Abstract
Cyprinid herpesvirus 3 (CyHV-3) infects koi and common carp and causes widespread mortalities. While the virus is a significant concern for aquaculture operations in many countries, in Australia the virus may be a useful biocontrol agent for pest carp. However, carp immune responses to CyHV-3, and the molecular mechanisms underpinning resistance, are not well understood. Here we used RNA-Seq on carp during different phases of CyHV-3 infection to detect the gene expression dynamics of both host and virus simultaneously. During acute CyHV-3 infection, the carp host modified the expression of genes involved in various immune systems and detoxification pathways. Moreover, the activated pathways were skewed toward humoral immune responses, which may have been influenced by the virus itself. Many immune-related genes were duplicated in the carp genome, and often these were expressed differently across the infection phases. Of particular interest were two interleukin-10 homologues that were not expressed synchronously, suggesting neo- or sub-functionalization. The carp immunoglobulin repertoire significantly diversified during active CyHV-3 infection, which was followed by the selection of high-affinity B-cells. This is indicative of a developing adaptive immune response, and is the first attempt to use RNA-Seq to understand this process in fish during a viral infection.
Cyprinid herpesvirus 3 (CyHV-3) is a large double-stranded DNA virus1 that was first recognized in the late 1990s2,3. The virus specifically infects koi and common carp (Cyprinus carpio) and causes widespread mortalities. Initial outbreaks of CyHV-3, for example, can result in population losses of 80–95%4,5. This is particularly concerning for the carp aquaculture industry, where CyHV-3 outbreaks result in substantial economic losses6. For example, an Indonesian outbreak in 2002 and 2003 is estimated to have cost farmers up to US$15 million5. CyHV-3 is thought to cause mortality by disrupting the osmoregulatory function of the host7, and by weakening the host immune system, thereby leaving the host more susceptible to infection by other viruses, bacteria or fungi8.
CyHV-3 belongs to the Alloherpesviridae family within the order Herpesvirales1. It has a genome size of 295 kb, which contains 156 putative open reading frames (ORFs), including two 22 kb terminal repeats9. CyHV-3 gene transcription follows a co-ordinated and sequential pattern during active host infection, similar to other herpesviruses10,11. Accordingly, the ORFs can be divided into 3 broad groups reflecting these events: 15 immediate-early; 112 early; 22 late; 7 remain unclassified10. Although the functional role of many CyHV-3 proteins is unclear, the characterised ORFs include various kinases, RING gene families, membrane glycoproteins and structural virion components, as well as pseudogenes and frameshifted ORFs1,8,9. A particularly interesting group of ORFs seems to have evolved to evade the host’s immune response. These include a functional Interleukin-10 (IL-10), known as khvIL-10, which is homologous to the carp’s own IL-10 and may mediate an immunosuppression mechanism12,13,14. Moreover, ORF12 and ORF4 are viral homologues of tumor necrosis factor receptors (TNFRs), which are often used by large DNA viruses to regulate host immune responses15,16.
Interestingly, CyHV-3 may be useful in some countries as a biocontrol agent. In Australia, for example, carp are an introduced invasive species that cause major disruptions to native ecosystems17,18. Indeed, carp are now the most abundant freshwater fish in south-east Australia, and in some instances comprise 90% of all fish biomass19. This results in the dislocation of native species, increased water turbidity, the loss of aquatic vegetation, and alterations in zooplankton and benthic invertebrate diversity18. Several characteristics of CyHV-3 suggest that it could be a suitable biocontrol agent for carp, including its lethality, specificity (only infects target species), and its rapid spread through populations8,17.
One aspect that needs further examination, however, is the development of the carp immune response to CyHV-3 infection, and subsequent mechanisms of carp resistance. A thorough understanding of carp immune responses and resistance mechanisms is required for controlling viral spread in aquaculture through the development of both CyHV-3 resistant carp and vaccines, and conversely, for facilitating viral spread in biocontrol programs by ensuring continued viral virulence in pest populations. Moreover, immunological responses can reveal if carp have been previously exposed to CyHV-3 and if they are potential carriers that may spread CyHV-3 to naïve fish.
Carp that survive a primary infection with CyHV-3 have superior resistance to subsequent infections, and mortalities are significantly reduced20,21. These fish may have greater innate resistance through pre-existing genetic factors, or alternatively, they may have mounted an effective adaptive immune response. Innate resistance of carp to CyHV-3 has been linked to single nucleotide polymorphisms (SNPs) in IL-10 genes22, specific genotypes of major histocompatibility class II genes23, and higher regulation of various immune factors24. In terms of adaptive immunity, carp have been shown to mount a cell-mediated immune response25, and to produce specific immunoglobulin (Ig) molecules in response to CyHV-321,26. Most studies to date have used serological methods to analyse the carp immune response, which can be challenging because immunological systems in fish are more primitive compared to mammals. For example, there are fewer Ig classes, no class switching, and the process of affinity maturation is much less efficient27.
