of exosomes. Traffic 3:321–330. doi: 10.1034/j.1600-0854.2002.30502.x. [DOI] [PubMed] [Google Scholar]
- 8.Lee YS, Pressman S, Andress AP, Kim K, White JL, Cassidy JJ, Li X, Lubell K, Lim Do H, Cho IS, Nakahara K, Preall JB, Bellare P, Sontheimer EJ, Carthew RW. 2009. Silencing by small RNAs is linked to endosomal trafficking. Nat Cell Biol 11:1150–1156. doi: 10.1038/ncb1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. 2009. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol 11:1143–1149. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- 10.Bache KG, Brech A, Mehlum A, Stenmark H. 2003. Hrs regulates multivesicular body formation via ESCRT recruitment to endosomes. J Cell Biol 162:435–442. doi: 10.1083/jcb.200302131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raiborg C, Bremnes B, Mehlum A, Gillooly DJ, D'Arrigo A, Stang E, Stenmark H. 2001. FYVE and coiled-coil domains determine the specific localisation of Hrs to early endosomes. J Cell Sci 114:2255–2263. [DOI] [PubMed] [Google Scholar]
- 12.Gruenberg J, Stenmark H. 2004. The biogenesis of multivesicular endosomes. Nat Rev Mol Cell Biol 5:317–323. doi: 10.1038/nrm1360. [DOI] [PubMed] [Google Scholar]
- 13.Lerner RS, Seiser RM, Zheng T, Lager PJ, Reedy MC, Keene JD, Nicchitta CV. 2003. Partitioning and translation of mRNAs encoding soluble proteins on membrane-bound ribosomes. RNA 9:1123–1137. doi: 10.1261/rna.5610403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jagannathan S, Hsu JC, Reid DW, Chen Q, Thompson WJ, Moseley AM, Nicchitta CV. 2014. Multifunctional roles for the protein translocation machinery in RNA anchoring to the endoplasmic reticulum. J Biol Chem 289:25907–25924. doi: 10.1074/jbc.M114.580688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tahbaz N, Kolb FA, Zhang H, Jaronczyk K, Filipowicz W, Hobman TC. 2004. Characterization of the interactions between mammalian PAZ PIWI domain proteins and Dicer. EMBO Rep 5:189–194. doi: 10.1038/sj.embor.7400070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tahbaz N, Carmichael JB, Hobman TC. 2001. GERp95 belongs to a family of signal-transducing proteins and requires Hsp90 activity for stability and Golgi localization. J Biol Chem 276:43294–43299. doi: 10.1074/jbc.M107808200. [DOI] [PubMed] [Google Scholar]
- 17.Barman B, Bhattacharyya SN. 2015. mRNA targeting to endoplasmic reticulum precedes ago protein interaction and microRNA (miRNA)-mediated translation repression in mammalian cells. J Biol Chem 290:24650–24656. doi: 10.1074/jbc.C115.661868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunckley T, Parker R. 1999. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J 18:5411–5422. doi: 10.1093/emboj/18.19.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z, Jiao X, Carr-Schmid A, Kiledjian M. 2002. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc Natl Acad Sci U S A 99:12663–12668. doi: 10.1073/pnas.192445599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steiger M, Carr-Schmid A, Schwartz DC, Kiledjian M, Parker R. 2003. Analysis of recombinant yeast decapping enzyme. RNA 9:231–238. doi: 10.1261/rna.2151403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muhlrad D, Decker CJ, Parker R. 1994. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes Dev 8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- 22.Anderson JS, Parker RP. 1998. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J 17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gould GW, Lippincott-Schwartz J. 2009. New roles for endosomes: from vesicular carriers to multi-purpose platforms. Nat Rev Mol Cell Biol 10:287–292. doi: 10.1038/nrm2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kinchen JM, Ravichandran KS. 2008. Phagosome maturation: going through the acid test. Nat Rev Mol Cell Biol 9:781–795. doi: 10.1038/nrm2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hyttinen JM, Niittykoski M, Salminen A, Kaarniranta K. 2013. Maturation of autophagosomes and endosomes: a key role for Rab7. Biochim Biophys Acta 1833:503–510. doi: 10.1016/j.bbamcr.2012.11.018. [DOI] [PubMed] [Google Scholar]
- 26.Sonnichsen B, De Renzis S, Nielsen E, Rietdorf J, Zerial M. 2000. Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of Rab4, Rab5, and Rab11. J Cell Biol 149:901–914. doi: 10.1083/jcb.149.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghosh J, Bose M, Roy S, Bhattacharyya SN. 2013. Leishmania donovani targets Dicer1 to downregulate miR-122, lower serum cholesterol, and facilitate murine liver infection. Cell Host Microbe 13:277–288. doi: 10.1016/j.chom.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shapira M, Zinoviev A. 2011. Leishmania parasites act as a Trojan horse that paralyzes the translation system of host macrophages. Cell Host Microbe 9:257–259. doi: 10.1016/j.chom.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 29.Murray HW. 2008. Accelerated control of visceral Leishmania donovani infection in interleukin-6-deficient mice. Infect Immun 76:4088–4091. doi: 10.1128/IAI.00490-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scheller J, Chalaris A, Schmidt-Arras D, Rose-John S. 2011. The pro- and anti-inflammatory properties of the cytokine interleukin-6. Biochim Biophys Acta 1813:878–888. doi: 10.1016/j.bbamcr.2011.01.034. [DOI] [PubMed] [Google Scholar]
