ABSTRACT
Repair of damaged DNA is critical for maintenance of genetic information. In eukaryotes, DNA double-strand breaks (DSBs) are recognized by the Ku70-Ku80 heterodimer, which then recruits proteins that mediate repair by nonhomologous end joining (NHEJ). Prolonged retention of Ku70/80 at DSBs prevents completion of repair, however, with ubiquitylation of Ku80 having been implicated in Ku70/80 dissociation from DNA. Here, we identify RNF126 as a ubiquitin ligase that is recruited to DSBs and ubiquitylates Ku80, with UBE2D3 serving as an E2 enzyme. Knockdown of RNF126 prevented Ku70/80 dissociation from DSBs and inhibited break repair. Attenuation of Ku80 ubiquitylation by replacement of ubiquitylation site lysines with arginine residues delayed Ku70/80 release from chromatin after DSB induction by genotoxic insults. Together, our data indicate that RNF126 is a novel regulator of NHEJ that promotes completion of DNA repair by ubiquitylating Ku80 and releasing Ku70/80 from damaged DNA.
KEYWORDS: DNA repair, Ku80, RNF126, nonhomologous end joining (NHEJ), ubiquitylation
INTRODUCTION
DNA damage poses a threat to living organisms because they rely on the genetic code to execute cellular functions. Genomic integrity is maintained in the face of DNA damage by the operation of dedicated DNA repair machinery. In eukaryotic cells, DNA double-strand breaks (DSBs), which constitute the most deleterious type of DNA damage, are repaired by two major, mechanistically distinct pathways: homologous recombination (HR) and nonhomologous end joining (NHEJ). HR replaces lost or damaged DNA sequence with high fidelity in a manner dependent on the presence of an intact sister chromatid as a template. HR thus functions only during S and G2 phases of the cell cycle, when a sister chromatid is available (1). In contrast, NHEJ is active throughout the cell cycle and repairs DSBs by directly ligating chromosome ends without the use of a DNA template. As a consequence of its mode of action, however, NHEJ is error prone (2).
The NHEJ pathway is initiated on recognition of DSBs by chromatin-remodeling factors and the Ku70-Ku80 heterodimer, the latter of which recruits additional factors to process the DSBs for repair. The Ku heterodimer forms a ring-shaped toroidal structure, through which the broken end of DNA is threaded. Given its abundance and the high affinity of Ku70/80 for DNA ends, a specific mechanism is thought to be required to dislodge the heterodimer from DNA after the recruitment of repair proteins in order to prevent its inhibition of repair completion and postrepair recovery (3). In Xenopus egg extracts, removal of Ku70/80 from DNA is dependent on Ku80 ubiquitylation, which occurs after loading of the heterodimer onto chromatin and induces not only the release of Ku80 from DNA but also its degradation by the proteasome (4). SCFFbxl12 mediates ubiquitylation of Ku80 in Xenopus eggs (5), but this mechanism is not likely conserved in mammalian cells (6). Instead, RNF8- and NEDD8-dependent ubiquitin ligases have been found to mediate Ku80 and Ku70 ubiquitylation, respectively, in mammalian cells (7, 8). Furthermore, RNF138 was shown to ubiquitylate Ku80 at S-G2 phases of the cell cycle (6). However, it has remained unclear whether these are the only ubiquitin ligases that target the Ku heterodimer and which residues of Ku80 and Ku70 are ubiquitylated, with the exception of a few sites whose mutation does not affect Ku release from damaged DNA in chromatin (8).
Ubiquitin ligases (E3s) are classified into two major families on the basis of their domain structure (9): RING (really interesting new gene) domain-type and HECT (homologous to E6-AP carboxyl terminus) domain-type ubiquitin ligases. Although the human genome is thought to encode more than 600 E3s or substrate recognition subunits of E3 complexes (10), most of these proteins remain to be investigated.
We now present evidence that the RING finger domain-containing protein RNF126 is a ubiquitin ligase for both Ku70 and Ku80. Comprehensive proteomics analysis identified Ku80 and the ubiquitin-conjugating enzyme (E2) UBE2D3 among RNF126 binding proteins. Furthermore, RNF126 was found to bind directly to Ku80 and Ku70 as well as to ubiquitylate both proteins both in vitro and in cells. RNF126 was found to be recruited to DSBs, and RNA interference (RNAi)-mediated knockdown of RNF126 inhibited the dissociation of Ku70/80 from chromatin as well as the DNA damage response and DSB repair, resulting in an increased susceptibility to DSB-induced cell death. Proteomics and structural analyses identified 19 lysine residues as ubiquitylation sites in Ku80, and the mutation of all of these sites inhibited the dissociation of Ku70/80 from chromatin and the DNA damage response. Collectively, our data reveal that RNF126 regulates NHEJ by mediating the ubiquitylation of Ku80 and thereby triggering the release of Ku70/80 from DSB sites and allowing completion of DNA repair.
RESULTS
RNF126 associates with the Ku70-Ku80 heterodimer.
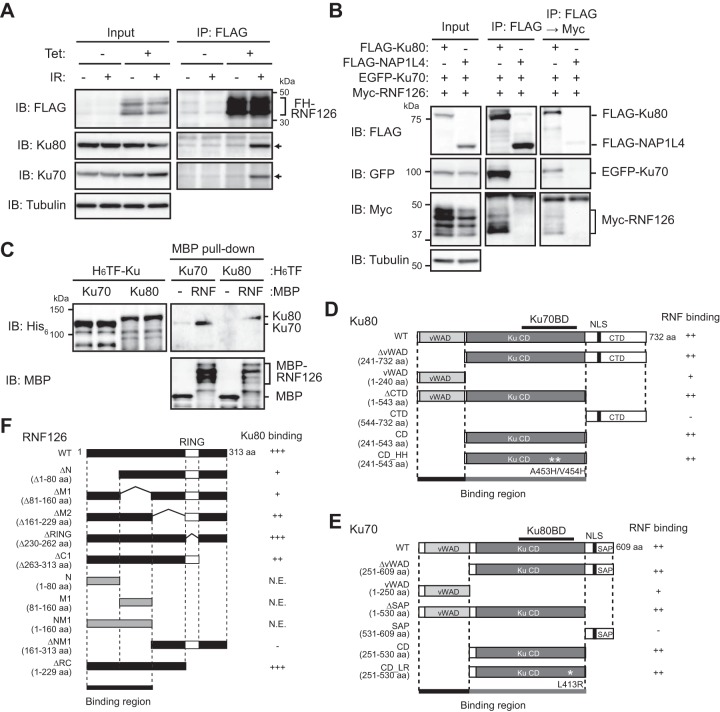
RNF126 has been identified as an uncharacterized protein that contains a zinc finger domain in its NH2-terminal region and a RING finger domain in its COOH-terminal region and which is conserved among vertebrates (see Fig. S1 in the supplemental material). The presence of a RING finger domain suggested that RNF126 functions as a ubiquitin ligase. To characterize the molecular function of RNF126, we searched for proteins with which it associates. Extracts of HEK293T cells expressing FLAG epitope-tagged human RNF126 at a low level were subjected to immunoprecipitation with antibodies to FLAG, and the resulting precipitates were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) to identify RNF126 binding proteins. The results of several independent experiments revealed that at least 30 proteins, including the E2 enzyme UBE2D3 (UbcH5C) and XRCC5 (Ku80), interacted with FLAG-RNF126 (Table S1). Among these proteins, we further examined Ku80 as a potential substrate of RNF126, given that Ku80 had been shown to be regulated by ubiquitylation. We first generated an HEK293 subline, Flp-In T-REx 293-RNF126, in which the expression of FLAG- and HA-tagged RNF126 (FH-RNF126) could be induced by Tet, given that prolonged overexpression of RNF126 was found to be cytotoxic (Fig. 1A). To validate the association of RNF126 with Ku80, we subjected extracts of the Tet-treated cells to immunoprecipitation analysis. Endogenous Ku80 was found to bind to FH-RNF126 in a manner dependent on prior exposure of the cells to gamma radiation (IR) in order to induce DNA DSBs (Fig. 1A). We also detected endogenous Ku70 in the FH-RNF126 immunoprecipitates (Fig. 1A), suggesting that the Ku heterodimer binds to RNF126. Indeed, serial immunoprecipitation analysis of HEK293T cells transiently overexpressing Ku70 and Ku80 as well as RNF126 confirmed this notion (Fig. 1B). Furthermore, pulldown assays revealed that recombinant RNF126 bound to recombinant Ku80 or Ku70 in vitro, suggesting that the interaction between RNF126 and Ku70 and Ku80 is direct (Fig. 1C). Reciprocal coimmunoprecipitation analysis with a series of deletion mutants of Ku80 or Ku70 showed that either the NH2-terminal vWAD domain or the Ku core domain (CD) of the Ku proteins is sufficient for binding to RNF126 (Fig. 1D and E; Fig. S2A and D). Similar experiments with a series of deletion mutants of RNF126 revealed that the NH2-terminal portion of RNF126 is responsible for binding to Ku80 (Fig. 1F; Fig. S3). Consistent with our finding that Ku80 and Ku70 each directly bind to RNF126 in vitro, mutation of the Ku70 binding site of Ku80 (A453H/V454H; HH) (11) or of the Ku80 binding site of Ku70 (L413R; LR) (12) did not affect the binding of either Ku protein to RNF126 (Fig. 1D and E; Fig. S2B, C, E, and F). In addition, incubation of cell lysates with either ethidium bromide or Benzonase did not interfere with the interaction between FLAG-RNF126 and endogenous Ku80 or Ku70 apparent in HEK293T cells treated with bleomycin to induce DNA DSBs (Fig. S2G), suggesting that this binding is not mediated by DNA. Together, these data indicated that RNF126 interacts with Ku80 and Ku70 directly, giving rise to formation of a trimeric complex in cells.