Here we used RNA-Seq to simultaneously examine the gene expression dynamics of CyHV-3 and carp during various infection phases. Apart from identifying components of the cell-mediated immune response, a molecular approach, rather than serological, may also improve sensitivity for the detection of specific anti-CyHV-3 Ig. Our aims were to improve the current understanding of CyHV-3 genome transcription, carp resistance and acquired immunity, and to improve early detection of carp that have been exposed to CyHV-3. This would have implications for both aquaculture and biocontrol programs. Three different phases of infection were used to explore immune development in carp, including acutely infected fish (acute phase), infected fish held at a low, non-permissive temperature for CyHV-3 replication (persistent phase), and infected fish reacclimatized to a permissive temperature after being held at the non-permissive temperature (reactivation phase). The persistent phase was not intended to mimic a latent infection, but rather a low-level persistent infection that could then be stimulated in the reactivation phase. We hypothesized that carp in the acute phase would show an early immune response, with some increase in Ig diversity. Infected carp in the persistent phase were expected to slowly develop a mature immune response in the face of suppressed CyHV-3 replication. Finally, carp in the reactivation phase were predicted to develop a stronger immune response due to reactivated virus replication, and greater Ig diversity with developing specificity, due to their previously acquired immunity.
Results
Genome mapping and replicate similarity
Carp immune responses and CyHV-3 open reading frame (ORF) expression were examined during three phases of infection: acute, persistent and reactivation. Fish from each of these phases were sampled, messenger RNA (mRNA) was extracted and RNA-Seq technology was used to produce several million high quality reads per sample (Table 1). After cleaning, a large proportion of the reads were successfully mapped to the carp genome, with good coverage for gene expression analyses (Table 1). The percentage of CyHV-3 reads in carp cells was highest in the acute and reactivated infections, as expected, although there was variability between the replicates (Table 1). For example, acute replicate 1 contained ~1.44% viral reads compared to ~0.02% in both other replicates. In the reactivated samples, replicate 3 had ~0.03% CyHV-3 reads, whereas the other replicates had an order of magnitude fewer. In both persistent and mock samples, low numbers of reads mapped to the CyHV-3 genome. The few CyHV-3 reads detected in the mock replicates may be the result of low-level cross-contamination, which is common in high-throughput sequencing studies.
Table 1. Read quality and mapping results for the RNA-Seq libraries.
| Sample | NCBI BioSample | Treatment | Total reads | Cleaned reads | Mapped to Cyprinus carpio genome | Mapped to CyHV-3 genome |
|---|---|---|---|---|---|---|
| 1 | SAMN04537393 | Acute | 28,645,158 | 25,048,054 | 18,741,223 (74.8%) | 360,418 (1.4389%) |
| 2 | SAMN04537394 | Acute | 31,309,358 | 28,452,933 | 21,568,752 (75.8%) | 5,434 (0.0191%) |
| 3 | SAMN04537395 | Acute | 33,802,985 | 30,992,654 | 23,530,831 (75.9%) | 5,337 (0.0172%) |
| 4 | SAMN04537396 | Persistent | 33,014,462 | 30,415,615 | 23,004,950 (75.6%) | 96 (0.0003%) |
| 5 | SAMN04537397 | Persistent | 32,699,016 | 30,139,958 | 22,157,303 (73.5%) | 101 (0.0003%) |
| 6 | SAMN04537398 | Persistent | 31,940,121 | 29,333,923 | 22,088,110 (75.3%) | 66 (0.0002%) |
| 7 | SAMN04537399 | Reactivated | 33,243,586 | 30,521,918 | 22,959,764 (75.2%) | 407 (0.0013%) |
| 8 | SAMN04537400 | Reactivated | 33,933,720 | 31,064,624 | 23,427,770 (75.4%) | 261 (0.0008%) |
| 9 | SAMN04537401 | Reactivated | 32,050,049 | 28,934,613 | 21,425,596 (74.0%) | 9,059 (0.0313%) |
| 10 | SAMN04537403 | Mock | 31,401,721 | 28,838,999 | 20,698,717 (71.8%) | 95 (0.0003%) |
| 11 | SAMN04537404 | Mock | 36,074,985 | 32,918,417 | 24,248,317 (73.7%) | 39 (0.0001%) |
*Cyprinus carpio genome is from www.carpbase.org and the CyHV-3 genome is GenBank #DQ657948.1.
Gene expression determined by RNA-Seq and qRT-PCR are highly correlated
To validate our approach, we compared the RNA-Seq expression levels from several representative CyHV-3 ORFs to expression levels determined by quantitative reverse transcription-PCR (qRT-PCR) using the same samples. Six ORFs that span the CyHV-3 genome and are transcribed at different stages of active infection were chosen for the comparison (Supplementary Fig. S1). ORF expression values determined by RNA-Seq and qRT-PCR were significantly correlated and had a Pearson’s r value greater than 0.90 for five of the six ORFs tested (Supplementary Fig. S1). The only exception was ORF78, which had a slightly lower correlation (Pearson’s r = 0.86, p = 0.059). These results suggest that RNA-Seq is as sensitive and accurate as qRT-PCR for determining viral gene expression in vivo, with the added benefit of characterizing expression over the entire genome. This means that not only can all viral ORFs be examined, but other genomic regions that may not be annotated easily, such as micro-RNAs and other non-coding RNAs, can also be studied.