- 31.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. 2007. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 32.Cremer T, Cremer C. 2001. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet 2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 33.Lerner RS, Nicchitta CV. 2006. mRNA translation is compartmentalized to the endoplasmic reticulum following physiological inhibition of cap-dependent translation. RNA 12:775–789. doi: 10.1261/rna.2318906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Djuranovic S, Nahvi A, Green R. 2012. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 336:237–240. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghosh S, Bose M, Ray A, Bhattacharyya SN. 2015. Polysome arrest restricts miRNA turnover by preventing exosomal export of miRNA in growth-retarded mammalian cells. Mol Biol Cell 26:1072–1083. doi: 10.1091/mbc.E14-11-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tat TT, Maroney PA, Chamnongpol S, Coller J, Nilsen TW. 2016. Cotranslational microRNA mediated messenger RNA destabilization. eLife 5:e12880. doi: 10.7554/eLife.12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mukherjee K, Ghoshal B, Ghosh S, Chakrabarty Y, Shwetha S, Das S, Bhattacharyya SN. 2016. Reversible HuR-microRNA binding controls extracellular export of miR-122 and augments stress response. EMBO Rep 17:1184–1203. doi: 10.15252/embr.201541930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuzuoglu-Ozturk D, Bhandari D, Huntzinger E, Fauser M, Helms S, Izaurralde E. 2016. miRISC and the CCR4-NOT complex silence mRNA targets independently of 43S ribosomal scanning. EMBO J 35:1186–1203. doi: 10.15252/embj.201592901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ozgur S, Basquin J, Kamenska A, Filipowicz W, Standart N, Conti E. 2015. Structure of a human 4E-T/DDX6/CNOT1 complex reveals the different interplay of DDX6-binding proteins with the CCR4-NOT complex. Cell Rep 13:703–711. doi: 10.1016/j.celrep.2015.09.033. [DOI] [PubMed] [Google Scholar]
- 40.Rouya C, Siddiqui N, Morita M, Duchaine TF, Fabian MR, Sonenberg N. 2014. Human DDX6 effects miRNA-mediated gene silencing via direct binding to CNOT1. RNA 20:1398–1409. doi: 10.1261/rna.045302.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Y, Boland A, Kuzuoglu-Ozturk D, Bawankar P, Loh B, Chang CT, Weichenrieder O, Izaurralde E. 2014. A DDX6-CNOT1 complex and W-binding pockets in CNOT9 reveal direct links between miRNA target recognition and silencing. Mol Cell 54:737–750. doi: 10.1016/j.molcel.2014.03.034. [DOI] [PubMed] [Google Scholar]
- 42.Mathys H, Basquin J, Ozgur S, Czarnocki-Cieciura M, Bonneau F, Aartse A, Dziembowski A, Nowotny M, Conti E, Filipowicz W. 2014. Structural and biochemical insights to the role of the CCR4-NOT complex and DDX6 ATPase in microRNA repression. Mol Cell 54:751–765. doi: 10.1016/j.molcel.2014.03.036. [DOI] [PubMed] [Google Scholar]
- 43.Eystathioy T, Chan EK, Tenenbaum SA, Keene JD, Griffith K, Fritzler MJ. 2002. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol Biol Cell 13:1338–1351. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sheth U, Parker R. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ingelfinger D, Arndt-Jovin DJ, Luhrmann R, Achsel T. 2002. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA 8:1489–1501. [PMC free article] [PubMed] [Google Scholar]
- 46.Brengues M, Teixeira D, Parker R. 2005. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shyu AB, Wilkinson MF, van Hoof A. 2008. Messenger RNA regulation: to translate or to degrade. EMBO J 27:471–481. doi: 10.1038/sj.emboj.7601977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Filipowicz W, Bhattacharyya SN, Sonenberg N. 2008. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9:102–114. [DOI] [PubMed] [Google Scholar]
- 49.Tamai K, Tanaka N, Nakano T, Kakazu E, Kondo Y, Inoue J, Shiina M, Fukushima K, Hoshino T, Sano K, Ueno Y, Shimosegawa T, Sugamura K. 2010. Exosome secretion of dendritic cells is regulated by Hrs, an ESCRT-0 protein. Biochem Biophys Res Commun 399:384–390. doi: 10.1016/j.bbrc.2010.07.083. [DOI] [PubMed] [Google Scholar]
- 50.Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, Xu C, Mason WS, Moloshok T, Bort R, Zaret KS, Taylor JM. 2004. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol 1:106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- 51.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. 2004. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 52.Kundu P, Fabian MR, Sonenberg N, Bhattacharyya SN, Filipowicz W. 2012. HuR protein attenuates miRNA-mediated repression by promoting miRISC dissociation from the target RNA. Nucleic Acids Res 40:5088–5100. doi: 10.1093/nar/gks148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mazumder A, Bose M, Chakraborty A, Chakrabarti S, Bhattacharyya SN. 2013. A transient reversal of miRNA-mediated repression controls macrophage activation. EMBO Rep 14:1008–1016. doi: 10.1038/embor.2013.149. [DOI] [PMC free article] [PubMed] [Google Scholar]