Abstract
Objectives:
Roughly half of hereditary nonpolyposis colorectal cancer (HNPCC) cases are Lynch syndrome and exhibit germ-line mutations in DNA mismatch repair (MMR) genes; the other half are familial colorectal cancer (CRC) type X (FCCTX) and are MMR proficient. About 70% of Lynch syndrome tumors have germ-line MLH1 or MSH2 mutations. The clinical presentation, histopathological features, and carcinogenesis of FCCTX resemble those of sporadic MMR-proficient colorectal tumors. It is of interest to obtain biomarkers that distinguish FCCTX from sporadic microsatellite stable (MSS) CRC, to develop preventive strategies.
Methods:
The tumors and adjacent normal tissues of 40 patients with HNPCC were assayed using the Illumina Infinium HumanMethylation27 (HM27) BeadChip to assess the DNA methylation level at about 27,000 loci. The germ-line mutation status of MLH1 and MSH2 and the microsatellite instability status in these patients were obtained. Genome-wide DNA methylation measurements of three groups of patients with general CRC were downloaded from public domain databases. Probes with DNA methylation levels that differed significantly between patients with sporadic MSS CRC and FCCTX were examined, to explore their potential as biomarkers.
Results:
We found that MSS HNPCC tumors were overwhelmingly hypomethylated compared with those from patient groups with other types of CRC, including germ-line MLH1/MSH2-mutated HNPCC and sporadic MSS CRC. Five gene-marker panels that exhibited a sensitivity of 100% and a specificity higher than 90% in both discovery and validation cohorts were proposed to distinguish MSS HNPCC tumors from sporadic MSS CRC.
Conclusions:
Our results warrant further investigation and validation. The loci identified here may become useful biomarkers for distinguishing between FCCTX and sporadic MSS CRC tumors.
Introduction
It is estimated that about half of the families with hereditary nonpolyposis colorectal cancer (HNPCC), as defined according to the Amsterdam II criteria,1 carry germ-line mutations in DNA mismatch repair (MMR) genes.2, 3 The resulting tumors are referred to as Lynch syndrome colorectal cancer (CRC). Among them, roughly 70% carry MLH1 and MSH2 mutations, and the remaining tumors are related to MSH6 and PMS2 mutations.4, 5, 6, 7, 8, 9, 10, 11 Previous studies have reported that HNPCC families with Lynch syndrome CRC are at a high risk of developing early-onset colorectal and endometrial cancers that are microsatellite instable (MSI).8, 12, 13, 14 For HNPCC families with Lynch syndrome CRC, intensive surveillance and prophylactic surgery or chemotherapy are suggested as potential preventive and therapeutic strategies; in addition, strategies for screening Lynch syndrome in patients with CRC have been proposed.15
In the other half of HNPCC families for which there is no evidence of the presence of germ-line mutations in MMR genes, tumors are microsatellite stable (MSS) and constitute an entity distinct from Lynch syndrome tumors; they are termed as familial CRC type X (FCCTX).16, 17, 18 As the clinical presentation and histopathological features of FCCTX mimic those of sporadic MMR-proficient tumors,16, 19 enormous efforts have been made to elucidate the genetic and epigenetic causes of FCCTX.20, 21, 22, 23, 24 It seems that FCCTX carcinogenesis resembles largely that occurring in sporadic MMR-proficient CRC, and it is speculated that most families with FCCTX are at increased risk of developing “sporadic tumors” by being more susceptible to environmental carcinogenic factors.21, 25
In fact, it was shown that MSS HNPCC tumors display a significantly lower degree of methylation at LINE-1, which is a marker of global hypomethylation, than do other subgroups of CRC,22, 23 especially sporadic MSS CRC. Moreover, global DNA hypomethylation has been associated with a poor prognosis, shorter survival, and younger age at onset of CRC, as well as with familial CRC cancer risk.23, 26, 27 It was also observed that the CpG island methylator phenotype (CIMP) is inexistent or rare among FCCTX,21, 22 whereas it is present among sporadic MMR-proficient CRC tumors.28, 29, 30, 31, 32 A recent comprehensive analysis of DNA methylation in CRC tumors, which was performed using the Illumina Infinium HumanMethylation27 (HM27) BeadChip, identified four DNA methylation-based subgroups: one CIMP-high (CIMP-H) group, one CIMP-low (CIMP-L) group, and two non-CIMP groups.33, 34
These advances motivated us to identify DNA methylation-based subgroups of HNPCC, to study CIMP among subgroups of HNPCC; and to compare DNA methylation levels between FCCTX and sporadic MSS CRC tumors, to identify differentially methylated probes. It was hoped that these loci would be useful for the identification of FCCTX-associated CRC. For this, we performed DNA methylation profiling in the tumors and adjacent non-tumor tissues of 40 patients with HNPCC using the HM27 BeadChip, and compared these DNA methylation profiles with those observed in sporadic MSS CRCs.33, 34
Methods
Collection of samples from patients with HNPCC
We adopted the Amsterdam criteria II to define HNPCC, as follows: (1) diagnosis of HNPCC-related cancers in three or more family members; (2) one affected relative should be a first-degree relative of the other two; (3) at least two successive generations should be affected; (4) cancer diagnosis in one or more relatives before the age of 50 years; (5) exclusion of familial adenomatous polyposis; and (6) verification of tumors by pathological examination.1, 35 A total of 135 families with HNPCC were referred from three regions of Taiwan: the Linkou Chang Gung Memorial Hospital, the MacKay Memorial Hospital, and the Cathay General Hospital in northern Taiwan; the Taichung Veterans General Hospital and the Kuang Tien Hospital in central Taiwan; and the Kaohsiung and Chiayi Chang Gung Memorial Hospitals in southern Taiwan. This study used both adenocarcinoma tumors and adjacent non-tumor tissues from 40 patients with HNPCC from these families; samples were selected based on their tissue quality. This study was approved by the Institutional Review Board of the National Health Research Institutes, and all participating patients provided written informed consent. Further information on the study population, including the mutations and frequencies of MLH1 and MSH2 and microsatellite stability status, were reported in our previous study,36 where tumors were classified as having a high frequency of microsatellite instability (MSI-H) if instability in two or more markers was observed in the reference panel (BAT25, BAT26, D2S123, D5S346, and D17S250). In the current study, a tumor was classified as MSS HNPCC if it was wild type for MLH1 and MSH2, and instability was observed in at most one of the five markers in the reference panel, which was in agreement with the findings of an earlier report.37 Using this definition, 10 tumors were classified as being MSS HNPCC.
DNA methylation assay and quality assessment
Genomic DNA was extracted from each fresh frozen tissue sample (both from tumors and adjacent normal tissues) using the DNeasy Blood & Tissue Kit (Qiagen Inc, Valencia, CA, USA), according to the manufacturer's specifications. Bisulfite conversion of genomic DNA was performed using the EZ DNA Methylation Kit (Zymo Research, Irvine, CA, USA), according to the manufacturer's instructions.
Bisulfite-converted genomic DNA (500 ng) from each tissue sample was used to examine DNA methylation levels with the HM27 BeadChip, according to the manufacturer's instructions. All samples were run using a 96-well plate format, to reduce batch effects. The HM27 BeadChip examines the DNA methylation level at 27,578 CpG sites in the promoter regions of 14,495 protein-coding genes and 110 microRNAs.38 The DNA methylation level at each CpG site was measured as a DNA methylated percentage and is referred to as the β-value henceforth (ranging from 0 to 1, with values close to 0 indicating a low level of DNA methylation and values close to 1 indicating a high level of DNA methylation). For quality assessment, the β-values obtained from the HM27 BeadChip were managed using the Methylation Module v1.1 implemented in BeadStudio software (Illumina, San Diego, CA, USA).