CyHV-3 gene expression follows a co-ordinated pattern across infection phases
Although absolute expression levels of CyHV-3 were much higher in acute replicate 1, the relative expression pattern was similar to the other acute replicates and to the reactivated samples (Fig. 1). For example, the most highly expressed ORFs were the same within each sample (Fig. 1). These included ORF11 (unknown function), ORF12 (tumor necrosis factor receptor), ORF31 (similar to eukaryotic PLAC8 proteins), ORF59 (predicted transmembrane protein), ORF81 (viral envelope, type III membrane protein with 4 transmembrane domains), ORF106 (predicted transmembrane protein), and ORF121 (function unknown)1,9. Interestingly, the largest peak in the raw coverage profiles was consistently located within the 22 kb terminal repeat region of the CyHV-3 genome (Supplementary Fig. S2). This peak was between ORF5 and ORF6, outside both protein coding regions, and appeared to be associated with the polyadenylation sites for each gene9.
Figure 1. Normalized gene expression from CyHV-3 open reading frames (ORFs).

Values were normalised to reads per kilobase of transcript per million mapped reads (RPKM) and log10 transformed to enable sample comparison.
To determine if the most highly expressed CyHV-3 ORFs were indeed the same in the different infection phases, and if any ORFs were significantly differently expressed, pairwise comparisons were conducted (Supplementary Fig. S3). Comparison of the acute and reactivation phases showed an approximately linear relationship, confirming the coordinated expression of ORFs in the CyHV-3 genome (Supplementary Fig. S3). CyHV-3 ORFs in the acute phase were clearly more highly expressed than in the other infection phases, and many of the expression values were significantly different. Several of the acute expression values were also found to be non-significant, despite appearing to have much higher levels. This is likely due to variability in the acute replicates (replicate 1 had far more CyHV-3 reads), reducing the power of the test to find significant differences.
CyHV-3 infection extensively modifies gene expression dynamics in carp
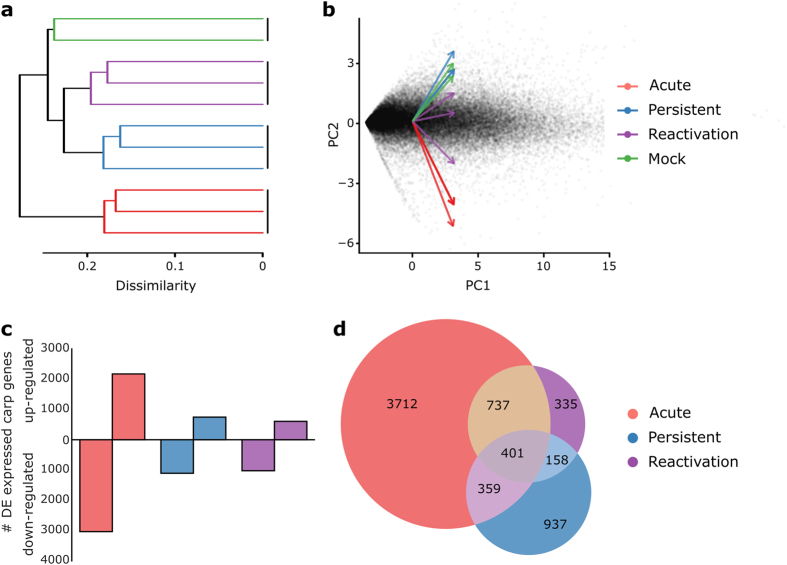
Replicate carp samples within each phase had similar gene expression profiles, providing high power for significance testing (Fig. 2a,b). Interestingly, the carp gene expression profiles changed in an approximately linear fashion from phases without active CyHV-3 replication, to reactivated samples, to samples in the acute infection phase (i.e., principal component 2 progressing from positive to negative values in Fig. 2b). Moreover, several hundred to several thousand carp genes were significantly up- or down-regulated in response to the CyHV-3 infection phases (Fig. 2c). The highest number of differentially expressed genes was seen in the acute phase, and more genes were down-regulated than up-regulated (Fig. 2c). A high proportion of differentially expressed genes in the reactivation phase were also differentially expressed in the acute phase, while in contrast, differentially expressed genes in the persistent phase were often unique to that group (Fig. 1d). The top differentially expressed genes in the acute phase were related to the detoxification of foreign substrates and immune responses, including nitric oxide synthase, microfibril-associated glycoprotein 4, cytoglobin 1, myeloperoxidase, glutathione S-transferase 3, a signal peptide and immune-responsive gene 1 (Table 2). In the persistent and reactivation phases, significant expression differences were also seen for immune related and detoxification related genes, although often they were down-regulated. These included single Ig IL-1-related receptor, cytosolic sulfotransferase, signal peptides, trans-1,2-dihydrobenzene-1,2-diol dehydrogenase, CXC chemokines and glutathione S-transferase 3 (Table 2).
Figure 2.
Similarities in carp gene expression profiles using Jensen-Shannon distances (a) and principal components (b), and significantly differently expressed carp genes in each phase (c) and common to different phases (d). In each plot the colours indicate the phase of infection. The numbers in (d) indicate significantly differently expressed genes in each phase and the overlapping segments indicate the number of genes differentially expressed in, and between, phases of infection. PC is an acronym for principal component.
