Abstract
Objectives:
More convenient and effective blood-based methods are believed to increase colorectal cancer (CRC) detection adoption. The effectiveness of methylated SPET9 for CRC detection has been reviewed in the newly published recommendation statement by US Preventive Services Task Force (USPSTF), while detailed instructions were not provided, which may be a result of insufficient evidence. Therefore, more evidence is needed to assist practitioners to thoroughly understand the utilization of this special maker.
Methods:
Based on the standard method, a systematic review and meta-analysis was performed. Quadas-2 was used to assess the methodological quality of studies. Relevant studies were searched and screened from PubMed, Embase and other literature databases up to June 1, 2016. Pooled sensitivity, specificity and diagnostic odds ratio were summarized by bivariate mixed effect model and area under the curve (AUC) was estimated by hierarchical summary receiver operator characteristic curve.
Results:
25 studies were included for analysis. The pooled sensitivity, specificity and AUC were 0.71, 0.92 and 0.88, respectively. Among the various methods and assays, Epipro Colon 2.0 with 2/3 algorithm was the most effective in colorectal cancer detection. Positive ratio of mSEPT9 was higher in advanced CRC (45% in I, 70% in II, 76% in III, 79% in IV) and lower differentiation (31% in high, 73% in moderate, 90% in low) tissue. However, this marker has poor ability of identifying precancerous lesions according to current evidence.
Conclusions:
mSEPT9 is a reliable blood-based marker in CRC detection, particularly advanced CRC. Epipro Colon 2.0 with 2/3 algorithm is currently the optimal method and assay to detect CRC.
Introduction
Colorectal cancer (CRC) is one of the most common malignant tumors and places an enormous burden on the society. It was estimated that 1.4 million new cases were diagnosed worldwide in 2012,1 of which, more developed countries accounted for the larger proportion. In contrast to incidence, mortality rates of CRC have been found to decrease in numerous countries, which most likely benefits from early detection.2 It is predicted that a total of 277,000 new CRC cases and 203,000 CRC-induced deaths in United States will be averted from 2013 to 2018 if National Colorectal Cancer Roundtable reaches the goal of increasing the prevalence of CRC screening to 80% by 2018.3 Although there are various guideline-recommended methods one can choose for detection, the compliance remains low. The data in 2013 showed that only about 57% of eligible adults adhered to screening recommendations provided by US Preventive Services Task Force (USPSTF).4 There are many reasons for low adoption for CRC detection. Obstacles specific to colonoscopy include aversion to bowel preparation, discomfort during the procedure, pre- and post-procedure time requirements, and costs.5 Guiac-based fecal occult blood tests or fecal immunochemical tests (FITs) are easier to be accepted. However, both methods continue to be underutilized and have relatively low diagnosis value.6 Since the currently utilized methods have various limitations and there is no other information available for detection, it is very important to introduce better and more patient-friendly approaches, especially blood testing, for detecting CRC.7
It is known that CRC occurs due to the genetic and epigenetic alterations of intestinal epidermal cells.8 Therefore, the determination of specific molecular markers targeting the changes may be a promising method for detecting early CRC. Aberrant methylation of tumor DNA sequences has been found in various genes, of which, methylated Septin 9 (mSEPT9) DNA is validated to be able to effectively diagnose CRCs from normal blood using real-time PCR.9 SEPT9, a member of the Septin family, has been found to function in cytokinesis and remodeling cytoskeletal.10 mSEPT9 was found to be correlated with carcinogenesis.10 Multiple research assays have been developed to identify mSEPT9 in circulating plasma by PCR amplification. A number of case–control studies, which encompassed thousands of clinical samples,9, 11, 12, 13 have been performed to verify the accuracy of mSEPT9 for CRC detection. In these studies, the sensitivity and specificity ranged from 69 to 79% and 82 to 99%, respectively. However, a prospective study (PRESPET NCT00855348) published later in 2014, which recruited almost 8000 samples, showed that the sensitivity was only 50.9%, lower than the expected data.14 Until then, it still lacked convincing evidence to translate such methods from research into clinical practice.
Given that determination of mSEPT9 in blood has a promising future for CRC screening, existing researches and guidelines still fall short of giving detailed instructions to improve clinical applications which may be a result of insufficient evidence or underestimated diagnostic value. There are various methods (MethyLight, MSP-DHPLC, MS-HRM) and assays used in detecting mSEPT9, most of which are claimed to have high value. Epi proColon itself has two generations of assays and three inspection methods. The limitations above may hinder the understanding of optimal utilization strategy until more accurate and detailed explanations are provided. Therefore, we have performed a systematic review and meta-analysis of the diagnostic accuracy of mSEPT9 in order to explore the optimal method and kit for CRC detection.
Methods
Criteria for considering studies for this review
We included all the primary studies which were performed to determine the diagnostic accuracy of the index test and compared them with the reference standard ones in CRC screening. The types of studies included cohort studies, cross-sectional studies and case–control studies from which we can extract data for true-positives (TP), true-negatives (TN), false-positives (FP), and false-negatives (FN). We excluded unpublished studies that were only reported in abstracts, or studies with inadequate data to construct a two-by-two table.
To estimate mSEPT9 in peripheral blood, the index test should be the methods and kits used, while the reference test should be colonoscopy. Any studies that estimated mSEPT9 in stools or other tissues were not included, neither were the ones using other comparator tests.
Search strategy
We searched the following literature databases for publications from their inception to 1 June 2016: Cochrane Central Register of Controlled Trials (CENTRAL), Cochrane Library, Medline via PubMed, EMBASE via embase.com, China National Knowledge Infrastructure Database (CNKI), Chinese Biomedical Literature Database (CBM), Chinese Scientific Journal Database (VIP database), and Wanfang database. To improve recall ratio in retrieval, the search strategy consisted of medical subject heading terms, keywords and free terms related to the marker (septin 9 or sept 9, etc.) combined with the disease (colorectal neoplasms, colon cancer, or rectum cancer, etc.). The search language was restricted to English and Chinese. (See Supplementary Information 1).
We manually retrieved and examined the reference lists of relevant articles for additionally eligible studies. We also searched OpenGrey.eu for potential grey studies and clinical trials registry platforms such as ICRTP for ongoing and recently completed ones.