FIG 1.
RNF126 interacts with the Ku70-Ku80 heterodimer. (A) Flp-In T-REx 293-RNF126 cells were exposed (or left unexposed) to Tet or gamma radiation (IR; 2 Gy), lysed, and subjected to immunoprecipitation (IP) with antibodies to FLAG. The resulting precipitates, as well as the original cell lysates (Input), were subjected to immunoblot (IB) analysis with antibodies to FLAG, Ku80, Ku70, or tubulin (loading control). The position of bands corresponding to FH-RNF126 is indicated by the bracket and that of Ku80 and Ku70 bands by the arrows. (B) Extracts of HEK293T cells transiently expressing Myc epitope-tagged mouse RNF126 and EGFP-tagged human Ku70 together with FLAG-tagged human Ku80 or NAP1L4 (negative control) were subjected to immunoprecipitation with antibodies to FLAG, and the resulting precipitates were subjected either directly to immunoblot analysis (middle) or to further immunoprecipitation with antibodies to Myc, followed by immunoblot analysis (right). (C) MBP-tagged mouse RNF126 or MBP alone (negative control) was subjected to a pulldown assay with His6 (H6)- and TF-tagged human Ku80 or Ku70, followed by immunoblot analysis with antibodies to His6 and MBP. (D) Domain structure of WT and mutant (ΔvWAD, vWAD, ΔCTD, CTD, CD, and CD_HH) forms of human Ku80. The ability of the Ku80 mutants to bind to mouse RNF126 in transiently transfected HEK293T cells is summarized (Fig. S2A to C). vWAD, von Willebrand factor type A domain; KuCD, Ku core domain; NLS, nuclear localization sequence; CTD, COOH-terminal domain; Ku70BD, Ku70 binding domain. (E) Domain structure of WT and mutant (ΔvWAD, vWAD, ΔSAP, SAP, CD, and CD_LR) forms of human Ku70. The ability of the Ku70 mutants to bind to mouse RNF126 in transiently transfected HEK293T cells is summarized (Fig. S2D to F). SAP, SAF-A/B, Acinus, and PIAS domains are shown. (F) Domain structure of WT and mutant (ΔN, ΔM1, ΔM2, ΔRING, ΔC1, N, M1, NM1, ΔNM1, and ΔRC) forms of mouse RNF126. The ability of the RNF126 mutants to bind to human Ku80 in transiently transfected HEK293T cells is summarized (Fig. S3). N.E., not expressed.
RNF126 ubiquitylates Ku80 and Ku70 in an IR-dependent manner.
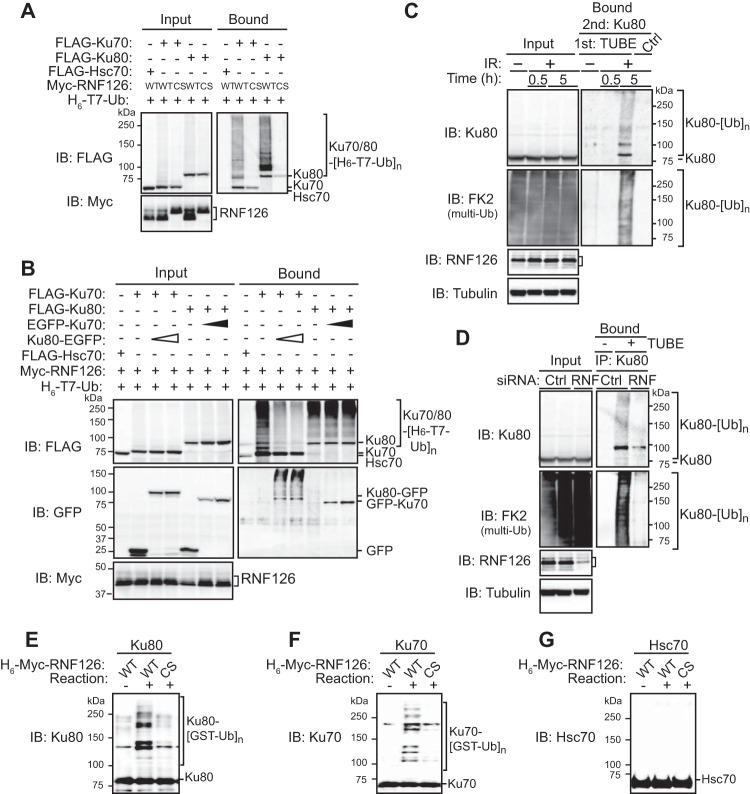
We next investigated the ubiquitin ligase activity of RNF126 toward Ku80 or Ku70. Overexpression of RNF126 induced the polyubiquitylation of separately overexpressed Ku80 and Ku70 in HEK293T cells (Fig. 2A). However, the C231S/C234S (CS) mutant of mouse RNF126, which would be expected to be inactive as a ubiquitin ligase because of the putative role of the mutated cysteine residues in zinc binding, did not induce polyubiquitylation of Ku80 or Ku70 (Fig. 2A). RNF126-dependent ubiquitylation of Ku80 and Ku70 was detected in cells expressing the K63R mutant of ubiquitin but not in those expressing the K48R mutant (Fig. S4A and B), suggesting that RNF126 promotes formation of a K48-linked ubiquitin chain on Ku proteins. Forced expression of Ku80 and Ku70 together revealed that polyubiquitylation of Ku70 was suppressed by Ku80 but not vice versa, indicating that RNF126 preferentially ubiquitylates Ku80 over Ku70 in the Ku heterodimer (Fig. 2B). To examine ubiquitylation of endogenous Ku80, we isolated ubiquitylated proteins from U2OS human osteosarcoma cells with the use of the tandem-repeated ubiquitin-binding entity (TUBE) system (13) and subjected them to immunoprecipitation with antibodies to Ku80 followed by immunoblot analysis with antibodies to ubiquitin. Although we did not detect ubiquitylation of endogenous Ku80 under basal culture conditions, pronounced ubiquitylation of Ku80 was apparent at 5 h after exposure of cells to IR (Fig. 2C), consistent with the previous observation that DSBs induce the ubiquitylation of Ku80 in Xenopus egg extracts (4). However, ubiquitylation of Ku80 was not detected 30 min after gamma irradiation (Fig. 2C), indicating that it is a late response to the generation of DSBs. Knockdown of RNF126 by RNAi with specific short interfering RNAs (siRNAs) reduced the extent of Ku80 ubiquitylation by >50% (Fig. 2D; Fig. S4C), suggesting that RNF126 is a key ubiquitin ligase that catalyzes Ku80 ubiquitylation in response to the induction of DSBs. An in vitro ubiquitylation assay revealed that recombinant RNF126 possesses E3 activity for Ku80 (Fig. 2E) and Ku70 (Fig. 2F) but not for Hsc70 (Fig. 2G) in the presence of a ubiquitin-activating enzyme (E1; Uba1), E2 (UBE2D3), glutathione S-transferase (GST)-tagged ubiquitin, and ATP (Fig. S4D and E), suggesting that modification of Ku proteins, such as by phosphorylation (14) or acetylation (15), is not required for their recognition and ubiquitylation by RNF126. Together, these data indicated that RNF126 is able to directly ubiquitylate both Ku proteins in vitro, whereas Ku80 within the Ku70-Ku80 heterodimer is preferentially ubiquitylated in response to the generation of DSBs in cells.
FIG 2.