We performed data preprocessing in a similar fashion as that described by Hinoue et al.,33 as follows. Based on Illumina's Infinium Assay for Methylation Protocol Guide, probes with a detection P value ≥0.05 for any one sample were excluded. In addition, a probe was excluded if the probe sequence contained single-nucleotide polymorphisms or copy-number variants, or if the probe sequence could not be uniquely mapped with a perfect match in the human genome sequence (hg18). For the former, we used the single-nucleotide polymorphisms collected from Han Chinese in Beijing, China (CHB) and Chinese in Metropolitan Denver, Colorado (CHD) in HapMap Phase III (release 3, Human Genome build 36, hg18) and the copy-number variants provided by the ASN population in the 1000 Genomes Project (version 20100804).39, 40 For the latter, we used the computer tools BLAT and MAQ to align probe sequences.41, 42 We also excluded probes located on chromosomes X and Y. As a result, 20,955 probes were selected for subsequent analysis.
To check the diagnosis based on tissue pathology, unsupervised hierarchical clustering and a principal component analysis were used. More specifically, the top 10% (2,758) most variable probes, as evaluated based on the s.d. of β-values over 80 samples, were used to generate dendrograms via hierarchical clustering, using Ward linkage and Euclidean distance for samples and Pearson's correlation distance for probes. Four tumors were clustered among normal tissues and were excluded from further study (see Supplementary Figure S1A,B online).
Statistical analysis
The Wilcoxon rank-sum test was used to identify differentially methylated probes between any two groups. To address the multiple comparison issue, we report the q-value using the q-value package in R.43, 44 A probe was claimed to be differentially methylated if the q-value was <0.05 and the difference of the median β-values between the two groups was >0.2. The recursively partitioned mixture model (RPMM) was used to identify the subgroups of HNPCC tumors using a level-weighted version of the Bayesian information criterion for split criteria, as implemented in the RPMM package.45 Heatmap representations with dendrograms and partitions were plotted using the function heatmap.3 in the GMD package. All data were analyzed using R statistical software (version 3.1.1, Vienna, Austria).
CRC data sets from the cancer genome atlas (TCGA) and gene expression omnibus
We downloaded three DNA methylation data sets from a public domain. The first included the DNA methylation data of all 194 CRC tumors resected from patients without a family history of CRC, for whom the microsatellite stability status was available from TCGA, and the DNA methylation data of the 32 matched normal tissues; these DNA methylation data were based on the HM27 BeadChip.34 All data were downloaded on 30 January 2015 and will be referred to as the TCGA data set in this study (its characteristics are reported in Supplementary Table S1). The second data set consisted of DNA methylation data for 129 CRC tumors and 29 matched normal tissues from gene expression omnibus (GSE25062), as published by Hinoue et al.33 The third data set consisted of 22 pairs of CRC tumors and adjacent normal tissues from gene expression omnibus (GSE17648), as published by Kim et al.46 For the first and second data sets, we followed the method of Hinoue and colleagues to exclude 4,484 probes with a sequence that contained single-nucleotide polymorphisms or could not be uniquely aligned to the human genome, probes with a detection P value≥0.05, and those located on chromosomes X and Y.33 Because the DNA methylation data in the third data set did not provide detection P values and because the third data set consisted of Korean individuals, in subsequent analyses we used the same probes as those that were retained for the data set of HNPCC described above. In summary, a total of 21,682 probes in the TCGA data set, 21,667 probes in GSE25062, and 20,955 probes in GSE17648 were obtained for subsequent analyses.
Data filtering and normalization were conducted in the same manner as that described by Hinoue et al.33, to minimize batch effects in the TCGA and GSE25062 data sets, separately.
Gene set enrichment analysis
We used a model-based gene set analysis (MGSA),47 which is a Bayesian model-based approach, to explore gene sets/biological pathways that are possibly affected by, or involved in, the differential methylation of probes between the patient groups of interest. In this analysis, the MSigDB collection C2.CGP v5.0,48 which is a collection of gene sets that represent several expression signatures of genetic and chemical perturbations reported in PubMed, was used to explore the target gene sets. The MGSA software was employed using the mgsa package in R/Bioconductor, and a threshold of posterior probability ≥0.5 was applied, as recommended by Bauer et al.47
Results
Sample of patients with HNPCC
The tumor and adjacent normal tissues of 40 patients with CRC from HNPCC families were obtained; their germ-line mutation status at MLH1 and MSH2 and microsatellite stability status were reported in an earlier publication.36 The DNA methylation levels in these 40 tumors and normal tissues were measured using the HM27 BeadChip. After data preprocessing, as described in the Materials and methods section, we obtained the β-values at 20,995 probes for 36 tumors and 40 normal tissues, which were used in subsequent analyses. We also followed the criteria of Hinoue et al.33 to classify 36 tumors as CIMP-H, CIMP-L, or non-CIMP tumors; the details of these criteria are provided in Supplementary Table S2. The clinical and genetic features of these 36 tumors are given in Table 1. In this study, patients with MSS HNPCC were analyzed to identify diagnostic DNA methylation gene-marker panels.
Table 1. Genetic and clinical characteristics of 36 HNPCC tumors.
| Characteristics | CIMP-H | CIMP-L | Neither | Fisher P value |
|---|---|---|---|---|
| Number of patients profiled | 8 | 7 | 21 | |
| Mean age at onset (range) | 47.54 (38.0–59.0) | 55.9 (42.0–68.9) | 51.8 (26.4–82.3) | 0.5177a |
| Gender | ||||
| Male | 6 | 1 | 14 | 0.0398 |
| Female | 2 | 6 | 7 | |
| Tumor location | ||||
| Right (proximal) colon | 3 | 2 | 4 | 0.2261 |
| Transverse colon | 2 | 5 | ||
| Left (distal) colon | 3 | 4 | 4 | |
| Rectum | 1 | 7 | ||
| No information | 1 | |||
| Clinical stage | ||||
| I/II | 4 | 4 | 14 | 0.5690 |
| III/IV | 4 | 3 | 6 | |
| No information | 1 | |||
| MLH1 mutation | ||||
| Mutant | 6 | 1 | 6 | 0.0458 |
| Wild type | 2 | 6 | 14 | |
| No information | 1 | |||
| MSH2 mutation | ||||
| Mutant | 1 | 1 | 4 | 1 |
| Wild type | 7 | 5 | 17 | |
| No information | 1 | |||
| MLH1+MSH2 mutation | ||||
| Mutant | 7 | 2 | 10 | 0.1223 |
| Wild type | 1 | 4 | 10 | |
| No information | 1 | 1 | ||
| MSI status | ||||
| MSI | 5 | 3 | 8 | 0.5252 |
| MSS | 3 | 3 | 13 | |
| No information | 1 | |||
ANOVA, analysis of variance; CIMP-H, CpG island methylator phenotype-high; CIMP-L, CIMP-low; HNPCC, hereditary nonpolyposis colorectal cancer; MSI, microsatellite instability; MSS, microsatellite stable.
ANOVA test.
Differentially methylated probes in HNPCC and general CRC
Considering probes that were differentially methylated between tumors and normal tissues at a false discovery rate of 0.05 (q-value<0.05), we found very similar patterns between HNPCC, the MLH1/MSH2-mutated subgroup of HNPCC, CRC, and the subgroups of CRC. For each of these groups or subgroups, hypermethylated probes were present to a greater extent in tumors compared with hypomethylated probes, <10% of the hypomethylated probes were located in CpG islands, and over 70% of the hypermethylated probes were located in CpG islands.49 In addition, ~90% of hypomethylated probes vs. ~97% of hypermethylated probes were located in promoter regions. The number of hypermethylated probes was always larger than that of hypomethylated probes, with the exception of the MSS HNPCC group (see Table 2 for details). In this sense, MSS HNPCC seems to be globally more hypomethylated than any of the other groups.