Data collections and analysis
Selection of studies
We created a database using Endnote X7 and uploaded all studies obtained from electronic searches and other sources to the database, excluding duplicates. Two researchers (SYM and CY) independently screened the searching results, including the titles, abstracts, and keywords. The articles that measured up to the inclusion criteria for this review were included for full-text screening. Disagreements were resolved by discussion or consulting with a third researcher (XS).
Data extraction and management
Two researchers (YM and YF) independently performed data extraction from the included studies. The authors were contacted when more information was needed. The key information was as follows:
General information about the studies, included first author' name, year, country, study type, etc.
Demographic information, including gender, ethnicity, age, CRC stage and differentiation, pathology types, and sample size.
Index test information included cut-off point, methods and kits used.
Outcomes included TP, FP, TN and TN.
Assessment of methodological quality
Another two researchers (YM and LY) independently assessed the quality of each study by using the Quality Assessment of Diagnostic Accuracy Studies-2(QUADAS-2) tool, which consisted of four domains: patient selection, index test, reference standard, and flow of patients and timing of the tests.15 All four domains were used to assess risk of bias and the first three domains were used to assess study applicability. Any disagreements were resolved by consensus or consulting the arbitrator (XS).
Statistical analysis and synthesis
We performed a bivariate mixed effect model to summarize the sensitivity, specificity, positive likelihood ratio, negative likelihood ratio, and diagnostic odds ratio (DOR) of mSEPT9 in CRC screening. We also conducted a hierarchical summary receiver–operator characteristic curve (HSROC) to estimate the area under the curve (AUC).We investigated potential heterogeneity by calculating the Cochran' Q statistic and I2 for other causes of heterogeneity. If the P value of the Q-test was ≥0.05 or the I2 value was ≤50%, it suggested that no significant heterogeneity existed.
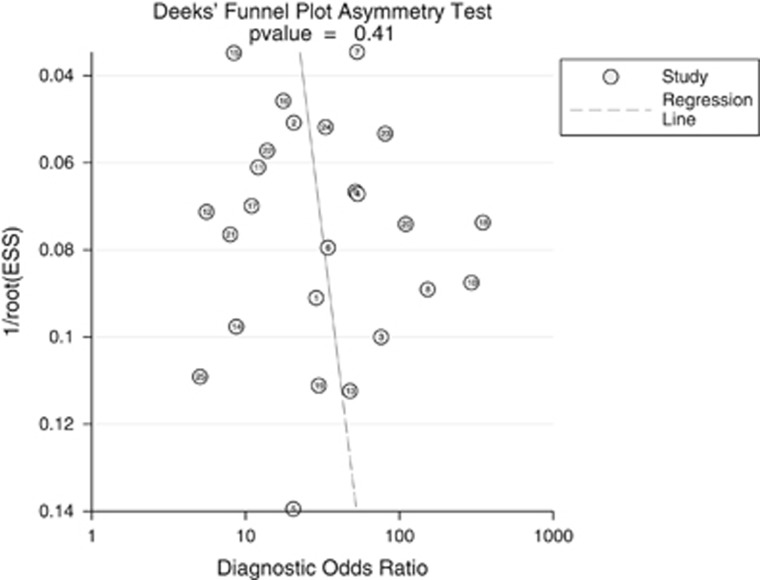
If significant heterogeneity existed, we investigated the causes of heterogeneity by performing subgroup analysis and meta-regression when sufficient studies were available. The following categorical covariates were used: assays or methods of index test, race, CRC stage and differentiation, pathology types, etc. Spearman correlation coefficients between sensitivity and 1-specificity were also estimated for the threshold effect. Furthermore, Deeks' funnel plot was used to estimate the risk of publication bias, and a P value <0.05 indicated high risk of bias.
Results
Search results
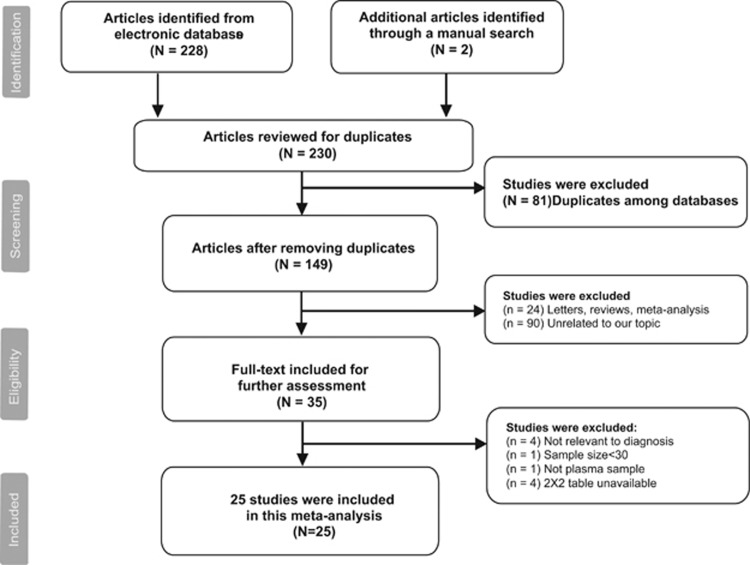
A total of 230 articles were initially retrieved using the search strategy above, of which, 228 were selected from electronic databases and two were identified through the manual screening of relevant articles in reference lists. One hundred and forty-nine articles were included for title and abstract screening after removing 81 duplications. Then, 24 were excluded due to inappropriate types and 90 were excluded for the reason that the studies were not related to our topic. As a result, 35 articles were suitable for full-text assessment. After full-text reading, 25 articles9, 11, 12, 13, 14, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35 were included in this meta-analysis. (See Figure 1).
Figure 1.
The flowchart of literature selection.
Characteristic of included studies
Table 1 outlines the characteristic of include studies. A total of 9927 samples from 25 studies were used in our meta-analysis, of which 2975 were CRCs and 6952 were adenoma, polyps or other colorectal diseases. The studies were conducted in seven countries from 2008 to 2016, including the United States, China, Germany, Hungary, Russia, Korea, and Denmark. Most of the studies were case–control studies in design, while four of them were prospective studies. Various types of methods and assays were employed, and Epipro Colon was utilized the most (18/25). Seventeen studies provided diagnostic results among TNM stages and four offered the data in different differentiations. FITs were used as combined methods to estimate the diagnostic accuracy in six studies.