RNF126 ubiquitylates Ku80 and Ku70 in an IR-dependent manner. (A) HEK293T cells transfected with expression vectors for the indicated proteins and incubated with the proteasome inhibitor MG132 (10 μM) for 5 h were lysed and subjected to Ni2+-agarose affinity chromatography under denaturing conditions for isolation of proteins conjugated to His6- and T7 epitope-tagged ubiquitin (H6-T7-Ub). The bound proteins as well as the original cell lysates were subjected to immunoblot analysis with antibodies to FLAG or to Myc. (B) Transfected HEK293T cells were assayed for in vivo ubiquitylation as described for panel A. (C) U2OS cells were exposed (or left unexposed) to IR (2 Gy), incubated with 10 μM MG132 for the indicated times, and then lysed for consecutive precipitation of ubiquitylated proteins with the TUBE system and immunoprecipitation with antibodies to Ku80. The first and second precipitates as well as the input cell lysates were subjected to immunoblot analysis. (D) U2OS cells transfected with RNF126 (RNF) or negative-control (Ctrl) siRNAs for 48 h were exposed to IR (2 Gy), incubated with 10 μM MG132 for 5 h, and then analyzed as described for panel C. (E to G) E1, E2, ATP, and GST-ubiquitin were incubated with purified recombinant Ku80 (E), Ku70 (F), or Hsc70 (G) as well as with His-6-Myc-tagged WT or CS mutant forms of RNF126 for assay of in vitro ubiquitylation by immunoblot analysis.
RNF126 promotes IR-dependent degradation of chromatin-associated Ku80.
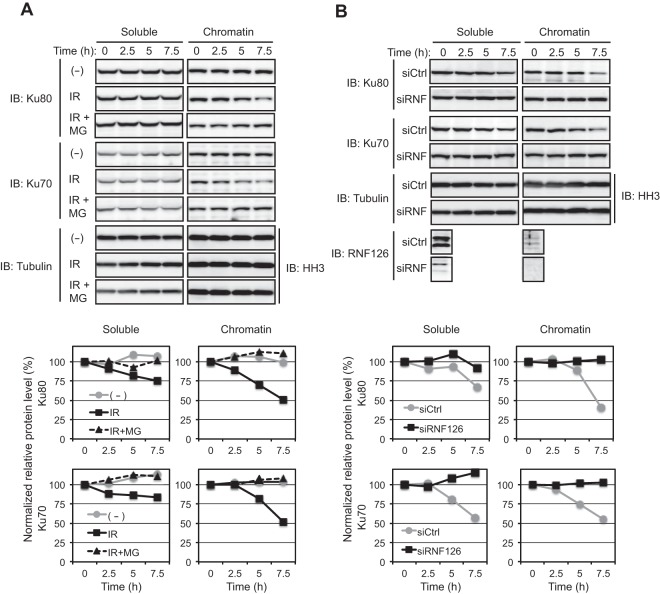
Polyubiquitylation of Ku80 induced by DNA with DSBs in Xenopus egg extracts was previously found to be followed by Ku80 degradation (4). We found that IR induced downregulation of Ku80 in U2OS cells, with this effect being more prominent in the chromatin fraction than in the soluble fraction, as well as being prevented by exposure of cells to the proteasome inhibitor MG132 (Fig. 3A), consistent with the notion that mammalian Ku80 is also degraded by the proteasome in response to the generation of DSBs. The abundance of Ku70 also decreased together with that of Ku80 in irradiated cells (Fig. 3A), but this effect might have been secondary to Ku80 degradation (16). Knockdown of RNF126 abolished the IR-induced degradation of Ku80 and Ku70 (Fig. 3B), further implicating RNF126 in the ubiquitylation and consequent degradation of these proteins in response to the generation of DSBs.
FIG 3.
RNF126 promotes IR-dependent degradation of Ku80 and Ku70 associated with chromatin. (A) U2OS cells were exposed (or left unexposed) to IR (2 Gy), incubated with cycloheximide (50 μg/ml) in the absence or presence of 10 μM MG132 (MG) for the indicated times, and then subjected to subcellular fractionation. Soluble and chromatin fractions were subjected to immunoblot analysis with antibodies to the indicated proteins. The relative amounts of Ku80 and Ku70 normalized by that of tubulin or histone H3 (HH3) at each time point were determined by measurement of band intensity. (B) U2OS cells transfected with RNF126 (siRNF) or control (siCtrl) siRNAs for 48 h were exposed to IR (2 Gy), incubated with cycloheximide for the indicated times, and then analyzed as described for panel A.
RNF126 accumulates at DSB sites in a Ku-dependent manner.
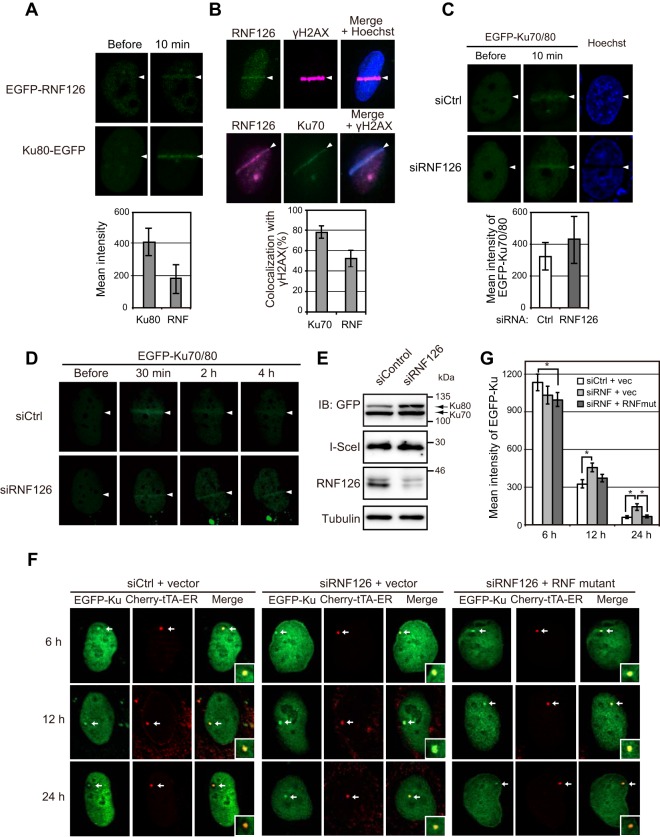
To elucidate the possible role of RNF126 in DNA repair, we examined the localization of RNF126 in U2OS cells exposed to bromodeoxyuridine or 8-methoxypsoralen as photosensitizers and then subjected them to laser microirradiation at 405 nm in order to induce DNA damage. Enhanced green fluorescent protein (EGFP)-tagged RNF126 accumulated at sites of irradiation, as did EGFP-tagged Ku80 (Fig. 4A). Endogenous RNF126 was also detected at sites of laser irradiation, where it colocalized with Ku70 and the DSB marker γH2AX (Fig. 4B). In Ku80-deficient CHO (XR-V15B) cells (17), however, we did not detect EGFP-RNF126 accumulation at laser-irradiated sites (Fig. S5A), suggesting that the Ku heterodimer is necessary for the recruitment of RNF126 to sites of DNA damage. In contrast, depletion of RNF126 had no effect on the recruitment of EGFP-Ku70/80 to such sites in U2OS cells (Fig. 4C), indicating that RNF126 is not required for localization of Ku to DSBs. However, dissociation of Ku from sites of DNA damage induced by laser microirradiation was delayed by RNF126 depletion (Fig. 4D).
FIG 4.
RNF126 accumulates at DSB sites and regulates dissociation of Ku70/80 from sites of DNA damage. (A) U2OS cells expressing EGFP-tagged RNF126 or Ku80 were monitored for EGFP fluorescence before and 10 min after laser microirradiation. Arrowheads indicate the path of irradiation. The mean intensity of EGFP fluorescence at the sites of DNA damage after irradiation was determined as the means ± SEM for 10 cells. (B) U2OS cells were subjected to immunofluorescence analysis of endogenous RNF126, γH2AX, and Ku70 at 10 min after laser microirradiation. Nuclei were stained with Hoechst 33342 as indicated. The proportion of cells showing colocalization of endogenous Ku70 or RNF126 with γH2AX was determined as the means ± SEM for 20 cells. (C) U2OS cells that had been transfected with RNF126 or control siRNAs, as well as with expression vectors for EGFP-tagged Ku70 and Ku80, were monitored for EGFP fluorescence before and 10 min after laser microirradiation. The mean intensity of EGFP fluorescence at sites of DNA damage after irradiation was determined as the means ± SEM for 10 cells. (D) U2OS cells that had been transfected as described for panel C were monitored for EGFP fluorescence before and at the indicated times after laser microirradiation. (E) Immunoblot analysis of U2OS/TRE/I-SceI-19 cells that had been infected with an empty lentivirus, transfected with RNF126 or control siRNAs, and then subjected to electroporation with expression vectors for EGFP-tagged Ku70 and Ku80, I-SceI, and Cherry-tTA-ER. See Fig. S6A in the supplemental material for similar analysis of corresponding cells infected with a lentivirus encoding an siRNA-resistant form of RNF126. (F) Fluorescence microscopic analysis of U2OS/TRE/I-SceI-19 cells that had been infected with a lentivirus encoding an siRNA-resistant form of RNF126 (RNF mutant) or the corresponding empty virus (vector), transfected with RNF126 or control siRNAs, and then subjected to electroporation with expression vectors for EGFP-tagged Ku70 and Ku80, I-SceI, and Cherry-tTA-ER. The cells were examined at 6, 12, and 24 h after electroporation and exposure to tamoxifen to induce translocation of Cherry-tTA-ER to the nucleus. Arrows indicate DSB sites marked by Cherry-tTA-ER, higher magnification views of which are shown in the insets of the merged images. (G) Mean fluorescence intensity of EGFP-Ku70/80 at DSB sites was determined as the means ± SEM for 20 cells treated as described for panel F. vec, vector. *, P < 0.05 (Student's t test).