Table 2. Number of differentially methylated probes between tumors and normal tissues in several subgroups of CRC at a false discovery rate of 0.05 (q-value<0.05).
| Data set |
HNPCC |
Sporadic CRCa |
CRCb | |||
|---|---|---|---|---|---|---|
| Subgroup | Overall | Lynch synd. | FCCTX | Overall | MSI-L/MSS | Overall |
| Samples | 36/40 | 19/20 | 10/11 | 194/32 | 171/25 | 129/29 |
| # Hypomethylation | 240 | 307 | 200 | 922 | 1,060 | 321 |
| In CGI (%) | 10 (4.2) | 9 (2.9) | 9 (4.5) | 96 (10.4) | 96 (9.1) | 11 (3.4) |
| In PR (%) | 212 (89.5) | 273 (89.5) | 176 (88.9) | 831 (91.5) | 961 (91.8) | 291 (91.5) |
| # Hypermethylation | 590 | 780 | 170 | 1,497 | 1,387 | 1,182 |
| In CGI (%) | 475 (80.5) | 591 (75.8) | 142 (83.5) | 1,039 (69.4) | 971 (70.0) | 874 (73.9) |
| In PR (%) | 520 (97.6) | 688 (96.5) | 151 (97.4) | 1,329 (96.4) | 1,230 (96.5) | 1,063 (97.3) |
CGI, CpG island; CRC, colorectal cancer; FCCTX, familial colorectal cancer type X; HNPCC, hereditary nonpolyposis colorectal cancer; MSI, microsatellite instability; MSS, microsatellite stable; PR, promoter region; TCGA, the cancer genome atlas.
Row 3 gives the number of tumors and normal tissues in each data set, with that for tumors placed in the front; row 4 reports the number of hypomethylated probes; row 5 lists the number (percentage) of hypomethylated probes located in CGIs; row 6 describes the number (percentage) of hypomethylated probes located in PRs, which are defined as being located within 1 kb upstream and 1 kb downstream from the transcription start site; rows 7–9 provide information that is similar to that given in rows 4–6, but for hypermethylated probes.
TCGA.
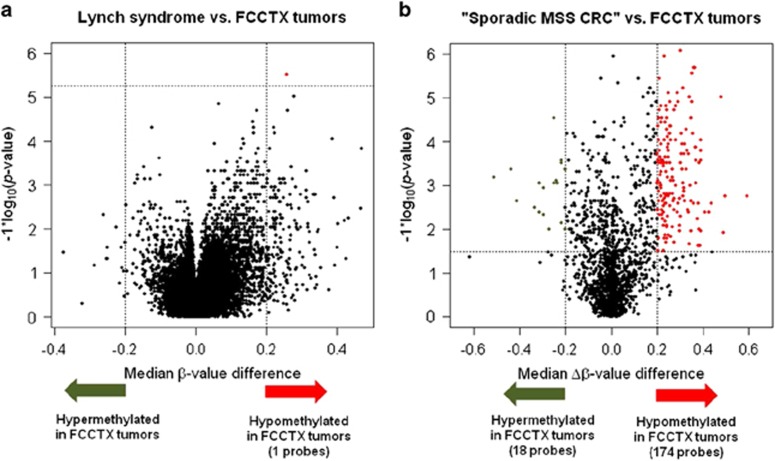
A similar observation was found in a volcano plot, which showed that the β-values in MSS HNPCC tumors were overwhelmingly lower than those detected in germ-line MLH1/MSH2-mutant tumors (Figure 1a). MSS HNPCC tumors were overwhelmingly hypomethylated, whereas only one probe (cg02927346) located in the promoter region of RASL10B showed a statistically significantly lower median of β-values in MSS HNPCC tumors with a difference larger than 0.2 (Figure 1a, red dot).
Figure 1.
Volcano plots show probes at which (a) the DNA methylation level or β-value was significantly lower in FCCTX compared with that observed for germ-line mutations; (b) the delta DNA methylation level or Δβ-value was significantly lower in FCCTX compared with “sporadic MSS CRC” in the TCGA data set. In (a), the x-axis of a point is the median β-value observed in tumors with a germ-line mutation at one probe minus that detected in tumors without the mutation at the same probe; the y-axis of that point gives the −log10 (P value), which compares the β-value in these two groups using the Wilcoxon rank-sum test. The x-axis of a point in (b) is the median Δβ-value at one probe in the “sporadic MSS CRC” from the TCGA data set minus that at the same probe in FCCTX; the y-axis of that point gives the −log10 (P value), which compares the Δβ-value in the two groups of interest using the Wilcoxon rank-sum test. The vertical and horizontal dotted lines, respectively, mark the threshold q-value (0.05) and the difference in the median β-values/Δβ-values (0.2). There were one and 174 probes that were significantly hypomethylated in tumors with FCCTX in a and b, respectively, marked by red dots.
Identification of diagnostic MSS HNPCC DNA methylation gene-marker panels
CpG sites showed little ethnical/dietary differences in β-values in the normal tissues
To minimize possible batch effects when comparing the HNPCC data set with the TCGA, GSE25062, or GSE17648 data sets, we considered a delta DNA methylation level, referred to as the Δβ-value, which was defined as the β-value observed in the tumor minus that recorded in the matched normal tissue. The Δβ-value was calculated for the 10 patients with MSS HNPCC, 32 patients with sporadic CRC from the TCGA data set, 29 GSE25062 patients, and 22 GSE17648 patients for whom the β-values in both the tumors and adjacent normal tissues were measured; hence, the Δβ-values were available for comparison. To reduce the ethnical or dietary disparities between Caucasian and Asian patients, we identified 2,070 probes that showed no differences in β-values among the normal tissues in the TCGA, GSE25062, and GSE17648 data sets (all P values >0.05 in TCGA vs. GSE25062, TCGA vs. GSE17648, and GSE25062 vs. GSE17648; see Supplementary Figure S2 online for details).
MSS HNPCC and “sporadic MSS CRC”
In this paper, a sporadic CRC was called MSIhMLH1 CRC if it was MSI and hypermethylated at MLH1; and non-MSIhMLH1 if it was not MSIhMLH1. We will refer to non-MSIhMLH1 as “sporadic MSS CRC”. Because in sporadic MSI CRC, the vast majority of hypermutated tumors are a result of MLH1 methylation and because in CIMP-H CRC, both histopathological and prognostic features associated with MLH1 methylation are directly related to MSI-H status,50, 51 we used “sporadic MSS CRC” tumors to look for biomarkers that identify MSS HNPCC tumors from sporadic MSS CRC tumors. Using GSE25062, we found that the MLH1 methylation level, as assessed using the MethyLight technology, was highly correlated with the β-value at cg00893636 in the HM27 BeadChip, with a correlation of 0.93. Based on this correlation and the fact that this probe is closest to the current RefSeq MLH1 transcription start sites, we considered a tumor as being methylated at MLH1 if the β-value was ≥0.1 at cg00893636.
Using the probe cg00893636, we found three MSIhMLH1 tumors in the TCGA data set (see column B and row 19 in Supplementary Table S1). Thus, we identified 29 “sporadic MSS CRC” tumors. In this study, we used the 10 MSS HNPCC patients and 29 “sporadic MSS CRC” patients as the discovery cohort for identifying diagnostic MSS HNPCC markers. Based on the Δβ-values calculated at these 2,070 probes, the volcano plot showed that MSS HNPCC tumors were overwhelmingly hypomethylated compared with the tumors with “sporadic MSS CRC” (Figure 1b). One hundred and seventy-four probes were identified as being differentially hypomethylated in MSS HNPCC tumors vs. “sporadic MSS CRC” tumors (Figure 1b, red dots; see Supplementary Table S3 online for a list of these probes and their gene annotations). Interestingly, although a previous study reported that MSS HNPCC tumors displayed a significantly lower global methylation than did sporadic MSS CRC tumors,22 we also identified 18 probes that showed significantly higher Δβ-values in MSS HNPCC tumors compared with those observed in “sporadic MSS CRC” tumors (Figure 1b, green dots; see Supplementary Table S4 online for a list of these probes and their gene annotations).