Table 1. Characteristic of included studies.
| NO | Study | Year | Country | Sample size | TP | TN | FP | FN | Cut-off value | Algorithm | Study type | Assay method | Kit used |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Yu D | 2015 | China | 123 | 57 | 46 | 7 | 13 | Ct<45. 0 | 2(3) | Case–control | RT-PCR | Epipro Colon 2.0 |
| 2 | Jin P | 2015 | China | 476 | 101 | 298 | 43 | 34 | Ct<45. 0 | 2(3) | Case–control | RT-PCR | Epipro Colon 2.0 |
| 3 | He Q | 2015 | China | 100 | 38 | 48 | 2 | 12 | PMR≥ 4% | NA | Case–control | MethyLight | NA |
| 4 | He N | 2015 | China | 281 | 54 | 196 | 9 | 22 | Ct<40. 5 | 2(3) | Case–control | RT-PCR | Epipro Colon 2.0 |
| 5 | Wang Z | 2012 | China | 56 | 25 | 18 | 2 | 11 | PMR≥1% | NA | Case–control | MS-HRM | NA |
| 6 | Li SJ | 2015 | China | 161 | 66 | 65 | 5 | 25 | NA | 2(3) | Case–control | RT-PCR | Epipro Colon 2.0 |
| 7 | Wu D | 2016 | China | 1031 | 223 | 697 | 43 | 68 | Ct<45. 0 | 1(1) | Prospective study | RT-PCR | Epipro Colon 2.0 |
| 8 | Kang Q | 2014 | China | 132 | 60 | 51 | 1 | 20 | NA | 2(3) | Case–control | RT-PCR | Epipro Colon 2.0 |
| 9 | Ding QQ | 2015 | China | 262 | 60 | 171 | 9 | 22 | Ct<45. 0 | 2(3) | Case–control | RT-PCR | Epipro Colon 2.0 |
| 10 | Warren JD | 2011 | US, Russia | 144 | 38 | 93 | 1 | 12 | Ct<45. 0 | 2(3) | Case–control | RT-PCR | Epipro Colon 1.0 |
| 2011 | USA, Russia | 144 | 45 | 83 | 11 | 5 | Ct<41. 0 | 1(3) | Case–control | RT-PCR | Epipro Colon 1.0 | ||
| 2011 | USA, Russia | 144 | 35 | 94 | 0 | 15 | Ct<45. 0 | 3(3) | Case–control | RT-PCR | Epipro Colon 1.0 | ||
| 11 | Johnson DA | 2014 | USA | 301 | 74 | 163 | 37 | 27 | NA | NA | Prospective study | RT-PCR | Epipro Colon 1.0 |
| 12 | Lee HS | 2013 | Korea | 197 | 37 | 87 | 9 | 64 | NA | 1(3) | Case–control | RT-PCR | Abbott Molecular |
| 13 | Lucia PC | 2014 | USA | 367 | 244 | 20 | 1 | 102 | NA | NA | Case–control | RT-PCR | Epipro Colon 1.0 |
| 14 | Marc T | 2010 | Germany | 161 | 24 | 98 | 30 | 9 | NA | 2(3) | Case–control | Heavy MethyLight | NA |
| 2010 | Germany | 161 | 27 | 81 | 47 | 6 | NA | 1(3) | Case–control | Heavy MethyLight | NA | ||
| 15 | Grutzmann R | 2008 | Germany | 831 | 193 | 403 | 50 | 185 | NA | 2(3) | Case–control | RT-PCR | Epipro Colon 1.0 |
| 16 | deVos T | 2009 | Germany | 514 | 138 | 282 | 45 | 49 | 3.4ug/L | 1(3) | Case–control | RT-PCR | Epipro Colon 1.0 |
| 2009 | Germany | 514 | 105 | 316 | 11 | 82 | 3.4ug/L | 2(3) | Case–control | RT-PCR | Epipro Colon 1.0 | ||
| 17 | Church TR | 2013 | Germany, USA | 1510 | 27 | 1331 | 126 | 26 | Ct<50 | 1(2) | Prospective study | RT-PCR | Epipro Colon 1.0 |
| 18 | Toth K | 2012 | Hungary | 184 | 73 | 91 | 1 | 19 | Ct<40. 5 | 2(3) | Case–control | RT-PCR | Epipro Colon 2.0 |
| 2012 | Hungary | 184 | 88 | 78 | 14 | 4 | Ct<40. 5 | 1(3) | Case–control | RT-PCR | Epipro Colon 2.0 | ||
| 19 | Toth K | 2014 | Hungary | 84 | 30 | 40 | 10 | 4 | PMR≥ 0.01% | NA | Case–control | RT-PCR | Epipro Colon 2.0 |
| 20 | Su XL | 2014 | China | 234 | 152 | 58 | 4 | 20 | MSP≥1% | NA | Case–control | MSP-DHPLC | NA |
| 21 | Potter NT | 2014 | USA | 1544 | 30 | 1182 | 318 | 14 | Ct<45. 0 | 1(3) | Prospective study | RT-PCR | Epipro Colon 2.0 |
| 22 | Lofton-Day C | 2008 | USA | 312 | 92 | 154 | 25 | 41 | NA | NA | Case–control | MethyLight | NA |
| 23 | He Q | 2010 | China | 352 | 136 | 164 | 6 | 46 | PMR≥ 4% | NA | Case–control | MethyLight | NA |
| 24 | Ørntoft MW | 2015 | Denmark | 470 | 93 | 282 | 60 | 35 | Ct<45. 0 | 1(3) | Case–control | RT-PCR | Epipro Colon 2.0 |
| 2015 | Denmark | 470 | 75 | 328 | 14 | 53 | Ct<45. 0 | 2(3) | Case–control | RT-PCR | Epipro Colon 2.0 | ||
| 25 | Ahlquist DA | 2011 | USA | 100 | 18 | 54 | 16 | 12 | Ct<45. 0 | 1(3) | Case–control | RT-PCR | Epipro Colon 1.0 |
MS-HRM, methylation sensitive high-resolution melting; MSP-DHPLC, methylation spective polymerase chain reaction; RT-PCR, real-time polymerase chain reaction.