RNF126 is required for release of the Ku heterodimer from DSB sites.
Dissociation of the Ku heterodimer is thought to be required for completion of the repair of DNA damage and postrepair recovery (3). Given that our data suggested that RNF126 mediates Ku ubiquitylation and thereby triggers Ku degradation in the late phase of DSB repair, we examined whether RNF126 might contribute to such dissociation of Ku from sites of DNA damage. We chose the cell line U2OS/TRE/I-SceI-19 to visualize the possible effect of RNF126 on Ku dissociation. This cell line stably harbors an array of ∼200 copies of a plasmid, pTRE/I-SceI, that contains the restriction site for I-SceI adjacent to Tet response elements (TREs) (18) (Fig. S5B). These cells were electroporated with plasmids encoding I-SceI and Cherry-tTA-ER (fluorescent Cherry protein tagged with the TRE binding domain of rTA and the hormone binding domain of the estrogen receptor). Tamoxifen was applied to the cell culture medium immediately after electroporation in order to induce translocation of Cherry-tTA-ER to the nucleus for visualization of the integrated pTRE/I-SceI array. Cutting of I-SceI sites by I-SceI induces DSBs at the sites of pTRE/I-SceI integration in the genome. We first confirmed the accumulation of EGFP-RNF126 at the I-SceI-induced DSB focus marked by Cherry-tTA-ER (Fig. S5C). We then found that knockdown of RNF126 prolonged the duration of EGFP-Ku70/80 localization at the DSB focus (Fig. 4E to G), suggesting that RNF126-mediated ubiquitylation of Ku plays a key role in its dissociation from sites of DNA damage. Importantly, this effect of RNF126 siRNA was attenuated by expression of an siRNA-resistant form of RNF126, confirming that it was due to RNF126 knockdown rather than to an off-target action (Fig. 4F and G; Fig. S6A).
RNF126 is required for completion of NHEJ.
Given that we found that RNF126 promotes dissociation of Ku80 from DSB sites by mediating its ubiquitylation, we next examined the contribution of RNF126 to completion of DSB repair, which is supported by Ku80 dissociation. We performed a neutral comet assay to measure DSBs remaining at various times after treatment of cells with bleomycin. Comet tails (moments) were significantly longer in RNF126-depleted U2OS cells at 4 h after bleomycin treatment than in cells transfected with a control siRNA (Fig. 5A), indicating that suppression of RNF126 function has a negative impact on DSB repair. This effect of RNF126 siRNA was also inhibited by expression of an siRNA-resistant form of RNF126 (Fig. S6B). Consistent with the notion that RNF126 is essential for completion of DSB repair, RNF126 knockdown also delayed the disappearance of the DSB marker γH2AX from U2OS cells exposed to IR (Fig. 5B). To examine the contribution of RNF126 to NHEJ specifically, we introduced DSBs in a human lung cancer cell clone (H1299dA3-1#1) that harbors a stably integrated DNA fragment with two recognition sites for the I-SceI endonuclease by transient transfection with an expression vector for I-SceI. Expression of I-SceI in this cell line induces DSBs at the I-SceI sites and consequent excision of a thymidine kinase (TK) gene by NHEJ, resulting in expression of an EGFP gene from an upstream cytomegalovirus (CMV) promoter (19). Thus, the efficiency of NHEJ can be assessed by measurement of EGFP expression. Depletion of RNF126 reduced the percentage of EGFP-positive cells by 30% to 50%, similar to the effect of Ku80 depletion (Fig. 5C). In accordance with the defective repair of DSBs by NHEJ in RNF126-depleted U2OS cells, these cells showed an increased sensitivity to IR and bleomycin treatment, both of which generate DSBs, but not to UV-C, which induces single-strand breaks (Fig. 5D). Again, the effect of RNF126 siRNA on the survival of cells exposed to IR was attenuated by expression of an siRNA-resistant form of RNF126 (Fig. S6C). These results suggested that RNF126-mediated dissociation of the Ku heterodimer from DSB sites is essential for completion of NHEJ, the disruption of which is detrimental to cells.
FIG 5.
RNF126 is required for completion of NHEJ. (A) U2OS cells were transfected with RNF126 or control siRNAs, treated (or not treated [NT]) with bleomycin, incubated for the indicated times, and then subjected to a single-cell neutral gel electrophoresis (comet) assay of DSBs. Representative comet images and quantitative analysis of tail moments for 750 cells from three independent experiments are shown. Quantitative data are means ± SEM. *, P < 0.05 (Student's t test). (B) U2OS cells transfected with RNF126 or control siRNAs were exposed (or left unexposed) to IR (2 Gy) and subjected to immunofluorescence analysis of γH2AX (red) at 0.5 or 24 h after irradiation. Nuclei were stained with Hoechst 33342 (blue). Representative images and quantitative analysis of the total intensity of γH2AX foci in a total of 750 cells before or at 1, 3, 6, 12, 24, and 36 h after irradiation are shown. Quantitative data are means ± SEM from three independent experiments. *, P < 0.05; **, P < 0.01 (Student's t test). (C) H1299dA3-1#1 cells expressing RNF126, Ku80, or control siRNAs were harvested 48 h after transfection with an expression vector for I-SceI [(+); (−), no transfection] for determination of the EGFP-positive cell fraction by flow cytometry. Quantitative data are means ± SEM from three independent experiments. **, P < 0.01; ***, P < 0.005 (Student's t test). The cells were also subjected to immunoblot analysis of the indicated proteins. (D) U2OS cells expressing RNF126 or control siRNAs were exposed to IR or UV-C radiation or treated with bleomycin before determination of colony-forming ability. Data are means ± SEM from three independent experiments. P values were <0.05 (*) and <0.01 (**) versus corresponding values for cells transfected with the control siRNA (Student's t test).
RNF126-dependent Ku80 ubiquitylation regulates NHEJ-mediated repair of DNA damage.