Table 2. Top 10 most differentially expressed carp genes in each phase of CyHV-3 infection compared to the mock controls.
| Gene ID | Function | Fold change (log2) | Adjusted p-value |
|---|---|---|---|
| Acute compared to Mock | |||
| cycg013101 | Nitric oxide synthase, inducible | 6.9 | 4.9−36 |
| cycg029192 | Microfibril-associated glycoprotein 4 | 6.4 | 4.9−24 |
| cycg038910 | Solute carrier family 2 | −4.9 | 5.9−24 |
| cycg049360 | Latexin | 3.9 | 7.5−24 |
| cycg047504 | Cytoglobin 1 | 5.9 | 3.2−23 |
| cycg025164 | Myeloperoxidase | 6.2 | 3.1−22 |
| cycg005547 | Glutathione S-transferase 3 | −4.5 | 4.1−22 |
| cycg018221 | Signal peptide | 6.3 | 5.7−22 |
| cycg036267 | Immune-responsive gene 1 | 6.6 | 1.2−21 |
| cycg049796 | Glutamate [NMDA] receptor-associated protein 1 | 4.3 | 2.8−21 |
| Persistent compared to Mock | |||
| cycg002335 | Insulin-like growth factor-binding protein | −5.4 | 4.4−18 |
| cycg027385 | Single Ig IL-1-related receptor | −2.3 | 4.4−18 |
| cycg041948 | Cytosolic sulfotransferase 3 | −3.7 | 5.1−17 |
| cycg003666 | Signal peptide | −6.1 | 5.2−17 |
| cycg010925 | Trans-1,2-dihydrobenzene-1,2-diol dehydrogenase | −4.2 | 1.0−16 |
| cycg043850 | Thymidine phosphorylase | 5.1 | 5.0−15 |
| cycg022138 | Natterin-4 | −5.6 | 9.2−15 |
| cycg008033 | Transmembrane protein C19orf77 | −2.6 | 4.3−14 |
| cycg046066 | Sulfate transporter | −3.3 | 8.4−14 |
| cycg041592 | Cocaine- and amphetamine-regulated transcript protein | 4.9 | 6.3−13 |
| Reactivated compared to Mock | |||
| cycg041089 | Sodium- and chloride-dependent creatine transporter 1 | 5.3 | 3.8−19 |
| cycg008033 | Transmembrane protein C19orf77 | −3.0 | 8.0−18 |
| cycg011420 | S-adenosylmethionine synthase isoform type-1 | −5.1 | 3.2−15 |
| cycg038910 | Solute carrier family 2 | −3.8 | 2.4−14 |
| cycg002359 | Myosin-11 | −2.8 | 1.2−12 |
| cycg003666 | Signal peptide | −4.6 | 6.5−12 |
| cycg042156 | UPI0001DE912A related cluster | −2.8 | 6.5−12 |
| cycg031384 | CXC chemokine receptor type 7 | −3.0 | 7.2−12 |
| cycg005547 | Glutathione S-transferase 3 | −3.2 | 1.2−11 |
| cycg030806 | Growth/differentiation factor 15 | 3.6 | 1.6−11 |
Carp humoral immune response predominates in acute CyHV-3 infections
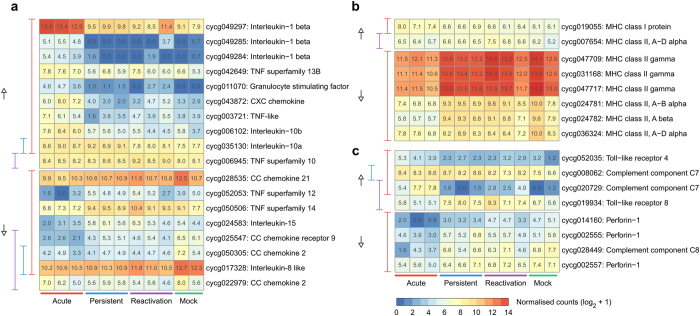
The immune responses of carp infected with CyHV-3 were examined more closely by analysing differentially expressed genes annotated with the gene ontology (GO) term “immune response” (GO:0006955; Fig. 3). When fish in the acute phase were compared to mock infected fish, several cytokine molecules were up-regulated, including interleukin-1 beta (IL-1b) homologues, interleukin-10 (IL-10) homologues, granulocyte stimulating factor, various chemokines and some, but not all, tumor necrosis factors (TNF) (Fig. 3a). On the other hand, interleukin-15 (Fig. 3a) and almost all major histocompatibility class II (MHC II) antigens were down-regulated during acute CyHV-3 infection, while one MHC class I antigen was up-regulated (Fig. 3b). Moreover, C7 components of the complement cascade were up-regulated, and various perforin genes, which are associated with cytotoxic T cells in a cell-mediated immune response, were down-regulated in the acute phase compared to the controls (Fig. 3c). The up-regulation of IL-10 and at least one component of the complement system, plus the blanket down-regulation of MHC II antigens and perforin molecules, suggests that carp in the acute phase of CyHV-3 infection predominantly mount a T Helper 2 (Th2) response, which stimulates the humoral immune system at the expense of a cell-mediated immune response.