Study quality
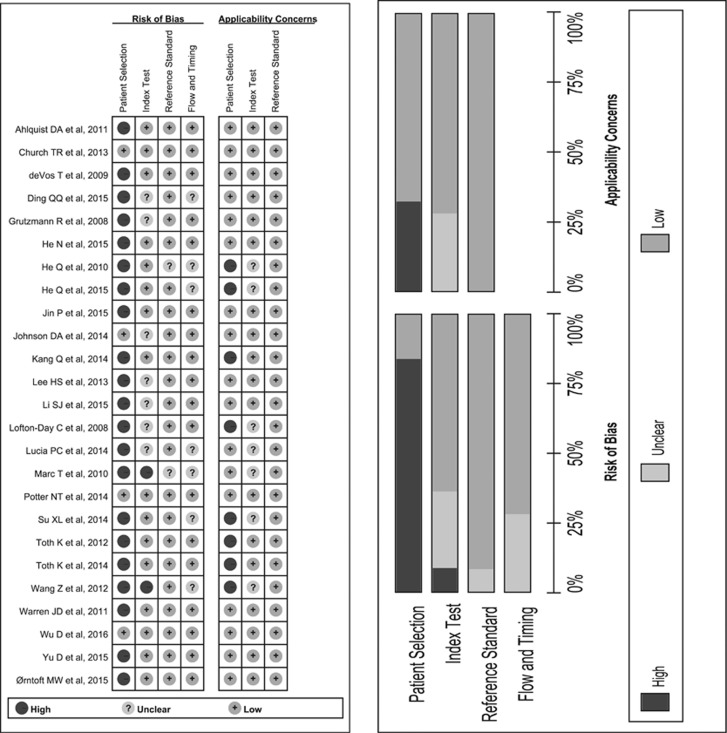
Figure 2 show the results of the quality appraisal of 25 studies that were included. Only two studies show a low risk of bias in all four domains of QUADAS 2. 21 studies inappropriately excluded “difficult-to-diagnose” patients, therefore the risk of bias of patient selection was rated as high. Seven studies had insufficient data about threshold setting and two selected their cut-off points by adjusting during their studies. As methylated SEPT9 is an objective index test, we omitted the signaling question about blinding the result of index test to reference one. Two studies offered insufficient data about blinding of reference standard, resulting in unclear risk in this domain. Seven studies showed unclear risk of flow and timing, because colonoscopy was examined before recruitment and intervals could not have been estimated.
Figure 2.
Methodological quality of included studies.
Eight studies showed high concern of applicability for the reason that they only enrolled healthy persons in control group. Seven studies had unclear concern because the threshold and assay were not interpreted in details. All of the studies showed low concern about reference standard.
Diagnostic accuracy and subgroup analysis
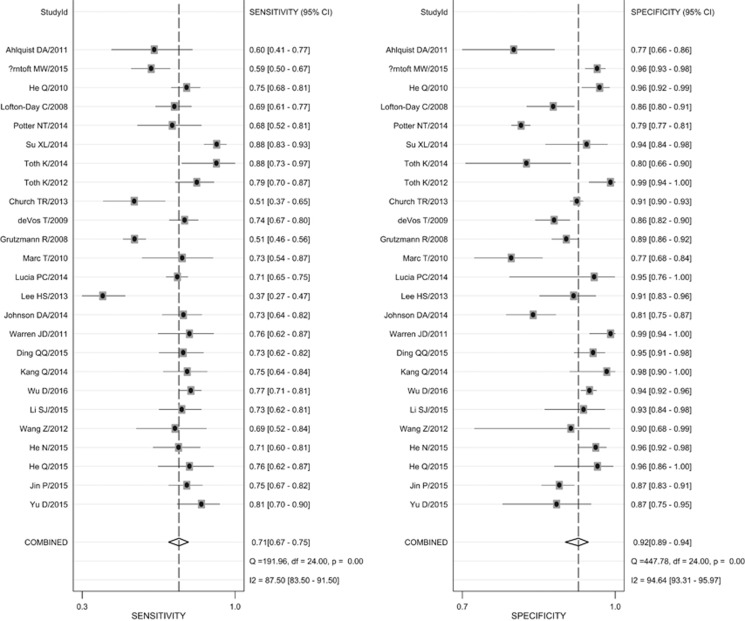
Spearman correlation coefficient was −0.310 and P value was 0.131. The proportion of heterogeneity likely due to threshold effect was 0.02, which meant there existed no significant threadhold effect among included 25 studies. Figure 3 indicates the forest plot of overall pooled sensitivity and specificity. According to the bivariate mixed effect model, the pooled sensitivity and specificity was 0.71 (95% confidence interval (CI): 0.67–0.75) and 0.92 (95%CI: 0.89–0.94), respectively. Figure 4 (Part A) shows the HSROC and its AUC (0.88, 95%CI: 0.85–0.91). The HSROC figure is symmetrical (Z=1.62 and P=0.105) and it presents significance in diagnostic value (λ=3.07).
Figure 3.
Forest plot of all included studies.
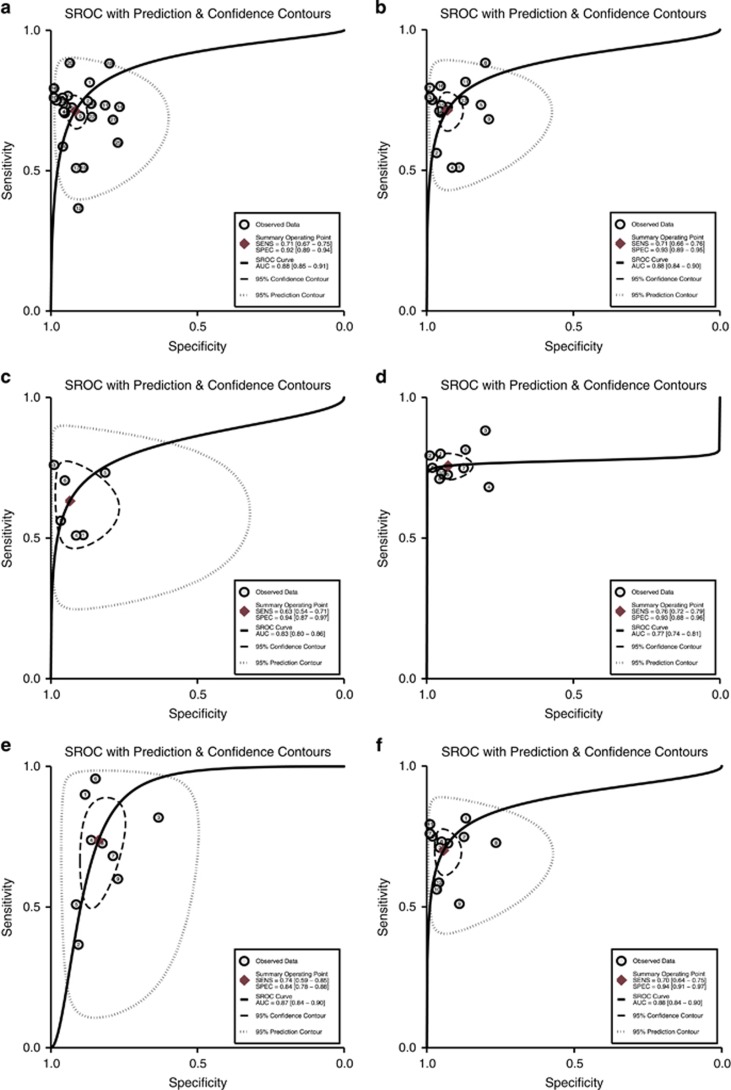
Figure 4.