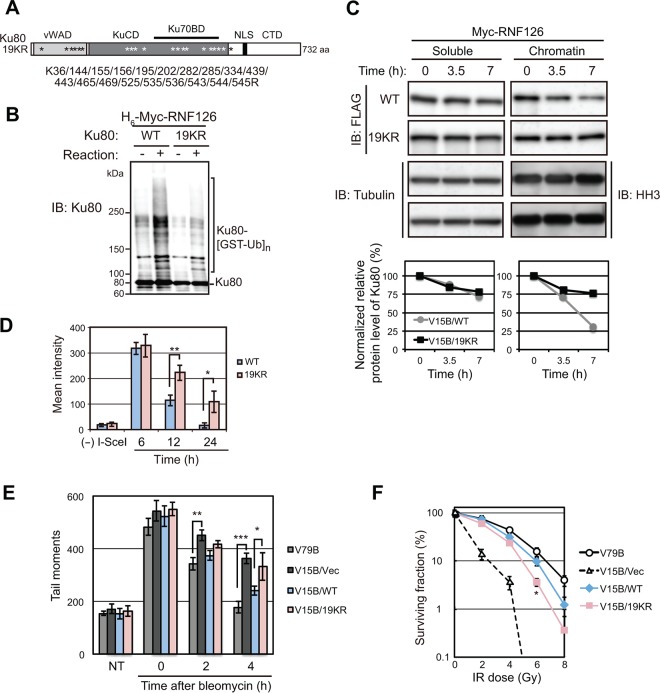
To elucidate the relevance of Ku80 ubiquitylation by RNF126 to NHEJ-mediated repair, we set out to identify the sites of Ku80 ubiquitylation by RNF126 in vivo with the use of proteomics analysis. Extracts of HEK293T cells expressing FLAG-tagged Ku80, Myc epitope-tagged RNF126, and His6-tagged ubiquitin were subjected to serial affinity purification with Ni2+-agarose for pulldown of ubiquitylated proteins and then with antibodies to FLAG. LC-MS/MS analysis revealed that 25 lysine residues of Ku80 were ubiquitylated in these cells. Application of CueMol2 software suggested that 19 of these 25 lysines are exposed on the surface of the Ku heterodimer (20) (Fig. 6A). The level of RNF126-mediated or IR-induced ubiquitylation of a Ku80 mutant [Ku80(19KR)] in which these 19 lysine residues are replaced with arginine (K36/144/155/156/195/202/282/285/334/439/443/465/469/525/535/536/543/544/545R) (Fig. 6A) was reduced to <50% of that of the wild-type (WT) protein in vitro (Fig. 6B) and in vivo (Fig. S7A), respectively, suggesting that these 19 lysines are the major ubiquitylation sites targeted by RNF126. To compare the effects of DSB induction on the stabilities of WT and 19KR mutant forms of Ku80, we introduced FLAG-tagged versions of these proteins in XR-V15B cells, which lack endogenous Ku80, by retroviral infection. The stable sublines V15B/FLAG-Ku80(WT) and V15B/FLAG-Ku80(19KR) expressed similar amounts of Ku70 and Ku80 in both soluble and chromatin fractions (Fig. S7B). Compared with the IR-induced downregulation of Ku80 in the chromatin fraction of V15B/FLAG-Ku80(WT) cells, the loss of FLAG-Ku80(19KR) was delayed (Fig. 6C), indicating that ubiquitylation of the 19 lysine residues plays a key role in the degradation of Ku80 induced by DSB generation. We also found that the release of Ku80(19KR) from DSBs marked with Cherry-tTA-ER was significantly delayed compared with that of Ku80(WT) in U2OS/TRE/I-SceI-19 cells (Fig. 6D; Fig. S7C). This experiment also revealed that Ku80(19KR) translocated to DSB sites. Consistent with these findings, V15B/FLAG-Ku80(19KR) cells manifested longer comet tails (moments) than V15B/FLAG-Ku80(WT) cells at 4 h after bleomycin treatment (Fig. 6E; Fig. S7D), indicative of a defect in completion of DSB repair. Finally, V15B/FLAG-Ku80(19KR) cells were found to be significantly more sensitive to IR than V15B/FLAG-Ku80(WT) cells (Fig. 6F), reinforcing the importance of Ku80 ubiquitylation by RNF126 for DSB repair.
FIG 6.
Ubiquitylation of Ku80 by RNF126 is required for DSB repair by NHEJ. (A) Schematic representation of the KR19 mutant of human Ku80, in which predicted RNF126 ubiquitylation sites are replaced with arginine. (B) In vitro ubiquitylation assay for WT and 19KR mutant forms of Ku80 performed as described for Fig. 2E. (C) XR-V15B cells stably expressing FLAG-Ku80(WT) or FLAG-Ku80(19KR) were electroporated with an expression vector for Myc epitope-tagged RNF126. At 24 h after electroporation, the cells were exposed to IR (2 Gy) and then incubated with cycloheximide (50 μg/ml) for the indicated times. Soluble and chromatin fractions were then prepared from the cells and subjected to immunoblot analysis with antibodies to the indicated proteins. The relative amounts of the Ku80 proteins normalized by the corresponding abundance of tubulin or histone H3 were determined by densitometry. (D) U2OS/TRE/I-SceI-19 cells electroporated with expression vectors for EGFP-Ku70, Cherry-tTA-ER, I-SceI, and EGFP-tagged Ku80(WT) or Ku80(19KR) were treated with tamoxifen and monitored for EGFP and Cherry fluorescence (Fig. S7C). Mean fluorescence intensity of EGFP-Ku70/80 at DSBs was measured for 20 cells at 6, 12, and 24 h after electroporation. Data are means ± SEM. *, P < 0.05; **, P < 0.01 (Student's t test). (E) XR-V15B cells stably expressing FLAG-tagged Ku80(WT) or Ku80(19KR) were treated (or not treated [NT]) with bleomycin (0.1 μg/ml) and then incubated for the indicated times, after which tail moments were measured by single-cell neutral gel electrophoresis (comet assay). V79B (Ku80+/+) cells and XR-V15B cells transfected with an empty vector (Vec) were analyzed as controls. Data are means ± SEM for a total of 500 cells examined in three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.005 (Student's t test). Representative results are shown in Fig. S7D in the supplemental material. (F) Colony-forming ability of XR-V15B cells stably expressing FLAG-tagged Ku80(WT) or Ku80(19KR) after exposure to the indicated doses of IR. Data are means ± SEM for three independent experiments. *, P value of <0.05 versus corresponding values for XR-V15B cells expressing FLAG-Ku80(WT) (Student's t test).
DISCUSSION
The Ku heterodimer, composed of Ku70 and Ku80, is an NHEJ initiating factor that binds to DNA ends generated by DSBs and recruits DNA repair proteins to DSB sites. It has been suggested that completion of NHEJ requires Ku removal from DSB ends after initiation of the NHEJ pathway, but the molecular mechanism of Ku dissociation has been largely unclear.
We have now identified a novel ubiquitin ligase, RNF126, that ubiquitylates Ku80 at DSB sites at a later stage of the NHEJ pathway and thereby induces Ku80 release from these sites, allowing completion of NHEJ (Fig. 7). Impairment of Ku80 ubiquitylation, either by depletion of RNF126 or mutation of Ku80 ubiquitylation sites, resulted in a delay in DSB repair and increased the sensitivity of cells to insults that generate DSBs.
FIG 7.
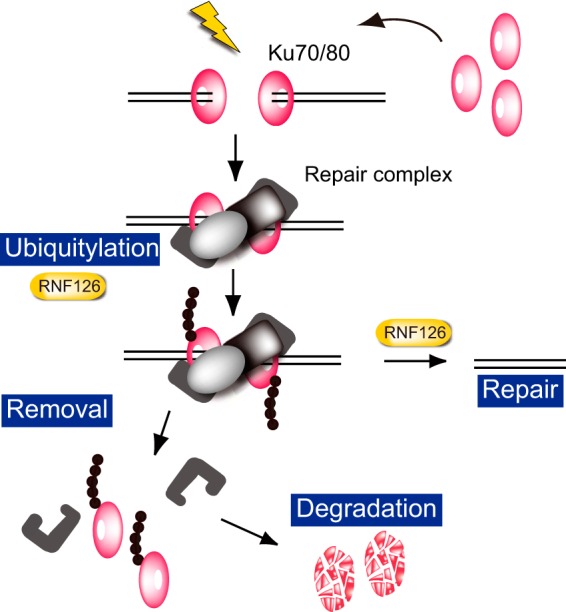
Regulation of Ku80 by RNF126. Model for the role of RNF126 in the ubiquitylation of Ku and its subsequent removal from DSB sites and degradation in the NHEJ pathway of DNA repair.
The importance of Ku80 ubiquitylation was first demonstrated in Xenopus egg extracts (4), with a NEDD8-dependent SCF-type ubiquitin ligase being suspected of mediating this reaction. SCFFbxl12 was subsequently shown to be responsible for such Ku80 ubiquitylation in the presence of DNA containing DSB ends (5). All of the components of SCFFbxl12 (CUL1, SKP1, ROC1, and Fbxl12) are conserved in mammals, suggesting that this ubiquitin ligase complex also targets mammalian Ku80 for ubiquitylation. However, the recent demonstration that Ku80 ubiquitylation induced by DNA damage was not affected by knockdown of Fbxl12 in mammalian cells suggests that SCFFbxl12 is not a major ubiquitin ligase for mammalian Ku80 (6). Other NEDD8-dependent ubiquitin ligases that mediate ubiquitylation of Ku might remain to be identified, as previously suggested (8). In addition, RNF8 was shown to ubiquitylate and thereby to induce the degradation of Ku80 in mammalian cells (7). However, we found that DSB-induced Ku80 ubiquitylation and release from chromatin were attenuated by RNF126 depletion in the presence of RNF8, suggesting that RNF8 and RNF126 have different roles in or cooperatively regulate Ku80 ubiquitylation related to dissociation from chromatin.
Different actions of RNF8 and RNF126 are also suggested by a difference in the time course of their localization to DSBs. Whereas RNF8 was found to localize to DSB sites as early as 30 s after the induction of DNA damage (21), we did not detect such an early response for RNF126. RNF126 may thus be recruited to DSBs to ubiquitylate Ku80 later than RNF8. Recruitment of RNF8 to DSB sites is accomplished by binding of its FHA domain to MDC1 that has been phosphorylated by ATM (21–23). Although the mechanism by which RNF126 translocates to DSB sites remains to be precisely determined, RNF126 recruitment is likely independent of the ATM-MDC1 pathway, given that RNF126 does not possess an FHA domain. We hypothesize that RNF8 and RNF126 target different lysine residues of Ku80 and that ubiquitylation on multiple sites (multiubiquitylation) leads to release of Ku80 from chromatin, whereas polyubiquitylation of any of these sites is required for Ku80 degradation. Consistent with this notion, we identified numerous ubiquitylation sites of Ku80. These two ubiquitin ligases might also contribute differentially to the processivity of Ku80 ubiquitylation. To investigate this possibility, we compared the ubiquitylation activities of RNF126 and RNF8 for Ku80. The level of Ku80 polyubiquitylation was higher in HEK293T cells overexpressing RNF126 than in those overexpressing RNF8 (data not shown), suggesting that RNF126 is a preferred E3 ligase for Ku80 polyubiquitylation after DSB induction. It is possible that RNF8 catalyzes an initial ubiquitylation of Ku80 and that the ubiquitin chain is then extended by RNF126. This scenario is supported by the recent observation that the zinc finger domain of RNF126 binds directly to ubiquitin (24).