Pathways involved in the genes which were observed hypomethylation in MSS HNPCC tumors
We used MGSA to explore the gene sets/pathways that are possibly associated with our observation that MSS HNPCC tumors were overwhelmingly hypomethylated compared with the “sporadic MSS CRC” tumors, based on these 174 probes (as mentioned above). Results from MGSA showed that among the three significantly enriched pathways detected here, the pathway “LOPES_METHYLATED_IN_COLON_CANCER_UP”, which represents genes that are methylated aberrantly in the HCT116 and Colo320 colon cancer cell lines, was the most significantly enriched (posterior probability=0.96; see Supplementary Table S5 online for a list of these three pathways).52 This pathway seemed to suggest that the genes that are hypomethylated in MSS HNPCC tumors compared with sporadic MSS CRC tumors were possibly hypermethylated in colon cancer cell lines.
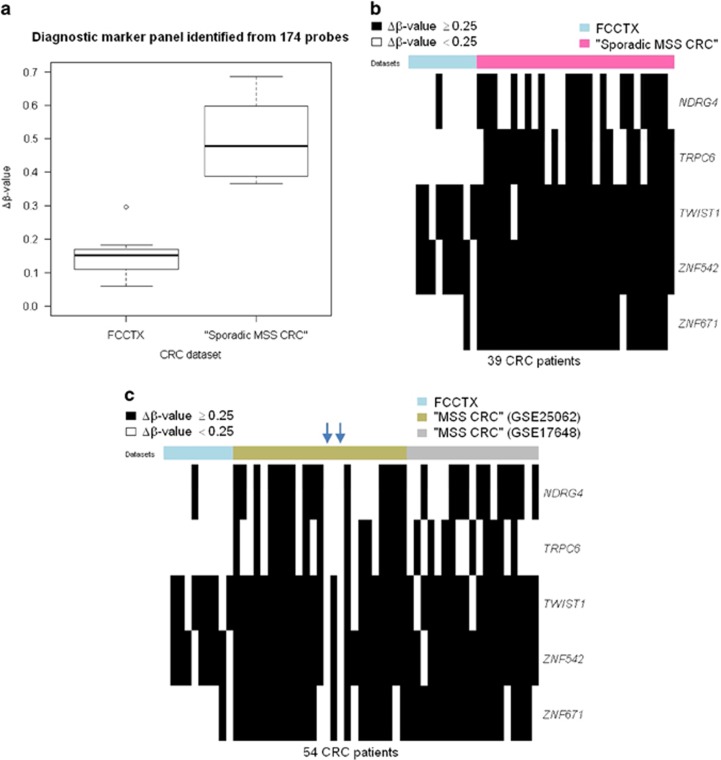
Furthermore, using a more stringent significance threshold (difference of median Δβ-values between tumors with MSS HNPCC and “sporadic MSS CRC” >0.3), we found 56 probes that can be used to develop a possibly more reliable set of DNA methylation makers for the identification of MSS HNPCC tumors. Based on these 56 probes, we proposed MSS HNPCC-defining marker panels consisting of five loci located in the promoter regions of NDRG4, TRPC6, TWIST1, ZNF542, and ZNF671 (see Supplementary Table S6 online). Using the condition that three or more loci with a Δβ-value<0.25 classify a sample as being MSS HNPCC, these marker panels distinguished patients with MSS HNPCC from those with “sporadic MSS CRC” with a sensitivity of 100% and a specificity of 100% (Figure 2a).
Figure 2.
Diagnostic biomarkers for patients with FCCTX. (a) Boxplot of the mean of the five Δβ-values, as defined by the β-value in the tumor minus that in the matched normal tissue, of patients in each data set. The Wilcoxon rank-sum test was used to compare the Δβ-value between patients with FCCTX and those with “sporadic MSS CRC” (P values=4.2e-06). (b,c) Heatmaps of five diagnostic biomarkers, based on the HM27 BeadChip. (b) Thirty-nine columns, 10 for patients with FCCTX and 29 for patients with “sporadic MSS CRC” from the TCGA data set; each row represents one of the five diagnostic biomarkers. (c) Fifty-four columns, 10 for patients with FCCTX and 25 and 19 for patients with ”MSS CRC” from the GSE25062 data set and GSE17648 data set, respectively; each row represents one of the five diagnostic biomarkers. A black bar indicates a Δβ-value ≥0.25 in (b and c). The blue arrow indicates patients with “MSS CRC” who were classified as MSS HNPCC based on Δβ-values <0.25 at all five diagnostic biomarkers.
Validation
To examine the performance of these marker panels, we analyzed the percentage of sporadic MSS CRC cases that could be selected by our marker panels. Because there is no microsatellite stability information for patients in the GSE25062 or GSE17648 data set, and because among CIMP-H CRC cases, both histopathological and prognostic features associated with the MLH1-methylated tumors are directly related to MSI status,51 we excluded CIMP-H and MLH1-methylated tumors in the GSE25062 or GSE17648 data set for the validation study; the resulting data set was called GSE25062MSS or GSE17648MSS. Based on the β-value calculated at the cg00893636 probe used for the identification of MLH1 methylation and the criteria used for identifying CIMP-H tumors proposed by Hinoue et al.,33 there were 25 tumors in GSE25062MSS and 19 tumors in GSE17648MSS. Thus, these 44 tumors, which are referred to as “MSS CRC”, were used as the validation cohort (see Supplementary Figure S3 online for the flow chart of the identification of diagnostic biomarkers). By applying the same criteria used in the discovery cohort, we identified sporadic MSS CRC patients with a specificity of 93.2% among the patients with “MSS CRC” or specificities of 88 and 100% in GSE25062MSS and GSE17648MSS, respectively. Figure 2c describes the Δβ-values at the five probes for each patient with MSS HNPCC and in the validation cohort.
DNA methylation-based HNPCC subgrouping
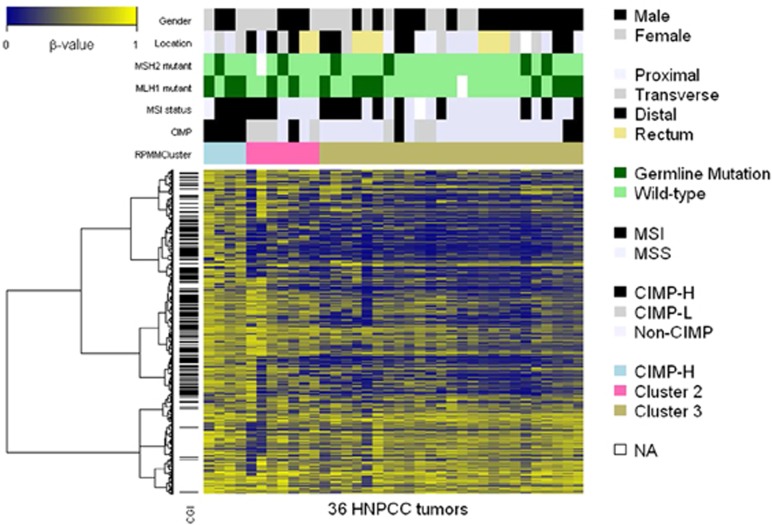
We applied RPMM to the DNA methylation level at the top 10% (2,758) most variable probes for unsupervised clustering of the 36 HNPCC tumors. A total of three clusters were identified and are referred to as clusters 1, 2, and 3. The heatmap of the DNA methylation level (β-value) of the 36 tumors and their genetic and clinical features are presented in Figure 3. We classified tumors as CIMP-H, CIMP-L, or non-CIMP tumors according to Hinoue et al.33 All tumors in cluster 1 were CIMP-H tumors and were either germ-line MLH1 or MSH2-mutated tumors. Therefore, here, we refer to this cluster as the CIMP-H cluster. We found that 57% of the tumors in cluster 2 were CIMP-L tumors and that 76% of the tumors in cluster 3 were neither CIMP-H nor CIMP-L tumors. Interestingly, half of the tumors in cluster 2 or 3 were germ-line MLH1/MSH2-mutated tumors (see Supplementary Table S7 online).