Hierarchical summary receiver–operator characteristic curve, HSROC. (a) Overall HSROC of all included studies; (b) HSROC of Epipro colon 1.0 and 2.0; (c) HSROC of Epipro colon 1.0; (d) HSROC of Epipro colon 2.0; (e) HSROC of Epipro colon with 1/3 algorithm; and (f) HSROC of Epipro colon with 2/3 algorithm.
Furthermore, subgroup analysis was therefore performed by ethnicity, study type, assay, tumor stage and differentiation, combined method and precancerosis (see Table 2). We also conducted subgroup analysis based on assay or method which was used in included studies and the results equaled that of Epipro Colon assay and other methods (MethyLight, MSP-DHPLC, MS-HRM, etc.) but differed between generation 1 and generation 2 Epipro colon assay. The pooled sensitivity was 0.76 and the specificity was 0.94 in the generation 2 assay, higher than that of generation 1.
Table 2. Subgroup analysis.
| Analyses | Sensitivity (95%CI) | Specificity (95%CI) | LR+ (95%CI) | LR− (95%CI) | DOR (95%CI) | AUC |
|---|---|---|---|---|---|---|
| Overall | 0.71 (0.67–0.75) | 0.92 (0.89–0.94) | 8.6 (6.2–11.8) | 0.31 (0.27–0.37) | 27 (18–42) | 0.88 |
| Ethnicity | ||||||
| Europe | 0.70 (0.51–0.83) | 0.94 (0.84–0.98) | 11.2 (4.1–30.4) | 0.32 (0.19–0.55) | 35 (10–120) | 0.90 |
| America | 0.71 (0.68–0.74) | 0.79 (0.78–0.81) | 3.4 (3.2–3.7) | 0.30 (0.33–0.41) | 9 (8–11) | 0.82 |
| Asia | 0.75 (0.71–0.78) | 0.94 (0.90–0.96) | 11.6 (7.7–17.5) | 0.27 (0.23–0.31) | 43 (27–68) | 0.79 |
| Study design | ||||||
| Case–control | 0.72 (0.67–0.76) | 0.92 (0.89–0.95) | 9.5 (6.6–13.7) | 0.31 (0.25–0.37) | 31 (19–50) | 0.89 |
| Cross-sectional | 0.69 (0.59–0.77) | 0.88 (0.80–0.93) | 5.7 (3.3–9.9) | 0.35 (0.26–0.48) | 16 (8–34) | 0.84 |
| Assay or method | ||||||
| Epipro Colon 1.0+2.0 | 0.71 (0.66–0.76) | 0.93 (0.89–0.95) | 10.2 (6.6–15.6) | 0.31 (0.26–0.37) | 33 (20–55) | 0.88 |
| Epipro Colon 1.0 | 0.63 (0.54–0.71) | 0.94 (0.87–0.97) | 9.8 (4.6–20.9) | 0.39 (0.31–0.50) | 25 (10–62) | 0.83 |
| Epipro Colon 2.0 | 0.76 (0.73–0.79) | 0.93 (0.88–0.96) | 10.4 (6.13–17.6) | 0.26 (0.23–0.30) | 39.60 (10–62) | 0.77 |
| MethyLight | 0.72 (0.67–0.77) | 0.91 (0.80–0.96) | 8.0 (3.3–19.3) | 0.30 (0.24–0.38) | 26 (9–76) | 0.78 |
| Algorithm for Epi proColon | ||||||
| 1/3 algorithm | ||||||
| 2/3 algorithm | 0.70 (0.64–0.75) | 0.94 (0.91–0.97) | 12.3 (7.3–20.8) | 0.32 (0.26–0.39) | 39 (21–72) | 0.88 |
| Stage | 0.74 (0.59–0.85) | 0.84 (0.78–0.88) | 4.5 (3.4–6.1) | 0.31 (0.19–0.51) | 14 (8–28) | 0.87 |
| Stage I | 0.45 (0.38–0.53) | 0.93 (0.90–0.95) | 6.4 (4.0–10.1) | 0.59 (0.50–0.68) | 11 (6–19) | 0.72 |
| Stage II | 0.70 (0.60–0.79) | 0.93 (0.90–0.95) | 10.0 (6.1–16.4) | 0.32 (0.23–0.45) | 31 (14–69) | 0.92 |
| Stage III | 0.76 (0.64–0.86) | 0.93 (0.90–0.95) | 10.8 (6.5–17.9) | 0.25 (0.15–0.41) | 43 (17–110) | 0.94 |
| Stage IV | 0.79 (0.69–0.87) | 0.93 (0.90–0.95) | 11.0 (7.3–16.6) | 0.22 (0.15–0.34) | 49 (24–101) | 0.92 |
| Differentiation | ||||||
| High | 0.31 (0.12–0.59) | 0.95 (0.93–0.96) | 6.1 (2.6–14.6) | 0.73 (0.51–1.04) | 8 (3–29) | 0.95 |
| Moderate | 0.73 (0.68–0.78) | 0.95 (0.93–0.96) | 14.5 (10.8–19.3) | 0.28 (0.23–0.34) | 51 (34–76) | 0.94 |
| Low | 0.90 (0.83–0.95) | 0.95 (0.93–0.96) | 17.8 (13.4–23.8) | 0.10 (0.06–0.19) | 173 (84–354) | 0.98 |
| Combined method | ||||||
| Sept 9+FIT (PT) | 0.94 (0.89–0.97) | 0.68 (0.56–0.78) | 2.9 (2.2–4.0) | 0.08 (0.04–0.15) | 36 (21–62) | 0.91 |
| Precancerosis | ||||||
| Adenoma | 0.15 (0.11–0.19) | 0.90 (0.85–0.94) | 1.5 (1.0–2.4) | 0.94 (0.89–1.00) | 2 (1–3) | 0.36 |
| Polyp | 0.05 (0.03–0.08) | 0.94 (0.90–0.97) | 0.83 (0.36–1.94) | 1.01 (0.96–1.06) | 0.82 (0.34–2.0) | 0.15 |
| Polyp/adenoma size | ||||||
| >1 cm | 0.23 (0.17–0.29 | 0.91 (0.89–0.93) | 2.56 (1.77–3.71) | 0.85 (0.78–0.92) | 3.01 (1.93–4.71) | 0.68 |
| ≤1 cm | 0.09 (0.06–0.14) | 0.91 (0.89–0.93) | 1.06 (0.66–1.70) | 0.99 (0.95–1.04) | 1.07 (0.64–1.79) | 0.51 |
AUC, area under the curve; CI, confidence interval; DOR, diagnostic odds ratio; FIT, fecal immunochemical test; LR+, positive likelihood ratio; LR−, negative likelihood ratio; PT, parallel test.