RNF126 was also recently shown to enhance HR-mediated DNA repair by upregulating the expression of BRCA1 in a manner independent of its ubiquitin ligase activity (25). This finding, together with our demonstration that RNF126 regulates NHEJ, suggests that RNF126 plays a central role in DSB repair.
In addition to its nuclear function, demonstrated in the present study, emerging evidence suggests that RNF126 targets multiple substrates in the cytosol for ubiquitylation and regulation. Polyubiquitylation of the epidermal growth factor receptor (EGFR) on the endosomal membrane by RNF126 was thus shown to promote its lysosomal sorting and degradation (24). RNF126 also regulates trafficking of another membrane protein, the mannose-6-phosphate receptor, from endosomes to the Golgi complex in a manner dependent on its RING finger domain, although the substrate of RNF126 in this regulation was not identified (26). Mislocalized proteins that contain a transmembrane domain but fail to enter the endoplasmic reticulum are also polyubiquitylated by RNF126 in the cytosol for subsequent degradation (27). These observations show that RNF126 regulates trafficking of membrane proteins at multiple steps in the cytosol, and combined with our present results, they suggest that nucleocytoplasmic shuttling of RNF126 plays a key role in regulation of its function.
Finally, further examination of the physiological functions of RNF126 awaits the generation of RNF126-deficient mice. Our immunoblot analysis of the tissue distribution of mouse RNF126 showed that the protein is highly expressed in testis and is present in smaller amounts in brain and thymus (data not shown), indicating that the effects of RNF126 ablation are prominent in these tissues. Of note, these tissues are especially vulnerable to gene mutations that disrupt the DNA damage response (28).
MATERIALS AND METHODS
Plasmid construction and mutagenesis.
Complementary DNAs encoding human RNF126, Ku70, Ku80, NAP1L4, UBE2D3 (UbcH5C), Hsc70, and ubiquitin, as well as mouse RNF126, were amplified from HeLa cells and NIH 3T3 cells, respectively, and cloned into p3xFLAG (Sigma), pcGN-myc, or EGFP-C1 (Clontech) for transient expression; into pMAL-c2 (New England BioLabs), pET30c (Qiagen), pGEX6P1 (GE Healthcare), or pCold TF DNA (TaKaRa) for recombinant protein production in bacteria; into pFastBac HTa (Life Technologies) for baculovirus production; or into pMX-puro (29) for retrovirus production. Point mutations and deletions were introduced by PCR-based mutagenesis. All constructs were verified by sequencing. The plasmids pCIneo-His6-T7-Ub and pGEX-6P-Ub were kindly provided by K. I. Nakayama (Kyushu University).
Cell culture, transfection, and retroviral and lentiviral infection.
HEK293T, U2OS, Flp-In T-REx 293, V79B, XR-V15B, U2OS/TRE/I-SceI-19, and HeLa cells were cultured under 5% CO2 at 37°C in Dulbecco's modified Eagle's medium (DMEM; Life Technologies) supplemented with 10% fetal bovine serum (Life Technologies). H1299dA3-1#1 cells were cultured under 5% CO2 at 37°C in RPMI 1640 (Life Technologies) supplemented with puromycin (2 μg/ml; Sigma-Aldrich). Vectors were introduced into HEK293T and HeLa cells by transfection with the use of FuGENE 6 (Promega), into Flp-In T-REx 293 and U2OS cells with FuGENE HD (Promega), into XR-V15B and U2OS/TRE/I-SceI-19 cells by electroporation (Microporator MP-100; Life Technologies), and into H1299dA3-1#1 cells with the use of Lipofectamine 2000 (Life Technologies).
Retroviruses encoding wild-type (WT) or mutant forms of Ku80 were generated with the use of Plat-E cells, which were cultured under 5% CO2 at 37°C in DMEM supplemented with puromycin (1 μg/ml) and blasticidin (10 μg/ml; Sigma-Aldrich) and transfected with the use of FuGENE 6. XR-V15B cells were infected with retroviruses as described previously (30), and the infected cells were subjected to selection with puromycin (2 μg/ml).
Lentiviruses encoding a small interfering RNA (siRNA)-resistant form of human RNF126 tagged with the FLAG epitope were generated by transfection of HEK293T cells with pCAG-HIVgp, pCMV-VSV-G-RSV-Rev, and CSII-EF-IRES-puro containing the cDNA for 3×FLAG-tagged siRNA-resistant RNF126 with the use of polyethylenimine MAX (Polysciences). U2OS/TRE/I-SceI-19 cells were infected with the resulting lentiviruses and then subjected to selection with puromycin (2 μg/ml).
RNAi.
Small interfering RNAs were described previously for Ku80 (18), were designed with the use of BLOCK-iT RNAi designer for human RNF126 (Stealth siRNA; http://rnaidesigner.lifetechnologies.com/rnaiexpress), or were obtained from Life Technologies for a negative-control duplex (medium GC duplex, no. 12935-300). To avoid off-target effects of siRNAs and to increase the reliability of gene suppression experiments, we used four siRNA duplexes targeted to human RNF126 mRNA (see Table S1 in the supplemental material), with siRNA 602 being used in most experiments. Cells were transfected with siRNA duplexes with the use of Lipofectamine RNAiMAX (Life Technologies). The efficiency of target protein knockdown was determined by immunoblot analysis.
Antibodies.
Polyclonal antibodies to RNF126 were generated in rabbits by standard procedures with His6-tagged mouse RNF126 (full length) as the antigen. Specific antibodies were purified from serum of injected rabbits by affinity chromatography with a glutathione S-transferase (GST) fusion protein of RNF126. Other antibodies used in this study are listed in Table S2.
Protein identification by LC-MS/MS analysis.
Proteins associated with FLAG-tagged human RNF126 in HEK293T cells were digested with Achromobacter protease I, and the resulting peptides were analyzed with a nanoscale LC-MS/MS (liquid chromatography and tandem mass spectrometry) system as described previously (31).
Immunoprecipitation and immunoblot analysis.
HEK293T, U2OS, HeLa, or Flp-In T-REx 293 cell lysates were incubated for 1 h at 4°C with Dynabeads Protein G (Veritas) and the indicated antibodies. The beads were washed three times with a solution containing 40 mM HEPES-NaOH (pH 7.9), 150 mM NaCl, 1 mM dithiothreitol, and 0.5% Triton X-100, and the immunoprecipitated proteins were subjected to immunoblot analysis. Immune complexes were detected with SuperSignal West Pico or Dura chemiluminescent substrate (Pierce).
Generation of Flp-In T-REx 293-RNF126 cells.
The stable inducible cell line Flp-In T-REx 293-RNF126 was established from Flp-In T-REx 293 cells (Life Technologies). The pcDNA5/FRT/FH-RNF126 vector was generated by subcloning cDNA for mouse RNF126 tagged with tandem FLAG and hemagglutinin (HA) epitopes at its NH2 terminus (FH-RNF126) into a modified pcDNA5/FRT vector (18). Flp-In T-REx 293 cells were transfected with pcDNA5/FRT/FH-RNF126 and pOG44 Flp-recombinase expression vector (Life Technologies) at a molar ratio of 1:9 and then subjected to selection with hygromycin. Hygromycin-resistant cell clones were picked up and expanded. For induction of FH-RNF126 expression, cells were treated with tetracycline (Tet; 0.5 μg/ml) for 20 to 24 h. The expression was verified by immunoblot analysis.
Production of recombinant proteins.
His6- and trigger factor (TF)-tagged Ku70, Ku80, or Hsc70, as well as maltose binding protein (MBP)-tagged mouse RNF126, were expressed in Escherichia coli strain BL21(DE3)pLysS (Novagen) and purified with the use of Ni2+-agarose or amylose-resin chromatography, respectively. Recombinant baculoviruses encoding His6-Myc-tagged WT or mutant forms of mouse RNF126 were generated with the use of the Bac-to-Bac HT expression system (Life Technologies), and the recombinant proteins expressed in Sf21 cells were purified by Ni2+-agarose chromatography as described previously (32).
In vitro binding assays.