Figure 3.
Heatmap representation of three clusters using unsupervised RPMM clustering, based on the top 10% (2,758) most variable probes. Each of the 36 columns represents one of the 36 HNPCC tumors. The clinical and genetic characteristics of each tumor are marked in color at the top of the heatmap. Three clusters referring to the CIMP-H cluster, cluster 2, and cluster 3 are indicated by blue, pink, and khaki bars, respectively, in the top panel. The clinical and genetic characteristics of three clusters are summarized in Supplementary Table S7. Each row presents β-values over 36 HNPCC tumors for one of the 2,758 most variable probes, and the probe located within a CpG island is marked by a horizontal black bar on the left of the heatmap, whereas the DNA methylation level (β-value) is shown by a color scale ranging from blue (low level of DNA methylation) to yellow (high level of DNA methylation).
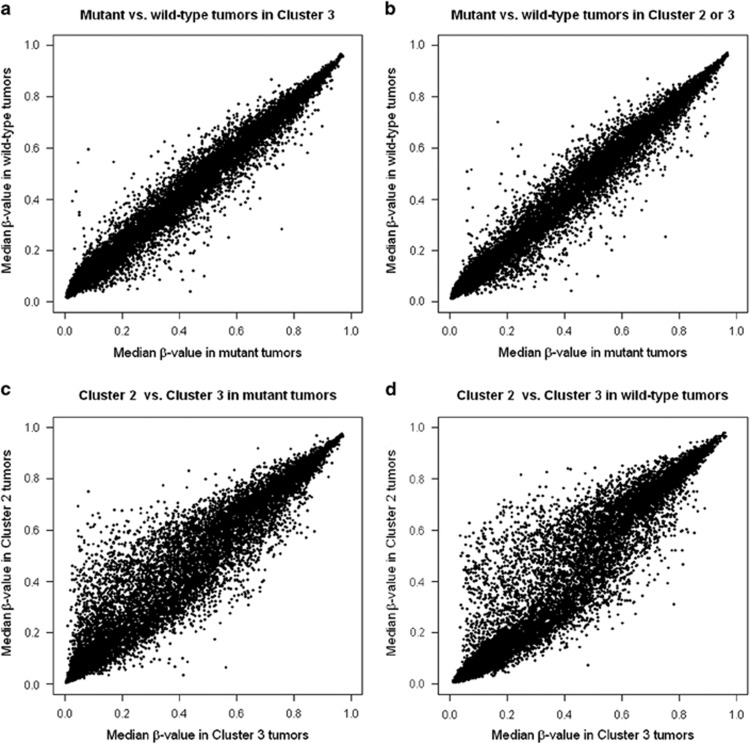
Furthermore, within clusters 2 and 3, there seemed to be little difference between the median β-values in germ-line MLH1/MSH2-mutated and wild-type tumors (Figure 4a). Conversely, within germ-line MLH1/MSH2-mutated tumors or wild-type tumors, the β-value in cluster 2 was generally higher than that detected in cluster 3 (Figure 4c).
Figure 4.
Scatter plots indicating that DNA hypermethylation occurs independently of the MLH1/MSH2 germ-line mutation status in HNPCC tumors, with the exclusion of the CIMP-H cluster. In each scatter plot, one point represents the median β-value at one probe in a subgroup of HNPCC tumors. The scatter plots represent 20,955 median β-values between the germ-line MLH1/MSH2 mutant and wild-type tumors within (a) cluster 3 and (b) clusters 2 and 3. Within both (c) germ-line MLH1/MSH2-mutant and (d) wild-type tumors, several probes showed higher median β-values in cluster 2 compared with cluster 3.
It is known that, in sporadic CRC, CIMP-H tumors are correlated with DNA hypermethylation of MLH1, as measured using the MethyLight technology.33, 53 Based on the β-value calculated at probe cg00893636, we studied the DNA methylation status of MLH1 in our HNPCC tumors and found that, using a threshold of β-value≥0.1, no tumors in HNPCC were MLH1-methylated tumors.
Discussion
Based on the DNA methylation data collected from 36 tumors and adjacent normal tissues from patients with CRC from HNPCC families, we identified probes that were differentially methylated between tumors and normal tissues. We found that similar percentages (<10%) of hypomethylated probes were located in CpG islands and similar percentages (over 70%) of hypermethylated probes were located in promoter regions across different subgroups of CRC, including HNPCC, subgroups of HNPCC, sporadic MSS CRC, and sporadic CRC. Our findings support earlier observations that indicate the presence of significant differences between the characteristics of genes that gain DNA methylation during tumorigenesis vs. those of gene exhibiting lose DNA methylation.54 For patients with MSS HNPCC, hypermethylated probes were present to a lesser extent in tumors compared with hypomethylated probes, suggesting that tumors with MSS HNPCC display a different DNA methylation profile compared with other subgroups of CRC, including Lynch syndrome, sporadic MSS CRC, and sporadic CRC.
We also identified three DNA methylation-based clusters using an unsupervised algorithm, RPMM.45 Every tumor in cluster 1 was a CIMP-H and MLH1/MSH2-mutated tumor. The majority of the tumors in cluster 2 were CIMP-H or CIMP-L tumors. The frequency and level of cancer-specific DNA hypermethylation were lowest in cluster 3. Based on cluster 2 or 3, the germ-line MLH1/MSH2 status seemed to be uncorrelated with DNA methylation level, suggesting that DNA methylation-based clusters, with the exclusion of the CIMP-H cluster, are not associated with germ-line mutations. The DNA methylation profiles observed within these subgroups showed that DNA hypermethylation occurred independently of the germ-line MLH1/MSH2 mutation status, suggesting that DNA methylation in HNPCC involves more complex tumorigenesis mechanisms.
In the Appendix, we classify the gene promoters that acquired cancer-specific DNA methylation into three categories based on DNA methylation profiles, and consider two additional categories of promoters that were constitutively methylated and unmethylated. The properties of these categories of gene promoters resembled those of general CRC in terms of their structural and sequence characteristics,33 including their proximity to Alu and LINE-1 elements and the trimethylation status of H3K4me3 and H3K27me3 in human ESCs. The finding that none of the 36 tumors were methylated at MLH1 seems to be a prominent feature of HNPCC.
We found that one probe in RASL10B was differentially hypomethylated in FCCTX tumors compared with the tumors with Lynch syndrome. RASL10B encodes a small GTPase with tumor-suppressor potential, and epigenetic silencing of this gene has been reported in human hepatocellular carcinoma cells and breast cancer;55, 56 most interestingly, the accumulation of aberrant methylation of RASL10B was reported in association with the development of sessile serrated adenomas/polyps,57 which are putative precursors of colon cancer with MSI.
Despite the similarities observed between sporadic MSS CRC and FCCTX tumors in terms of clinical presentation, histopathological features, and carcinogenesis, as discussed in the literature and as shown in our DNA methylation profiling, we identified 174 CpG sites that were not differentially methylated between normal tissues from the TCGA data set, the Netherlands/Ontario study (GSE25062), and the Korean study (GSE17648),33, 46, 58 but were significantly hypomethylated in FCCTX tumors compared with “sporadic MSS CRC” tumors in terms of Δβ-value. Taking advantage of this large set of hypomethylated probes, we proposed the diagnostic DNA methylation gene-marker panels to identify FCCTX patients among patients with sporadic MSS CRC. These marker panels consisted of five loci that were located in the promoter regions of NDRG4, TRPC6, TWIST1, ZNF542, and ZNF671 and had 100% sensitivity and 100% specificity in our discovery cohort, based on the criterion that the DNA methylation of three or more biomarkers with a Δβ-value<0.25 qualifies a patient as having FCCTX.