Figure 3. Significantly differentially expressed carp genes that are annotated with the gene ontology term “immune response” (GO:0006955).
The genes have been further categorized into cytokines (a), major histocompatibility classes (b) or other immune system components (c). For better visualisation, raw counts have been normalised (log2 + 1). The bars on the left side of each heatmap indicate which genes were significantly differentially expressed in each infection phase (red = acute, blue = persistent, violet = reactivation) compared to the mock controls, and the arrows indicate an up- or down-regulation. TNF is an acronym for tumor necrosis factor.
In contrast, fish in the persistent and reactivation phases altered the expression of far fewer immune-related genes. For example, they did not alter the expression of IL-1b homologues, granulocyte stimulating factor or most TNF transcripts, compared to mock infected fish (Fig. 3a). Moreover, MHC I and II antigens in both the persistent and reactivation phases were expressed in similar levels to the mock controls (Fig. 3b). In the persistent phase, however, an IL-10 homologue (IL-10a; cycg035130) was up-regulated compared to the controls, while another was unchanged (IL-10b; cycg0006102) (Fig. 3a). These differences in IL-10 homologue expression across CyHV-3 infection phases have interesting implications for this important anti-inflammatory cytokine and these were investigated further.
Differential interleukin-10 homologue expression
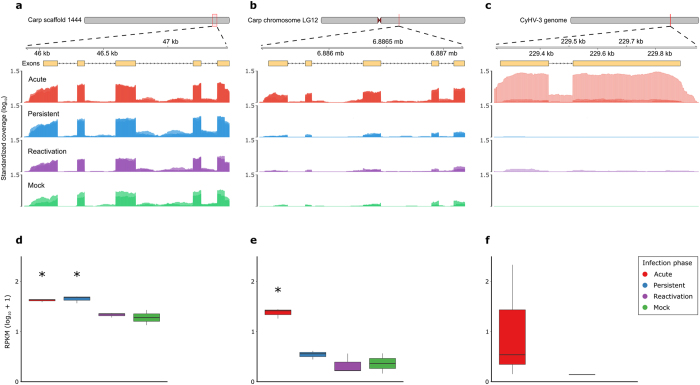
We analysed the carp genome for the presence of duplicated immune genes using reciprocal best match BLAST searches, and found numerous duplicated genes involved in a wide range of different immune pathways (Supplementary Fig. S4). Although the expression of duplicated genes was moderately correlated (Supplementary Fig. S4, inset), many gene pairs were differentially expressed during CyHV-3 infection (Supplementary Fig. S4). These included major histocompatibility antigens, tumor necrosis factors and various cytokine molecules (Supplementary Fig. S4). Of particular interest were expression changes in IL-10, which is an important modulator of immune responses and is frequently analysed in other carp experiments13,22,28. In the carp genome, two copies of IL-10, designated IL-10a (cycg035130; Fig. 4a,c) and IL-10b (cycg006102; Fig. 4b,d), are present22,29. These homologues were detected in the carp genome assembly30, and were also confirmed by PCR amplification and Sanger sequencing (GenBank accessions KX964677 and KX964678). The two IL-10 homologues have high amino acid identity (84%) and similar gene splicing organization, with each containing 5 exons (Fig. 4a,b). Moreover, the expression of the exons was highly conserved across the two genes (Fig. 4a,b). Despite the high sequence conservation and splicing similarity, however, the homologues were expressed differently during different phases of infection. For example, IL-10b was only significantly up-regulated in the acute phase (Fig. 4e), while IL-10a was up-regulated in both the acute and persistent phases and had a greater baseline expression level (Fig. 4d). The CyHV-3 genome also contains a captured homologue of carp IL-10, which shares 24% and 25% amino acid identity with carp IL-10a and IL-10b, respectively (Fig. 4c,f). Significantly, in acute replicate 1, viral IL-10 was expressed at more than double the rate of the carp’s own IL-10 homologues (Fig. 4).
Figure 4.
Genomic location, gene splicing and expression differences for interleukin-10 homologues in carp, IL-10a (cycg035130; a,d) and IL-10b (cycg006102; b,e), and the CyHV-3 homologue (c,f). Exons are indicated by yellow boxes and standardized coverage is the depth per million total mapped reads in each sample library (log10 scale; a,b,c). Reads per kilobase per million mapped reads (RPKM) in d, e and f indicate the combined expression signal over the whole gene and are normalized for library and gene size. Asterisks indicate a significant difference compared to the mock infected fish (d,e).