Recombinant MBP-RNF126 (500 ng; produced in bacteria) or MBP alone was mixed with 500 ng of recombinant His6-TF-tagged Ku70 or Ku80 (produced in bacteria), and the mixture was incubated for 2 h at 4°C with rotation before precipitation of MBP-RNF126 or MBP with amylose resin. The resulting precipitates were then subjected to immunoblot analysis with antibodies to His6 and MBP.
Ubiquitylation assays and identification of ubiquitylation sites.
For analysis of in vivo ubiquitylation, FLAG- or enhanced green fluorescent protein (EGFP)-tagged Ku70 or Ku80 and His6-T7-tagged ubiquitin were expressed in HEK293T cells. After treatment with MG132, the cells were harvested and ubiquitylated proteins were isolated with the use of Ni2+-agarose (Ni-NTA; Qiagen) under denaturing conditions (6 M urea, 40 mM Tris-HCl [pH 7.5], 25 mM imidazole). The eluted proteins were subjected to immunoblot analysis with antibodies to FLAG or GFP. Ubiquitylation of endogenous Ku80 was analyzed with TUBE technology (LifeSensors). U2OS cells left unexposed or exposed to ionizing radiation (IR; 2 Gy) were treated with 10 μM MG132 and then lysed in a solution containing 20 mM Na2HPO4, 20 mM NaH2PO4 (pH 7.2), 50 mM NaF, 5 mM tetrasodium pyrophosphate, 10 mM β-glycerophosphate, 2 mM EDTA, 1 mM dithiothreitol, and 1% Nonidet P-40. Polyubiquitylated proteins were precipitated in the presence of 100 μM isopeptidase inhibitor with glutathione beads conjugated with GST-TUBE2 (LifeSensors). Proteins eluted from the beads with glutathione were subjected to immunoprecipitation with antibodies to Ku80 followed by immunoblot analysis with antibodies to Ku80 and to multiubiquitin.
For identification of ubiquitylated sites of Ku80, FLAG-tagged Ku80 was expressed with Myc epitope-tagged RNF126 and His6-tagged ubiquitin in HEK293T cells. Ubiquitylated proteins were isolated from cell extracts with Ni2+-agarose, and FLAG-Ku80 was then purified from these proteins by affinity chromatography with antibodies to FLAG (M2) and subjected to SDS-polyacrylamide gel electrophoresis. The band corresponding to ubiquitylated FLAG-Ku80 was excised and subjected to in-gel digestion with trypsin. The resulting peptides were then analyzed with a nanoscale LC-MS/MS system (Orbitrap Velos with ETD; Thermo Fisher Scientific). The ubiquitylated peptides of Ku80 were identified from the raw data with the use of the MASCOT search engine (Matrix Science).
For in vitro ubiquitylation assays, His6-TF-tagged Ku80, Ku70, and Hsc70 were produced in bacteria and the His6-TF tags were cleaved with HRV 3C protease (Novagen). His6-tagged UBE2D3 and GST-tagged ubiquitin were also produced in and purified from bacteria. His6-Myc-tagged RNF126 was prepared from baculovirus-infected Sf21 cells. Ku80, Ku70, or Hsc70 substrate (50 ng) was incubated for 30 min at 26°C with 50 ng of Uba1 (Boston Biomedical), 100 ng of His6-UBE2D3, 200 ng of His6-Myc-RNF126, and 3 μg of GST-ubiquitin in a final volume of 10 μl containing 40 mM HEPES-NaOH (pH 7.9), 60 mM potassium acetate, 2 mM dithiothreitol, 5 mM MgCl2, 0.5 mM EDTA, 10% glycerol, and 1.5 mM ATP. The reaction mixture was then subjected to immunoblot analysis with antibodies to each substrate.
Cycloheximide chase analysis and subcellular fractionation.
U2OS cells (for analysis of endogenous Ku70/80) or XR-V15B cells expressing FLAG-tagged WT or 19KR mutant forms of Ku80 and Myc epitope-tagged human RNF126 were cultured with cycloheximide (50 μg/ml) for various times, after which soluble and chromatin fractions were prepared (33) and subjected to immunoblot analysis with antibodies to Ku70, Ku80, or FLAG.
Immunofluorescence analysis.
Cells were fixed in methanol-acetone (1:1, vol/vol) for 10 min at −20°C and probed with the indicated antibodies. Immune complexes were detected with antibodies to rabbit immunoglobulin G (IgG) (labeled with Alexa Fluor 488 or 556), antibodies to mouse IgG (Alexa Fluor 556), or antibodies to goat IgG (Alexa Fluor 488) (Molecular Probes). Nuclear DNA was stained with Hoechst 33342 (Life Technologies). Fluorescence images were acquired with a BZ9000 fluorescence microscope (Keyence) or DeltaVision microscopy imaging system (GE Healthcare).
Laser microirradiation.
Cells were treated with 10 μM 5-bromo-2-deoxyuridine for 24 h or with 100 μM 8-methoxypsoralen for 3 h before irradiation. The radiation dose was fixed at 800 μW (500 scans, each of 1.6 μW). At least 10 cells were irradiated in each experiment.
Visualization of proteins at DSBs.
RNF126 at DSBs was visualized as described previously (18, 34). U2OS/TRE/I-SceI-19 cells were infected with a lentivirus encoding an siRNA-resistant form of RNF126 (or with the corresponding empty virus), transfected with siRNAs, and then electroporated with plasmids encoding Cherry-tTA-ER (fluorescent Cherry protein tagged with the Tet response element [TRE] binding domain of rTA and the hormone binding domain of the estrogen receptor) and EGFP-tagged test proteins, as well as with pCMV-3xNLS-I-SceI (kindly provided by M. Jasin). Tamoxifen was added to the culture medium immediately after electroporation in order to induce translocation of Cherry-tTA-ER to the nucleus. After incubation for 6 h or the indicated times, the cells were left unexposed or were exposed (Fig. 4F) for 90 s to CSK+R buffer (10 mM PIPES-NaOH [pH 7.0], 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 0.7% Triton X-100, and 0.3 mg/ml RNase A [MP Biomedicals]), fixed, and examined to detect the localization of EGFP-tagged proteins with a BZ9000 fluorescence microscope or DeltaVision microscopy imaging system.
Comet and γH2AX assays.
DSBs were quantified with a neutral comet assay (CometAssay kit; Trevigen) as described previously (18). After treatment with bleomycin (0.1 μg/ml; Wako) in DMEM for 2 h, U2OS cells were washed with phosphate-buffered saline, incubated in fresh DMEM for the indicated times, isolated by exposure to trypsin, mixed with molten agarose, and transferred to a microscope slide. The slides were immersed in a lysis solution for electrophoresis and stained with SYBR green I. Average tail moments were determined for 150 cells per sample with the use of Cellomics ArrayScan VTI (Comet V3 BioApplication; Thermo Fisher Scientific).
The total intensity of phosphorylated histone H2A.X (γH2AX) foci in siRNA-transfected U2OS cells negative for reactivity with antibodies to CENP-F (for elimination of cells in S-G2 phases of the cell cycle) was measured with the use of a Cellomics ArrayScan VTI (Spot Detector V3 BioApplication) after staining with antibodies to γH2AX (35).
NHEJ assay.
H1299dA3-1#1 cells (1 × 104 per well in a 24-well plate), which harbor two I-SceI sites located 1.3 kb apart between a cytomegalovirus (CMV) promoter and the EGFP gene, were transfected with siRNAs 24 h before transfection with the I-SceI expression plasmid pCMV-3xNLS-I-SceI. The cells were isolated by exposure to trypsin 48 h after transfection with the I-SceI plasmid, washed with phosphate-buffered saline, and analyzed with a flow cytometer (LSRFortessa; BD Biosciences). The proportion of EGFP-positive cells was determined for 1 × 104 cells with the use of FACSDiva software (BD Biosciences).
Survival assay.
About 500 U2OS cells were plated per 10-cm dish. For bleomycin treatment, the cells were incubated with the drug for 24 h, washed twice, and cultured in fresh DMEM for 2 weeks. For irradiation, cells were exposed to IR or UV-C at 12 h after plating and then cultured for 2 weeks. The cells were fixed and stained with 4% Giemsa solution for assessment of colony formation. The colonies with >50 cells per colony were counted.
Statistical analysis.
Quantitative data are presented as means ± standard errors of the means (SEM) and were analyzed with Student's t test. A P value of <0.05 was considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank H. Ogiwara and T. Kohno for providing the H1299dA3-1#1 cell line; M. Jasin and H. Tauchi for providing the plasmid pCMV-3xNLS-I-SceI; H. Miyoshi for providing the plasmids pCAG-HIVgp and pCMV-VSV-G-RSV-Rev; K. I. Nakayama for providing the plasmids pCIneo-His6-T7-Ub and pGEX-6P-Ub; H. Nakatsumi for providing the plasmid CSII-EF-IRES-puro; L. Li, S. Nakajima, R. Watanabe, and N. Ishii for technical assistance with DNA damage experiments; K. Murayama for help with structural analysis of the Ku70-Ku80 complex; T. Kitamura for providing pMX-puro and Plat-E cells; the Biomedical Research Core of Tohoku University Graduate School of Medicine for technical support; and other laboratory members for discussion.