According to previous studies, four genes in our marker panels were reported as being hypermethylated in tumors with CRC compared with normal tissues. For example, Okada et al.59 reported that TWIST-1 is frequently hypermethylated in CRC tumors and its methylation may be a biomarker for the noninvasive detection of CRC using stool samples. Similarly, Melotte et al.60 considered hypermethylation in the promoter of NDRG4, which is a putative tumor-suppressor gene, as a potential biomarker for the same purpose. Gevaert et al.61 also reported that ZNF542 and ZNF671 were highly hypermethylated across 10 cancer types, including CRC, using the novel computational algorithm MethylMix. Interestingly, all five genes were markedly differentially hypermethylated in “sporadic MSS CRC” tumors compared with matched normal tissues; however, with the exception of NDRG4, which was not differentially methylated between FCCTX tumors and matched normal tissues (P value >0.05), these genes were less hypermethylated in tumors with FCCTX. These observations were also consistent with the top pathway identified by the gene set enrichment analysis. In contrast, among the 18 genes that showed higher Δβ-value in FCCTX, many were hypomethylated in tumor, compared with the matched normal tissues in both FCCTX and "sporadic MSS CRC" (see Supplementary Table S4 online). Etiologically, these results imply that the molecular pathways that are involved in the carcinogenesis of FCCTX and sporadic MSS CRC are epigenetically distinct. This novel finding warrants additional investigation regarding the detailed mechanistic insights.
We validated the performance of our diagnostic biomarkers using the patients with “MSS CRC” that were classified in the Netherlands/Ontario and Korean studies, because there was no microsatellite stability information in these two data sets and also because among CIMP-H CRC cases, both histopathological and prognostic features associated with the MLH1-methylated tumors are directly related to MSI status.51 Validation studies identified sporadic MSS CRC patients with specificities of 88 and 100% in the Netherlands/Ontario study and Korean study, respectively. Interestingly, we found that, in the Netherlands/Ontario study, there were two patients whose tumors were classified as MSS HNPCC with Δβ-values<0.25 at five diagnostic biomarkers, whereas in the Korean study, no patient showed these characteristic (see blue arrow in Figure 2c). We wonder if the higher specificity detected in the Korean study stemmed from the fact that no patients in the Korean study had clinically apparent polyposis syndrome or Lynch syndrome.46 This result suggests that there is a good chance that our diagnostic biomarkers will perform satisfactorily when applied to sporadic MSS CRC.
We followed the practice in this area to consider panels with five markers. In fact, there were 18 sets of five-marker panels with 100% sensitivity and >90% specificity in the discovery and validation cohorts (see Supplementary Table S10 online for a list of these sets). Specifically, among these 18 sets, another two sets had not only the same specificity (93.2%) as the original mark panel but the same DNA methylation profiles in the two patients indicated by the two blue arrows in Figure 2c as the original mark panel (data not shown). We chose a conservative set of maker panels to demonstrate the possibility of distinguishing FCCTX patients from patients with sporadic MSS CRC. It would be appropriate to explore other biomarkers in the future.
As this is one of the few studies that were specifically designed to investigate genome-wide DNA methylation profiling in HNPCC and one of the first studies to propose molecular biomarkers to discriminate FCCTX patients from patients with sporadic MSS CRC, we are encouraged by our interesting findings. However, the recruitment of additional patients and the analyses of their DNA methylation status at other CpG sites would be beneficial.
In summary, our findings demonstrated that a differential global DNA methylation pattern not only seemed to exist between patients with FCCTX and sporadic MSS CRC, but also led to the development of diagnostic DNA methylation gene-marker panels aimed at discriminating patients with FCCTX from those with sporadic MSS CRC. It is hoped that further validation studies will render these marker panels useful for the screening of FCCTX tumors in sporadic MSS CRC patients, which may lead to useful preventive strategies for the family members of patients with FCCTX.
Study Highlights
Acknowledgments
We would like to thank the patients and their relatives for their participation in this study. Dr Chen is supported by the Taiwan Bioinformatics Institute Core Facility. Dr Tsai is supported in part by a grant from the National Health Research Institutes (PH-100-PP-14). Part of the work is supported by a MOST grant from the Taiwan Bioinformatics Core project (103-2319-B-400-001).
Appendix
Characteristics of gene promoters in CIMP-associated HNPCC subgroups
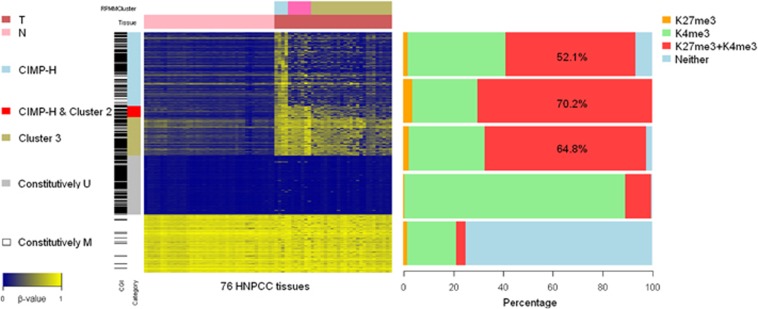
To explore further the characteristics of CIMP-associated HNPCC subgroups, we identified cluster-specific gene promoters in the CIMP-H cluster, cluster 2, and cluster 3. We identified 630 unique gene promoters that showed significant CIMP-H cluster-specific DNA hypermethylation (differential DNA hypermethylation in CIMP-H cluster vs. cluster 3, and a P value >0.05 for cluster 2 vs. cluster 3). We identified 97 unique gene promoters that showed DNA hypermethylation in both the CIMP-H cluster and cluster 2 compared with cluster 3 (differential DNA hypermethylation in CIMP-H cluster vs. cluster 3, and cluster 2 vs. cluster 3). Finally, we identified 324 unique gene promoters that acquired cancer-specific DNA hypermethylation in cluster 3 (differential DNA hypermethylation in cluster 3 tumors vs. cluster 3 normal tissues). For comparison, we selected two sets of gene promoters, each including 500 elements. The probes in one set were all constitutively methylated, and those in the other were all constitutively unmethylated, both in tumors and normal tissues (β-value >0.75 for constitutively methylated probes; β-value<0.05 for constitutively unmethylated probes). A heatmap of DNA methylation levels for these probes, which were classified into five categories is presented in Appendix Figure A1.
Figure A1.
Heatmap and proportion of ESC histone modification associated with unique gene promoters identified in the CIMP-associated HNPCC subgroups. In the panel shown on the left, a total of 76 columns represent 76 HNPCC tissues, including 40 normal tissues and 36 tumors, which are indicated as a light-red and dark-red bar, respectively, at the top of the heatmap. The RPMM-based clusters, CIMP-H cluster, cluster 2, and cluster 3, which are indicated by blue, pink, and khaki bars, respectively, correspond to those shown at the top of Figure 3. For each row, each gene promoter in five categories is indicated by five color bars in the left panel, as follows: (1) 630 CIMP-H cluster-specific DNA hypermethylated gene promoters, indicated by the blue bar; (2) 97 DNA hypermethylated gene promoters in both the CIMP-H cluster and cluster 2, indicated by the red bar; (3) 324 cancer-specific DNA hypermethylated gene promoters in cluster 3, indicated by the khaki bar; (4) 500 constitutively methylated gene promoters in both tumors and normal tissues, indicated by the gray bar; and (5) 500 constitutively unmethylated gene promoters in both tumors and normal tissues, indicated by the white bar. The panel on the right represents the proportions of H3K4me3 and H3K27me3 corresponding to the five categories, in the order shown on the left panel of the heatmap, based on a published data set.