In addition, data was further extracted and analyzed by the groups of disease stages and combined methods. The pooled sensitivity, specificity, LR+, LR−, DOR, and AUC are 0.79, 0.93, 11.0, 0.22, 49, and 0.92 in stage IV, respectively, which shows the highest diagnosis value, followed by stages III, II, and I. Similarly, CRC cases with low differentiation were more likely detected than moderate and high one. Three studies combined mSPET9 with FIT in parallel tests to estimate diagnosis accuracy and the results showed higher sensitivity (0.94) and lower specificity (0.68) than using mSPET9 alone. There was not enough data to combine carcinoembryonic antigen (CEA) or other methods in testing diagnostic accuracy. Twelve studies provided the details about results in adenomas and polyps. The pooled sensitivity was 0.15 and 0.05 in adenomas and polyps, respectively, both indicating low positive ratio of mSPET9 detection. Moreover, the pooled sensitivity was 0.23 for larger size (large than 1 cm) polyps or adenomas, which is higher than smaller ones (0.09; see Table 2).
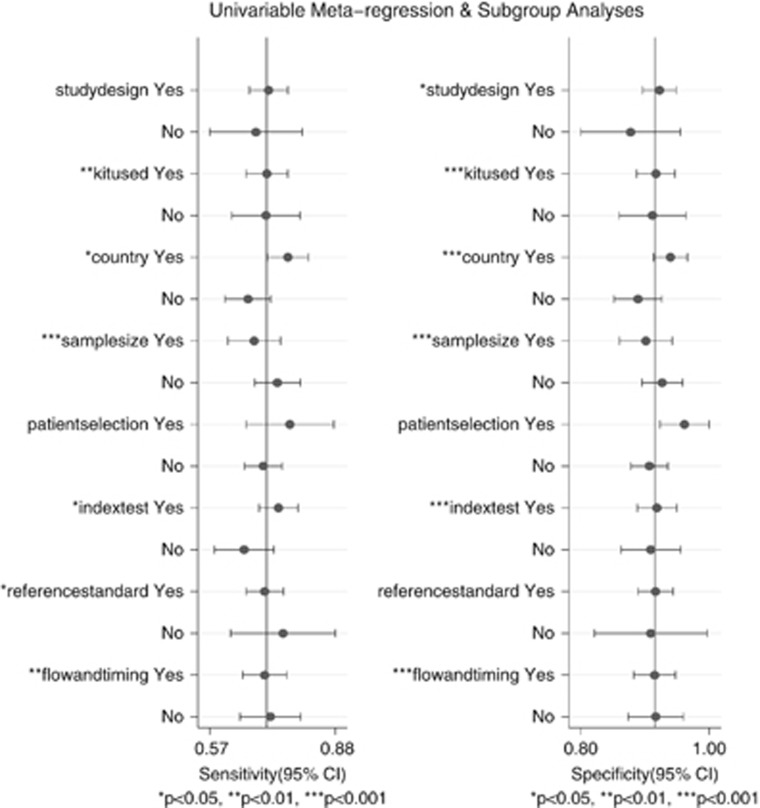
Since Figure 3 indicates significant heterogenity of sensetivity and specificity after computing the Cochran' Q statistic and I2 (both P value <0.05), meta-regression was therefore conducted to trace the causes. The result shows that study types, kits used (Epipro colon or not), country (Asia or not), sample size (> or <300) and risk of bias of included studies all lead to the heterogeneity of sensitivity and specificity in a single variable model, of which whether the studies were performed in Asian countries or not was significant in joint model (P=0.01; see Figure 5).
Figure 5.
Deek's funnel plot of all included studies.
Figure 6 presents symmetry in Deeks' funnel plot (P=0.41) and indicates that there exists no significant publication bias in the included studies.
Figure 6.
Meta-regression plot in a single variable model.