CyHV-3 infection increases carp immunoglobulin repertoire and V-region selection pressure
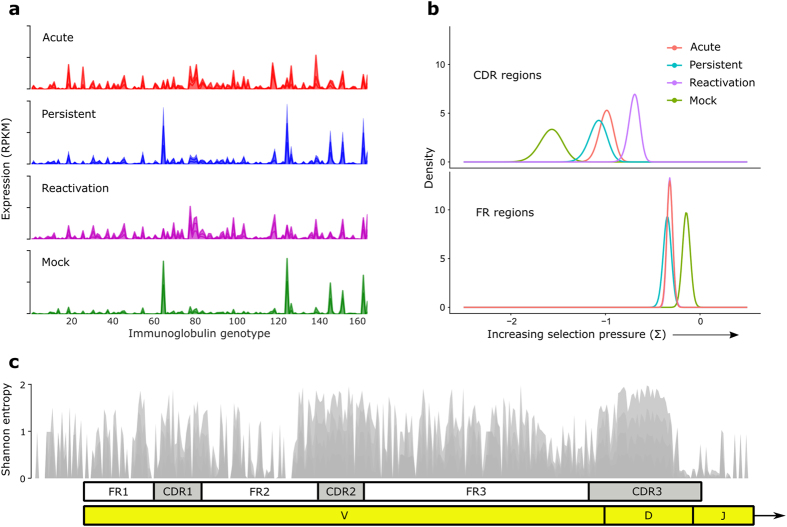
The developing immune response of carp in response to CyHV-3 infection was further scrutinized by examining the immunoglobulin (Ig) repertoire in each phase of infection (Fig. 5). In the absence of infection (mock) or in the face of a low level active infection (persistent phase), carp expressed remarkably similar Ig genotypes with similar expression levels (Fig. 5a). In contrast, fish in the acute and reactivation phases expressed a significantly greater diversity of Ig genotypes (p < 0.05), and did not highly express the mock Ig genotypes (Fig. 5a). Moreover, somatic mutation patterns revealed that Ig transcripts from the acute and reactivation groups were under higher selection pressure in complementarity determining regions (CDR) and lower selection pressure in framework regions (FR). This pattern was reversed for Ig transcripts in mock fish (Fig. 5b). In fact, fish that were subjected to the longest periods of infection with CyHV-3, i.e., in the reactivation phase, had the highest overall selection pressure in CDR regions (Fig. 5b). A global alignment of all carp Ig transcripts indicated that the highest nucleotide diversity was located within CDRs, particularly CDR3 (Fig. 5c), confirming that these regions are an important source of diversity for antigen matching.
Figure 5.
Carp immunoglobulin (Ig) repertoire in different phases of infection (a), selection pressure on complementarity determining regions (CDR) and framework regions (FR) of carp Ig molecules (b), and nucleotide diversity (as measured by Shannon entropy) in a multiple alignment of carp Ig transcripts (c). Selection pressure in (b) was estimated using somatic mutation patterns and the Bayesian estimation of Antigen-driven SELectIoN (BASELINe) algorithm (Yaari et al. 2012). Variable (V), diversity (D) and joining (J) regions of the Ig transcript in (c) are annotated.
Discussion
We used RNA-Seq on carp infected with CyHV-3 to analyse genome-wide expression changes in both the virus and host, including CyHV-3 transcription patterns, carp immune responses, expression differences in immune-related homologues, and carp Ig diversity. Three different phases of CyHV-3 infection were used to gain a more comprehensive understanding of the interplay between virus and host, including acute (initial severe infection), persistent (infected but little or no CyHV-3 replication) and reactivation (active infection following persistent phase).
CyHV-3 ORFs were consistently expressed in the same relative proportions across the sample replicates and infection phases, suggesting a sequential and coordinated expression pattern, as previously reported10. A number of the highly expressed viral ORFs have eukaryotic counterparts, suggesting a role in evasion of the host immune response. These include ORF31, which has homology to eukaryotic PLAC8 proteins, and ORF12, a viral homologue of tumor necrosis factor receptor (TNFR) SF11,15. Tumor necrosis factors (TNFs) are inflammatory cytokines that induce several immunological responses, including apoptosis of infected cells31. Virus-encoded TNFRs counteract TNF activity of the host by functioning as decoy receptors32. Through this act of mimicry the virus can modulate the host’s immune response and ensure its own survival15,16. In concordance with the results here, CyHV-3 TNFR was found to be one of the most abundant proteins in cell culture33, suggesting that this is a major immune avoidance mechanism for the virus. A disproportionately large number of the highly expressed CyHV-3 ORFs contained transmembrane domains, demonstrating the importance of signalling and cross membrane molecule transport to CyHV-3 functioning. ORF81, which is part of the viral envelope, is a type III membrane protein with 4 transmembrane domains and was the first CyHV-3 protein to be functionally characterised34. This protein was found to be abundant in CyHV-3 cell culture, supporting the high expression results seen here, and has been recombined into a plasmid for use in a vaccine experiment due to its high immunogenicity35. The high abundance and immunogenicity of ORF81 makes this an important genomic region in the study of viral-host co-evolution, particularly for studies examining the development of host resistance and the feasibility of CyHV-3 as a biocontrol agent for carp.