To explore whether retrotransposons are more frequently resistant to DNA hypermethylation in CIMP-associated CpG islands, we calculated the distances from these probes to the nearest Alu and LINE-1 repetitive elements, which were provided by the website of UCSC Genome Bioinformatics website, using the RepeatMasker software (http://www.repeatmasker.org). We found that distances in all three clusters were significantly larger than those observed in the constitutively methylated or constitutively unmethylated set, which was not involved in cancer-specific DNA methylation changes (all P values<0.01, Wilcoxon rank-sum test; details are provided in Supplementary Table S8).
Next, we examined the proportion of H3K4me3 and H3K27me3 in ESCs using a previously published data set.62 We found that, in each CIMP-associated HNPCC subgroup, more than 50% of gene promoters were occupied by both H3K4me3 and H3K27me3; that constitutively unmethylated genes were enriched for H3K4me3 but not for H3K27me3; and that constitutively methylated genes were not enriched for chromatin states with H3K4me3 or H3K27me3. The fraction of genes that coincided with ESC bivalent domains was substantially higher for the genes that underwent cancer-specific DNA methylation than it was for those that were constitutively methylated or unmethylated. These findings are similar to those obtained for CRC,33 suggesting that the characteristics of the CIMP-associated subgroups are not specific to sporadic cases; rather, they seem to be general features of CRC with or without hereditary cases.
Technical validation using pyrosequencing
To assess the reliability of the calculation of DNA methylation levels based on the HM27 BeadChip technology, a technical validation was performed using pyrosequencing with Qiagen PyroMark assays (Qiagen Inc). Based on the involvement of their corresponding genes in CRC, we validated the DNA methylated percentages at seven loci located in ADHFE1, APC, MLH1, and POMC in 12 HNPCC patients (24 samples), and explored the correlations between the two platforms.
In detail, genomic DNA samples (0.5–1 ng) were bisulfite converted using the EZ DNA Methylation Kit (Zymo Research). A DNA fragment was amplified by PCR from bisulfite-converted genomic DNA. The PCR products were purified and sequenced using a PyroMark Q24 System (Qiagen), according to the protocols suggested by the manufacturer. The percentage of DNA methylation at each probe was quantified using Pyro Q-CpG software, version 1.0.9 (Biotage, Uppsala, Sweden). The reverse primers and forward primers used for PCR amplification and the sequencing primers used for pyrosequencing are provided in Supplementary Table S9. All correlations between the two platforms were confirmed to have high consistency (0.77–0.99; Supplementary Figure S4).
Footnotes
Supplementary Information accompanies this paper on the Clinical and Translational Gastroenterology website (http://www.nature.com/ctg)
Guarantor of the article: I-Shou Chang, PhD.
Specific author contributions: Chung-Hsing Chen performed data analysis, assisted in data collection, interpreted the results, and drafted the manuscript. Shih Sheng Jiang conceptualized and designed the study and assisted in data analysis, and interpreted the results. Ling-Ling Hsieh and Reiping Tang conceptualized and designed the study and assisted in participant recruitment, cohort maintenance, and acquisition of data. Chao A. Hsiung conceptualized and designed the study and assisted in data analysis. Hui-Ju Tsai conceptualized, designed, and supervised the study and raised funding for the study. I-Shou Chang conceptualized, designed, and supervised the study, raised funding for the study, and drafted the manuscript.
Financial support: Chen is supported by the Taiwan Bioinformatics Institute Core Facility. Tsai is supported in part by a grant from the National Health Research Institutes (PH-100-PP-14). Part of the work is supported by a MOST grant from the Taiwan Bioinformatics Core project (103-2319-B-400-001).
Potential competing interests: None.
Supplementary Material
References
- Vasen HF, Watson P, Mecklin JP et al. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology 1999; 116: 1453–1456. [DOI] [PubMed] [Google Scholar]
- Heinimann K, Muller H, Weber W et al. Disease expression in Swiss hereditary non-polyposis colorectal cancer (HNPCC) kindreds. Int J Cancer 1997; 74: 281–285. [DOI] [PubMed] [Google Scholar]
- Syngal S. Hereditary nonpolyposis colorectal cancer: a call for attention. J Clin Oncol 2000; 18: 2189–2192. [DOI] [PubMed] [Google Scholar]
- Lin KM, Shashidharan M, Thorson AG et al. Cumulative incidence of colorectal and extracolonic cancers in MLH1 and MSH2 mutation carriers of hereditary nonpolyposis colorectal cancer. J Gastrointest Surg 1998; 2: 67–71. [DOI] [PubMed] [Google Scholar]
- Kolodner RD, Tytell JD, Schmeits JL et al. Germ-line msh6 mutations in colorectal cancer families. Cancer Res 1999; 59: 5068–5074. [PubMed] [Google Scholar]
- Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med 2003; 348: 919–932. [DOI] [PubMed] [Google Scholar]
- Nakagawa H, Lockman JC, Frankel WL et al. Mismatch repair gene PMS2: disease-causing germline mutations are frequent in patients whose tumors stain negative for PMS2 protein, but paralogous genes obscure mutation detection and interpretation. Cancer Res 2004; 64: 4721–4727. [DOI] [PubMed] [Google Scholar]
- Bonadona V, Bonaiti B, Olschwang S et al. Cancer risks associated with germline mutations in MLH1, MSH2, and MSH6 genes in Lynch syndrome. JAMA 2011; 305: 2304–2310. [DOI] [PubMed] [Google Scholar]
- Beck NE, Tomlinson IPM, Homfray T et al. Genetic testing is important in families with a history suggestive of hereditary non-polyposis colorectal cancer even if the Amsterdam criteria are not fulfilled. Br J Surg 1997; 84: 233–237. [PubMed] [Google Scholar]
- Chung DC, Rustgi AK. The hereditary nonpolyposis colorectal cancer syndrome: genetics and clinical implications. Ann Intern Med 2003; 138: 560–570. [DOI] [PubMed] [Google Scholar]
- Palomaki GE, McClain MR, Melillo S et al. EGAPP supplementary evidence review: DNA testing strategies aimed at reducing morbidity and mortality from Lynch syndrome. Genet Med 2009; 11: 42–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RJ, McPhillips M, Meldrum CJ et al. Hereditary nonpolyposis colorectal cancer in 95 families: differences and similarities between mutation-positive and mutation-negative kindreds. Am J Hum Genet 2001; 68: 118–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aaltonen LA, Peltomaki P, Leach FS et al. Clues to the pathogenesis of familial colorectal cancer. Science 1993; 260: 812–816. [DOI] [PubMed] [Google Scholar]
- Lindor NM, Petersen GM, Hadley DW et al. Recommendations for the care of individuals with an inherited predisposition to Lynch syndrome: a systematic review. JAMA 2006; 296: 1507–1517. [DOI] [PubMed] [Google Scholar]
- Kastrinos F, Syngal S. Screening patients with colorectal cancer for Lynch syndrome: what are we waiting for? J Clin Oncol 2012; 30: 1024–1027. [DOI] [PubMed] [Google Scholar]
- Llor X, Pons E, Xicola RM et al. Differential features of colorectal cancers fulfilling Amsterdam criteria without involvement of the mutator pathway. Clin Cancer Res 2005; 11: 7304–7310. [DOI] [PubMed] [Google Scholar]
- Mueller-Koch Y, Vogelsang H, Kopp R et al. Hereditary non-polyposis colorectal cancer: clinical and molecular evidence for a new entity of hereditary colorectal cancer. Gut 2005; 54: 1733–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindor NM, Rabe K, Petersen GM et al. Lower cancer incidence in Amsterdam-I criteria families without mismatch repair deficiency: familial colorectal cancer type X. JAMA 2005; 293: 1979–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klarskov L, Holck S, Bernstein I et al. Hereditary colorectal cancer diagnostics: morphological features of familial colorectal cancer type X versus Lynch syndrome. J Clin Pathol 2012; 65: 352–356. [DOI] [PubMed] [Google Scholar]