Discussion
Recently, USPSTF updated its recommendations and initially reviewed the evidence on the efficacy of detection CRCs with mSEPT9.36 In our systematic review, we estimated that the pooled sensitivity and specificity was 0.71 and 0.92, respectively, proving to be reliable for CRC detection. The results were apparently higher than those in PRESEPT study,14 which may owed to recruiting early asymptomatic CRC patients for analysis. The systematic review also performed stage and differentiation-related analysis in detection, and Table 2 presents an apparent positive correlation between the detection rates of CRC and stage degrees. The results indicates that advanced stage CRCs are easier to be detected by mSEPT9 than early stage. The trend was similarly observed in tumor differentiation. Low-differentiation CRCs has much higher sensitivity than high differentiation ones. The results showed Asia Group had higher sensitivity than other continents. However, the results from Korea18 showed obvious lower sensitivity (0.363). The discrepancy might have occurred due to the potential racial differences and kit variations.37
In our subgroup analysis, we tried to explore the optimal method and assay for mSEPT9. 20 studies investigated the accuracy of Epi proColon and only four of included studies focused on other assay kits (mainly using the MytheLight method). Both assays presented similar results, but the Epipro Colon was found to be described in details and thus easier for clinicians to operate. The second generation of Epipro Colon has received approval from the US Food and Drug Administration38 and was reported to have resolved many technical hurdles and improved in several aspects, such as employing a novel bisulfite DNA conversion and purification technology39 as well as a new real-time PCR reaction.13 Two different types of algorithms were applied for Epicolon Colon in the studies and the results were different in sensitivity and specificity. Sensitivity was high using a 1/3 algorithm test but the specificity was low. Although sensitivity was low using a 2/3 algorithm test, it had a high true negative rate. Since it is more important to improve the capability in excluding non-cancer samples and avoiding the rate of misdiagnosis, 2/3 algorithm is recommended for CRCs detection.
As a first blood-based detection method recommended for CRC, can mSEPT9 really improve compliance? The data results from a German research ensured the practicability, in which 83% of patients were willing to accept mSEPT9 test, which is higher than colonoscopy (37%) and stool test (15%).40
Even though the systematic review concluded an encouraging result of mSPET9 in CRC detection, it still has several limitations. First of all, FIT is currently widely used in CRC screening. However, due to lack of appropriate studies for further analysis, we did not provide further information about sensitivity and specificity in comparison between mSPET9 and FIT. Secondly, despite the diagnostic value of detecting advanced stage CRCs (III–IV), the analysis that were focused on early stage of CRC (Stage I) and adenomas or polyps showed low sensitivity. It turned out the diagnostic value of mSEPT9 may, to some degree, be limited in precancerous lesions and CRC in Stage I. However, mSEPT9 was shown to have low misdiagnosis rate and sensitivity may be improved when combined with FIT. Thirdly, as different methods were used for detecting mSEPT9, we did not subgroup analyze the optimal threshold for every method other than Epipro Colon. Three different cut-off points were used for this assay, of which Ct<45.0 was the most utilized. The sensitivity was 0.70 when Ct<45.0 was used, slightly lower than Ct<41.0, indicating Ct<45.0 may be more sensitive for utilization. But it still need further study to verify it as the best threshold. Fourthly, this meta-analysis did not include any language other than Chinese and English. Restriction in languages may bring about a potential risk of publication bias. In terms of methodological quality, most studies that were included were case–control in design and excluded “difficult-to-diagnose” patients, which may lead to a risk of bias in patient selection and overestimation of diagnostic accuracy.18 Finally, although it was reported that mSEPT9 could be employed as a predictor of CRC recurrence, metastasis and survival,18, 41 there is insufficient data for synthesis in our meta-analysis in order to draw robust conclusions about the value as a follow-up marker.
In conclusion, our systematic review suggests that mSEPT9 can be used as an effective marker for blood-based CRC detection. Based on current evidence, the second generation Epipro Colon (Epigenomics) could be used as the optimal assay kit with 2/3 algorithm. In addition, the review revealed that a larger sample size and more prospective studies were needed to further verify the diagnostic value of mSEPT9.
Study Highlights
Acknowledgments
Trial Register Number: CRD42016042457.
Guarantors of the article: Mingwei Yu, MD; Ganlin Zhang, MD; and Xiaomin Wang, PhD.
Specific author contributions: Jiayun Nian, MD and Xu Sun, MD contributed to the study design, data extraction and interpretation, and drafting and final approval of the manuscript. Su Yang Ming, MD and Chen Yan, MD contributed to selection of studies and final approval of the manuscript. Yunfei Ma, MD and Ying Feng, MD contributed to data extraction and final approval of the manuscript. Lin Yang, MD contributed to study appraisal and final approval of the manuscript.
Financial support: The Beijing Municipal Science and Technology Plan Projects (NO. D161100005116005); Natural Science Foundation of China (NO. 81473643).
Potential competing interests: None.
Footnotes
Supplementary Information accompanies this paper on the Clinical and Translational Gastroenterology website (http://www.nature.com/ctg)
Supplementary Material
References
- Torre LA, Bray F, Siegel RL et al. Global cancer statistics, 2012. Cancer J Clin 2015; 65: 87–108. [DOI] [PubMed] [Google Scholar]
- Edwards BK, Ward E, Kohler BA et al. Annual report to the nation on the status of cancer, 1975-2006, featuring colorectal cancer trends and impact of interventions (risk factors, screening, and treatment) to reduce future rates. Cancer 2010; 116: 544–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meester RG, Doubeni CA, Zauber AG et al. Public health impact of achieving 80% colorectal cancer screening rates in the United States by 2018. Cancer 2015; 121: 2281–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedewa SA, Sauer AG, Siegel RL et al. Prevalence of major risk factors and use of screening tests for cancer in the United States. Cancer Epidemiol Biomarkers Prev 2015; 24: 637–652. [DOI] [PubMed] [Google Scholar]
- Jones RM, Devers KJ, Kuzel AJ et al. Patient reported barriers to colorectal cancer screening: a mixed-methods analysis. Am J Prev Med 2010; 38: 508–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman DA. Clinical practice. Screening for colorectal cancer. N Engl J Med 2009; 361: 1179–1187. [DOI] [PubMed] [Google Scholar]