We have no conflicts of interest to declare.
This work was supported by grants 20570175 and 25891003 from the Japan Society for the Promotion of Science.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00347-16.
REFERENCES
- 1.Heyer WD, Ehmsen KT, Liu J. 2010. Regulation of homologous recombination in eukaryotes. Annu Rev Genet 44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lieber MR. 2010. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem 79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Postow L. 2011. Destroying the ring: freeing DNA from Ku with ubiquitin. FEBS Lett 585:2876–2882. doi: 10.1016/j.febslet.2011.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Postow L, Ghenoiu C, Woo EM, Krutchinsky AN, Chait BT, Funabiki H. 2008. Ku80 removal from DNA through double strand break-induced ubiquitylation. J Cell Biol 182:467–479. doi: 10.1083/jcb.200802146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Postow L, Funabiki H. 2013. An SCF complex containing Fbxl12 mediates DNA damage-induced Ku80 ubiquitylation. Cell Cycle 12:587–595. doi: 10.4161/cc.23408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ismail IH, Gagne JP, Genois MM, Strickfaden H, McDonald D, Xu Z, Poirier GG, Masson JY, Hendzel MJ. 2015. The RNF138 E3 ligase displaces Ku to promote DNA end resection and regulate DNA repair pathway choice. Nat Cell Biol 17:1446–1457. doi: 10.1038/ncb3259. [DOI] [PubMed] [Google Scholar]
- 7.Feng L, Chen J. 2012. The E3 ligase RNF8 regulates KU80 removal and NHEJ repair. Nat Struct Mol Biol 19:201–206. doi: 10.1038/nsmb.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown JS, Lukashchuk N, Sczaniecka-Clift M, Britton S, le Sage C, Calsou P, Beli P, Galanty Y, Jackson SP. 2015. Neddylation promotes ubiquitylation and release of Ku from DNA-damage sites. Cell Rep 11:704–714. doi: 10.1016/j.celrep.2015.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Metzger MB, Hristova VA, Weissman AM. 2012. HECT and RING finger families of E3 ubiquitin ligases at a glance. J Cell Sci 125:531–537. doi: 10.1242/jcs.091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li W, Bengtson MH, Ulbrich A, Matsuda A, Reddy VA, Orth A, Chanda SK, Batalov S, Joazeiro CA. 2008. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling. PLoS One 3:e1487. doi: 10.1371/journal.pone.0001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koike M, Yutoku Y, Koike A. 2014. Impact of amino acid substitutions in two functional domains of Ku80: DNA-damage-sensing ability of Ku80 and survival after irradiation. J Vet Med Sci 76:51–56. doi: 10.1292/jvms.13-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin S, Weaver DT. 1997. Double-strand break repair by Ku70 requires heterodimerization with Ku80 and DNA binding functions. EMBO J 16:6874–6885. doi: 10.1093/emboj/16.22.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hjerpe R, Aillet F, Lopitz-Otsoa F, Lang V, England P, Rodriguez MS. 2009. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep 10:1250–1258. doi: 10.1038/embor.2009.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan DW, Ye R, Veillette CJ, Lees-Miller SP. 1999. DNA-dependent protein kinase phosphorylation sites in Ku 70/80 heterodimer. Biochemistry 38:1819–1828. doi: 10.1021/bi982584b. [DOI] [PubMed] [Google Scholar]
- 15.Cohen HY, Lavu S, Bitterman KJ, Hekking B, Imahiyerobo TA, Miller C, Frye R, Ploegh H, Kessler BM, Sinclair DA. 2004. Acetylation of the C terminus of Ku70 by CBP and PCAF controls Bax-mediated apoptosis. Mol Cell 13:627–638. doi: 10.1016/S1097-2765(04)00094-2. [DOI] [PubMed] [Google Scholar]
- 16.Mayeur GL, Kung WJ, Martinez A, Izumiya C, Chen DJ, Kung HJ. 2005. Ku is a novel transcriptional recycling coactivator of the androgen receptor in prostate cancer cells. J Biol Chem 280:10827–10833. doi: 10.1074/jbc.M413336200. [DOI] [PubMed] [Google Scholar]
- 17.Errami A, Smider V, Rathmell WK, He DM, Hendrickson EA, Zdzienicka MZ, Chu G. 1996. Ku86 defines the genetic defect and restores X-ray resistance and V(D)J recombination to complementation group 5 hamster cell mutants. Mol Cell Biol 16:1519–1526. doi: 10.1128/MCB.16.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lan L, Ui A, Nakajima S, Hatakeyama K, Hoshi M, Watanabe R, Janicki SM, Ogiwara H, Kohno T, Kanno S, Yasui A. 2010. The ACF1 complex is required for DNA double-strand break repair in human cells. Mol Cell 40:976–987. doi: 10.1016/j.molcel.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 19.Ogiwara H, Ui A, Otsuka A, Satoh H, Yokomi I, Nakajima S, Yasui A, Yokota J, Kohno T. 2011. Histone acetylation by CBP and p300 at double-strand break sites facilitates SWI/SNF chromatin remodeling and the recruitment of non-homologous end joining factors. Oncogene 30:2135–2146. doi: 10.1038/onc.2010.592. [DOI] [PubMed] [Google Scholar]
- 20.Walker JR, Corpina RA, Goldberg J. 2001. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 21.Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. 2007. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 22.Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. 2007. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, Pelletier L, Jackson SP, Durocher D. 2007. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith CJ, Berry DM, McGlade CJ. 2013. The E3 ubiquitin ligases RNF126 and Rabring7 regulate endosomal sorting of the epidermal growth factor receptor. J Cell Sci 126:1366–1380. doi: 10.1242/jcs.116129. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Deng O, Feng Z, Du Z, Xiong X, Lai J, Yang X, Xu M, Wang H, Taylor D, Yan C, Chen C, Difeo A, Ma Z, Zhang J. 2015. RNF126 promotes homologous recombination via regulation of E2F1-mediated BRCA1 expression. Oncogene 35:1363–1372. doi: 10.1038/onc.2015.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith CJ, McGlade CJ. 2014. The ubiquitin ligase RNF126 regulates the retrograde sorting of the cation-independent mannose 6-phosphate receptor. Exp Cell Res 320:219–232. doi: 10.1016/j.yexcr.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 27.Rodrigo-Brenni MC, Gutierrez E, Hegde RS. 2014. Cytosolic quality control of mislocalized proteins requires RNF126 recruitment to Bag6. Mol Cell 55:227–237. doi: 10.1016/j.molcel.2014.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ciccia A, Elledge SJ. 2010. The DNA damage response: making it safe to play with knives. Mol Cell 40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morita S, Kojima T, Kitamura T. 2000. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther 7:1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- 30.Hosogane M, Funayama R, Nishida Y, Nagashima T, Nakayama K. 2013. Ras-induced changes in H3K27me3 occur after those in transcriptional activity. PLoS Genet 9:e1003698. doi: 10.1371/journal.pgen.1003698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Natsume T, Yamauchi Y, Nakayama H, Shinkawa T, Yanagida M, Takahashi N, Isobe T. 2002. A direct nanoflow liquid chromatography-tandem mass spectrometry system for interaction proteomics. Anal Chem 74:4725–4733. doi: 10.1021/ac020018n. [DOI] [PubMed] [Google Scholar]
- 32.Kamura T, Maenaka K, Kotoshiba S, Matsumoto M, Kohda D, Conaway RC, Conaway JW, Nakayama KI. 2004. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev 18:3055–3065. doi: 10.1101/gad.1252404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishida N, Hara T, Kamura T, Yoshida M, Nakayama K, Nakayama KI. 2002. Phosphorylation of p27Kip1 on serine 10 is required for its binding to CRM1 and nuclear export. J Biol Chem 277:14355–14358. doi: 10.1074/jbc.C100762200. [DOI] [PubMed] [Google Scholar]
- 34.Britton S, Coates J, Jackson SP. 2013. A new method for high-resolution imaging of Ku foci to decipher mechanisms of DNA double-strand break repair. J Cell Biol 202:579–595. doi: 10.1083/jcb.201303073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beucher A, Birraux J, Tchouandong L, Barton O, Shibata A, Conrad S, Goodarzi AA, Krempler A, Jeggo PA, Lobrich M. 2009. ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J 28:3413–3427. doi: 10.1038/emboj.2009.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.