- Ku CS, Cooper DN, Wu M et al. Gene discovery in familial cancer syndromes by exome sequencing: prospects for the elucidation of familial colorectal cancer type X. Mod Pathol 2012; 25: 1055–1068. [DOI] [PubMed] [Google Scholar]
- Bellido F, Pineda M, Sanz-Pamplona R et al. Comprehensive molecular characterisation of hereditary non-polyposis colorectal tumours with mismatch repair proficiency. Eur J Cancer 2014; 50: 1964–1972. [DOI] [PubMed] [Google Scholar]
- Goel A, Xicola RM, Nguyen TP et al. Aberrant DNA methylation in hereditary nonpolyposis colorectal cancer without mismatch repair deficiency. Gastroenterology 2010; 138: 1854–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S, Nishihara R, Lochhead P et al. Prospective study of family history and colorectal cancer risk by tumor LINE-1 methylation level. J Natl Cancer Inst 2013; 105: 130–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S, Lochhead P, Chan AT et al. Molecular pathological epidemiology of epigenetics: emerging integrative science to analyze environment, host, and disease. Mod Pathol 2013; 26: 465–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Valentin M, Therkildsen C, Da Silva S et al. Familial colorectal cancer type X: genetic profiles and phenotypic features. Mod Pathol 2015; 28: 30–36. [DOI] [PubMed] [Google Scholar]
- Colussi D, Brandi G, Bazzoli F et al. Molecular pathways involved in colorectal cancer: implications for disease behavior and prevention. Int J Mol Sci 2013; 14: 16365–16385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antelo M, Balaguer F, Shia JR et al. A high degree of LINE-1 hypomethylation is a unique feature of early-onset colorectal cancer. Plos One 2012; 7: e45357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jover R, Nguyen TP, Perez-Carbonell L et al. 5-Fluorouracil adjuvant chemotherapy does not increase survival in patients with CpG island methylator phenotype colorectal cancer. Gastroenterology 2011; 140: 1174–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosho K, Irahara N, Shima K et al. Comprehensive biostatistical analysis of CpG island methylator phenotype in colorectal cancer using a large population-based sample. PLoS One 2008; 3: e3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samowitz WS, Albertsen H, Herrick J et al. Evaluation of a large, population-based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology 2005; 129: 837–845. [DOI] [PubMed] [Google Scholar]
- Ogino S, Goel A. Molecular classification and correlates in colorectal cancer. J Mol Diagn 2008; 10: 13–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barault L, Charon-Barra C, Jooste V et al. Hypermethylator phenotype in sporadic colon cancer: study on a population-based series of 582 cases. Cancer Res 2008; 68: 8541–8546. [DOI] [PubMed] [Google Scholar]
- Hinoue T, Weisenberger DJ, Lange CP et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res 2012; 22: 271–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas N.. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012; 487: 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umar A, Risinger JI, Hawk ET et al. Testing guidelines for hereditary non-polyposis colorectal cancer. Nat Rev Cancer 2004; 4: 153–158. [DOI] [PubMed] [Google Scholar]
- Tang R, Hsiung C, Wang JY et al. Germ line MLH1 and MSH2 mutations in Taiwanese Lynch syndrome families: characterization of a founder genomic mutation in the MLH1 gene. Clin Genet 2009; 75: 334–345. [DOI] [PubMed] [Google Scholar]
- Sanchez-de-Abajo A, de la Hoya M, van Puijenbroek M et al. Molecular analysis of colorectal cancer tumors from patients with mismatch repair—Proficient hereditary nonpolyposis colorectal cancer suggests novel carcinogenic pathways. Clin Cancer Res 2007; 13: 5729–5735. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Le J, Barnes B et al. Genome-wide DNA methylation profiling using Infinium (R) assay. Epigenomics 2009; 1: 177–200. [DOI] [PubMed] [Google Scholar]
- Altshuler DM, Gibbs RA, Peltonen L et al. Integrating common and rare genetic variation in diverse human populations. Nature 2010; 467: 52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler DM, Durbin RM, Abecasis GR et al. A global reference for human genetic variation. Nature 2015; 526: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res 2002; 12: 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 2008; 18: 1851–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD. A direct approach to false discovery rates. J R Stat Soc Ser B Stat Methodol 2002; 64: 479–498. [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA 2003; 100: 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Christensen BC, Yeh RF et al. Model-based clustering of DNA methylation array data: a recursive-partitioning algorithm for high-dimensional data arising as a mixture of beta distributions. BMC Bioinformatics 2008; 9: 365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YH, Lee HC, Kim SY et al. Epigenomic analysis of aberrantly methylated genes in colorectal cancer identifies genes commonly affected by epigenetic alterations. Ann Surg Oncol 2011; 18: 2338–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer S, Gagneur J, Robinson PN. GOing Bayesian: model-based gene set analysis of genome-scale data. Nucleic Acids Res 2010; 38: 3523–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci USA 2002; 99: 3740–3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology 2010; 138: 2073–U87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Bae JM, Cho NY et al. Distinct features between MLH1-methylated and unmethylated colorectal carcinomas with the CpG island methylator phenotype: implications in the serrated neoplasia pathway. Oncotarget 2016; 7: 14095–14111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes EC, Valls E, Figueroa ME et al. Kaiso contributes to DNA methylation-dependent silencing of tumor suppressor genes in colon cancer cell lines. Cancer Res 2008; 68: 7258–7263. [DOI] [PubMed] [Google Scholar]
- Weisenberger DJ, D Siegmund K, Campan M et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet 2006; 38: 787–793. [DOI] [PubMed] [Google Scholar]
- Selamat SA, Chung BS, Girard L et al. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res 2012; 22: 1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin ZY, Chuang WL. Genes responsible for the characteristics of primary cultured invasive phenotype hepatocellular carcinoma cells. Biomed Pharmacother 2012; 66: 454–458. [DOI] [PubMed] [Google Scholar]
- Zou HF, Hu LD, Li JX et al. Cloning and characterization, of a novel small monomeric GTPase, RasL10B, with tumor suppressor potential. Biotechnol Lett 2006; 28: 1901–1908. [DOI] [PubMed] [Google Scholar]
- Inoue A, Okamoto K, Fujino Y et al. B-RAF mutation and accumulated gene methylation in aberrant crypt foci (ACF), sessile serrated adenoma/polyp (SSA/P) and cancer in SSA/P. Br J Cancer 2015; 112: 403–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzny DM, Bainbridge MN, Chang K et al. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012; 487: 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, Suehiro Y, Ueno K et al. TWIST1 hypermethylation is observed frequently in colorectal tumors and its overexpression is associated with unfavorable outcomes in patients with colorectal cancer. Genes Chromosomes Cancer 2010; 49: 452–462. [DOI] [PubMed] [Google Scholar]
- Melotte V, Lentjes MHFM, van den Bosch SM et al. N-Myc downstream-regulated gene 4 (NDRG4): a candidate tumor suppressor gene and potential biomarker for colorectal cancer. J Natl Cancer Inst 2009; 101: 916–927. [DOI] [PubMed] [Google Scholar]
- Gevaert O, Tibshirani R, Plevritis SK. Pancancer analysis of DNA methylation-driven genes using MethylMix. Genome Biol 2015; 16: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 2008; 4: e1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.