In the carp host, the greatest number of gene expression changes were seen in the acute phase of CyHV-3 infection, with over 5,000 genes significantly up- or down-regulated compared to mock infected fish. Interestingly, fewer gene expression changes were seen for fish in the reactivation phase (approximately 1,600 genes up- or down-regulated) despite these fish also experiencing a severe infection. This discrepancy may have arisen because fish in the reactivation phase had been pre-exposed to CyHV-3, possibly leading to the development of a more focused immune response. This is consistent with other studies, which show that following initial expression changes, many genes return to control levels after several days of infection13,24,36. In another study that used high-throughput sequencing to assemble a carp transcriptome, more than 20,000 differentially expressed genes were detected during CyHV-3 infection37. This is significantly more than detected here, which probably relates to the different bioinformatics techniques used, i.e., transcriptome assembly versus genome mapping, and different experimental designs. Interestingly however, they also found that more carp genes were down-regulated than up-regulated in response to CyHV-3, which likely reflects the suppression of broad metabolic pathways in favour of a targeted immune response.
Many of the most differentially expressed carp genes in this experiment have also been detected in previous reports. For example, nitric oxide synthase (NOS), which was highly expressed in the kidney of infected carp here, is also highly expressed in carp skin, intestine, spleen and gills during CyHV-3 infection36,38. The up-regulation of NOS can control the replication of many viruses through inhibition of viral protease activity39, and may be a general host defence mechanism. Microfibril-associated glycoprotein 4 (MFAP4) was also very highly expressed in carp in the acute phase of CyHV-3 infection. Interestingly, Rakus et al. found that MFAP4 was more highly expressed in carp with greater resistance to CyHV-3, indicating its importance to carp immunity24. Indeed, Niu et al. suggested that MFAP4 is the teleost homologue of ficolins, which can recognize pathogens, activate the complement system, initiate opsonisation, and trigger other pathogen clearing mechanisms40. The up-regulation of both myeloperoxidase (MPO) and cytoglobin during active CyHV-3 infection may be interrelated. For example, MPO is expressed by granulocytes to catalyse the production of acids and kill pathogens41, however, this can also cause oxidative damage to host cells, which in turn, may stimulate the production of cytoglobin as a protective measure42. Similarly, glutathione S-transferase (GST) is a well-known antioxidant enzyme43. In this study, however, it was down-regulated in response to CyHV-3 infection. This may be related to the many roles of GSTs, which include the suppression of apoptosis by inhibition of c-Jun NH2-terminal kinase (JNK) and p3844. Thus, the down-regulation of GST in acutely infected fish would allow apoptosis of infected cells. Overall, carp infected with CyHV-3 alter the expression of a broad group of genes involved in general immune responses, clearing pathogens and dealing with oxidative stress induced by the virus directly, or as a consequence of other detoxification pathways.
An examination of specific adaptive immune responses in carp revealed differences between acutely infected carp, and those in the persistent and reactivation groups. Specifically, fish in the acute phase up-regulated interleukin-1 beta (IL-1b) homologues, interleukin-10 (IL-10) homologues and components of the complement system, while most major histocompatibility class II (MHC II) antigens and perforin molecules were down-regulated. This suggests that a humoral, as opposed to cell-mediated, immune response was mounted by carp in the late stages of acute CyHV-3 infection. For example, an increase in IL-10 expression can down-regulate cytokines involved in T Helper 1 (Th1) responses, MHC II antigens and other macrophage stimulatory molecules (cell-mediated responses), in addition to catalysing the proliferation of B-cells and antibody production (humoral responses)45. Indeed, in the current study, greater IL-10 expression in infected carp corresponded with decreases in MHC II antigen and perforin expression. Moreover, several components of the complement cascade, which can help clear immune complexes and pathogens in a humoral response46, were up-regulated in acutely infected carp. This predominance of humoral immune responses may be unexpected during the latter stages of CyHV-3 infection, where CyHV-3 particles have presumably become established within cells. Interestingly, the type of immune response mounted by carp might be influenced by the virus itself. For example, CyHV-3 encodes a functional IL-10 homologue12,14,33,47, which in one acute replicate was expressed at double the rate of the carp’s own IL-10 homologues. This would artificially increase local or systemic IL-10 concentrations, further encouraging a humoral immune response and suppression of cell-mediated immunity, thus allowing intracellular virus to survive and to potentially develop latency. On the other hand, carp in the persistent and reactivation phases expressed a range of genes that suggested a balance between humoral and cell-mediated immune responses. This divergence may be attributed to longer periods of infection in the persistent and reactivation phases, allowing for a more targeted and mature response.
The carp genome has been studied with particular interest due to a relatively recent whole genome duplication (WGD) event (~8.2 million years ago), which allows researchers to examine the initial consequences of genome duplication on gene function30,48,49,50. For example, WGD often produces extensive genetic redundancy, which results in gene deletions or functional divergence. Interestingly however, Li et al. found that recently duplicated carp genes have mostly been retained, although approximately half of the duplicates were not expressed synchronously, suggesting functional divergence or sub-functionalization49. In this study, we found that many immune-related genes from a broad group of pathways were also duplicated. Although the expression of duplicated genes was moderately correlated, a large number of homologues showed significant dissociation of expression, including genes that are central immune components, such as tumor necrosis factors, interleukins and various other cytokines. This has important implications for understanding carp immune responses.