- Pratt VM. Are we ready for a blood-based test to detect colon cancer? Clin Chem 2014; 60: 1141–1142. [DOI] [PubMed] [Google Scholar]
- Tariq K, Ghias K. Colorectal cancer carcinogenesis: a review of mechanisms. Cancer Biol Med 2016; 13: 120–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lofton-Day C, Model F, Devos T et al. DNA methylation biomarkers for blood-based colorectal cancer screening. Clin Chem 2008; 54: 414–423. [DOI] [PubMed] [Google Scholar]
- Hall PA, Russell SE. The pathobiology of the Septin gene family. J Pathol 2004; 204: 489–505. [DOI] [PubMed] [Google Scholar]
- Warren JD, Xiong W, Bunker AM et al. Septin 9 methylated DNA is a sensitive and specific blood test for colorectal cancer. BMC Med 2011; 9: 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth K, Sipos F, Kalmár A et al. Detection of methylated SEPT9 in plasma is a reliable screening method for both left- and right-sided colon cancers. PLoS One 2012; 7: e46000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deVos T, Tetzner R, Model F et al. Circulating methylated SEPT9 DNA in plasma is a biomarker for colorectal cancer. Clin Chem 2009; 55: 1337–1346. [DOI] [PubMed] [Google Scholar]
- Church TR, Wandell M, Lofton-Day C et al. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut 2014; 63: 317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- QUADAS-2 Group Whiting PF, Rutjes AW, Westwood ME et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med 2011;155:529–536. [DOI] [PubMed] [Google Scholar]
- Johnson DA, Barclay RL, Mergener K et al. Plasma Septin9 versus fecal immunochemical testing for colorectal cancer screening: a prospective multicenter study. PLoS One 2014; 9: e98238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth K, Wasserkort R, Sipos F et al. Detection of methylated septin 9 in tissue and plasma of colorectal patients with neoplasia and the relationship to the amount of circulating cell-free DNA. PLoS One 2014; 9: e115415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Hwang SM, Kim TS et al. Circulating methylated septin 9 nucleic Acid in the plasma of patients with gastrointestinal cancer in the stomach and colon. Transl Oncol 2013; 6: 290–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Carbonell L, Balaguer F, Toiyama Y et al. IGFBP3 methylation is a novel diagnostic and predictive biomarker in colorectal cancer. PLoS One 2014; 9: e104285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grützmann R, Molnar B, Pilarsky C et al. Sensitive detection of colorectal cancer in peripheral blood by septin 9 DNA methylation assay. PLoS One 2008; 3: e3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tänzer M, Balluff B, Distler J et al. Performance of epigenetic markers SEPT9 and ALX4 in plasma for detection of colorectal precancerous lesions. PLoS One 2010; 5: e9061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su XL, Wang YF, Li SJ et al. High methylation of the SEPT9 gene in Chinese colorectal cancer patients. Genet Mol Res 2014; 13: 2513–2520. [DOI] [PubMed] [Google Scholar]
- Potter NT, Hurban P, White MN et al. Validation of a real-time PCR-based qualitative assay for the detection of methylated SEPT9 DNA in human plasma. Clin Chem 2014; 60: 1183–1191. [DOI] [PubMed] [Google Scholar]
- He Q, Chen HY, Bai EQ et al. Development of a multiplex MethyLight assay for the detection of multigene methylation in human colorectal cancer. Cancer Genet Cytogene 2010; 202: 1–10. [DOI] [PubMed] [Google Scholar]
- Wu D, Zhou G, Jin P et al. Detection of colorectal cancer using a simplified SEPT9 gene methylation assay is a reliable method for opportunistic screening. J Mol Diagn 2016; 18: 535–545. [DOI] [PubMed] [Google Scholar]
- Ahlquist DA, Taylor WR, Mahoney DW et al. The stool DNA test is more accurate than the plasma septin 9 test in detecting colorectal neoplasia. Clin Gastroenterol Hepatol 2012; 10: 272–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ørntoft MB, Nielsen HJ, Ørntoft TF et al. Danish Study Group on Early Detection of Colorectal Cancer. Performance of the colorectal cancer screening marker Sept9 is influenced by age, diabetes and arthritis: a nested case-control study. BMC Cancer 2015; 15: 819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Zhang XH, Lu XX. Study on diagnostic value of SEPT9 gene methylation in serum for colorectal cancer. Chin J Chin Lab Sci 2015; 33: 687–689. [Google Scholar]
- Jin P. Role of Mismatch Repair Protein in Estradiol-induced Colon Cancer Cells Apoptosis and Performance of Methylated SPET9 Test in Detecting Colorectal Neoplasm. Chongqing: Third Military Medical University, 2015. [Google Scholar]
- He Q, Wang M, Zhou JW et al. Detection of ALX4 and SEPT9 methylation in human colorectal cancer by Multiplex MethyLight assay. J Sun Yat-Sen Univ (Med Sci) 2015; 36: 657–662. [Google Scholar]
- He N The Study on the Application Value of Serum Methylated SEPT9 for Optimizing Screening Scheme of Colorectal Cancer. Fourth Military Medical University: Xi'an, China, 2015.
- Wang Z, Chen JC, He Q et al. MS-HRM detecting free methylation SEPT9 in early diagnosis of colorectal cancer. Guangdong Med J 2012; 33: 1732–1734. [Google Scholar]
- Li SJ, Liu YG, Wang J et al. The study of peripheral methylation SEPT9 in screening colorectal cancer. Chin J Gen Surg 2015; 124: 1756–1760. [Google Scholar]
- Kang J, Jin P, Yang L et al. The study of methylation SEPT9 in peripheral DNA for colorectal cancer screening. Natl Med J China 2014; 94: 3839–3841. [Google Scholar]
- Ding QQ, Zhang H, Xu HC et al. Study of methylation SEPT9 for colorectal cancer screening in aged population. Chin J Geriatr 2015; 34: 1348–1350. [Google Scholar]
- US Preventive Services Task ForceBibbins-Domingo K, Grossman DC, Curry SJ et alUS Preventive Services Task Force. Screening for Colorectal Cancer: US Preventive Services Task Force Recommendation Statement. JAMA 2016;315:2564–2575. [DOI] [PubMed] [Google Scholar]
- Li Y, Song L, Gong Y et al. Detection of colorectal cancer by DNA methylation biomarker SEPT9: past, present and future. Biomark Med 2014; 8: 755–769. [DOI] [PubMed] [Google Scholar]
- Premarket approval (PMA) for Epi proColon.US Food and Drug Administration. Available at: http://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfPMA/pma.cfm?id=P130001. Accessed 21 April 2016.
- Tetzner R, Dietrich D, Distler J. Control of carry-over contamination for PCR-based DNA methylation quantification using bisulfite treated DNA. Nucleic Acids Res 2007; 35: e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler A, Geiger S, Keil A et al. Improving compliance to colorectal cancer screening using blood and stool based tests in patients refusing screening colonoscopy in Germany. BMC Gastroenterol 2014; 14: 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tham C, Chew M, Soong R et al. Postoperative serum methylation levels of TAC1 and SEPT9 are independent predictors of recurrence and survival of patients with colorectal cancer. Cancer 2014; 120: 3131–3141. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.