One such homologue pair was IL-10a and IL-10b, which showed particularly large expression differences. Specifically, both homologues were up-regulated during the acute infection phase, but only IL-10a was up-regulated during the persistent phase. In addition, IL-10a had a higher baseline expression level in mock infected carp and during the persistent phase compared to IL-10b. This suggests either sub-functionalization (both homologues retain some functionality), neo-functionalization (new homologue functionality), or that the homologues are induced by different processes. Recently, Piazzon et al. also found that carp IL-10a was more highly expressed under baseline conditions, and that IL-10b was up-regulated during an infection with spring viraemia of carp (SVCV)29. This is similar to our findings with CyHV-3, except that we also saw an up-regulation of IL-10a during infection. Moreover, Piazzon et al. reported that the two IL-10 homologues have a similar structure and exert similar bioactivity on phagocytes29. They suggested that the duplicates have undergone sub-functionalization, and are expressed differently due to differences in their promoter regions. Future studies focusing on further clarifying the nature of this divergence would be particularly interesting because IL-10 is an important regulator of broader immune responses, as previously discussed22,51,52. In another study that examined IL-10 homologue expression, Benedicenti et al. found that the homologues were expressed similarly in Atlantic salmon with amoebic gill disease, although they did not examine different phases of infection51. At a nucleotide level, modifications of the host IL-10a homologue sequence have been associated with greater resistance of carp to CyHV-322. In this study, however, all of the wild Australian fish had identical IL-10 homologue sequences, suggesting that greater resistance due to SNP diversity in IL-10a22 may not affect this population. We suggest, however, that there may be selection for rare SNPs that confer resistance if CyHV-3 were to be released in Australia. These results highlight the need for greater study into homologue functionality of carp genes, including their influence on resistance and immune function. Moreover, studies examining immune responses in carp using PCR based techniques should be aware of the presence of homologues and design primers accordingly.
A successful humoral immune response relies on the production of a large diversity of B-cells with numerous receptors that are capable of matching a correspondingly large variety of antigen epitopes. The subsequent production of specific Ig can then be detected using serological techniques. In fish, however, the humoral response is relatively primitive, which can result in non-specific Ig production, longer response times, and variable levels of Ig production26,27. Consequently, often populations of carp, rather than individuals, are required for detection of CyHV-3-specific antibodies with ELISA-based protocols26. Moreover, cross-reactivity between CyHV-3 and CyHV-1 using ELISA can be problematic21. A potential alternative to these serological techniques is the use of high-throughput sequencing to assemble individual Ig transcripts, and then to identify the specific transcripts of interest. This approach may be more sensitive and specific, as well as providing the nucleotide sequence and information on gene and isoform expression53. This technique is relatively novel, particularly for fish, although it has been used to examine the antibody repertoire in healthy zebrafish54,55.
Here we used high throughput sequencing in fish to follow the changes in Ig repertoire during different phases of CyHV-3 infection. We found a highly reproducible pattern of Ig genotypes expressed by carp with no, or low levels of, CyHV-3 replication, i.e., all mock infected fish and fish in the persistent phase. In contrast, fish in the acute and reactivation phases produced a significantly greater diversity of Ig genotypes, suggesting the development of an adaptive immune response. Moreover, Ig transcripts from the acute and reactivation phases were under higher selection pressure in complementarity determining regions (CDR) and lower selection pressure in framework regions (FR), suggesting that clonal selection of high-affinity B-cells was occurring in these fish56,57. This can be inferred because clonal selection is manifested through fewer changes in regions that maintain structural integrity, i.e., FRs, but more changes in antigen recognition sites, i.e., CDRs56,57. Moreover, carp that were infected with CyHV-3 for the longest time, i.e., in the reactivation phase, had the highest overall selection pressure in CDR regions, further suggesting a clonal B-cell selection process. Only a small number of other studies have examined the selection pressure dynamics on fish Ig molecules, and these have produced inconsistent results27. For example, Yang et al. did not find any difference in selection pressure on catfish FRs and CDRs and suggested that clonal selection was inefficient in bony fish58. On the other hand, Magadán-Mompó et al. found higher frequencies of mutations in CDRs compared to FRs in medaka59, and this has also been seen in zebrafish60 and Nile tilapia61. Overall, our results indicate that Ig repertoire diversification was followed by clonal selection of high-affinity B-cells in carp infected with CyHV-3. In this experiment, however, only fish that succumbed to CyHV-3 infection were sampled and, thus, we could not detect a successful immune response or associated antibodies. Future studies will use carp that survive infection with CyHV-3 in order to detect CyHV-3-specific antibodies.
In summary, carp acutely infected with CyHV-3 altered the expression of thousands of genes, which were often involved in typical innate or adaptive immune response, or detoxification pathways. Interestingly, the carp predominantly mounted a humoral immune response, despite the probable establishment of CyHV-3 in cells. An intriguing possible explanation is that the virus itself has evolved to tip the balance of the host away from a cell-mediated response, and toward a humoral response, through the expression of a captured IL-10 homologue, thereby favouring virus survival. Moreover, many immune-related carp genes were found to be duplicated, and many of these showed dissociation of expression. Of particular interest were two IL-10 homologues that were expressed differently across different phases of infection, suggesting neo- or sub-functionalization. Finally, the carp Ig repertoire was examined during different phases of CyHV-3 infection. Ig diversification was detected during acute and reactivation phases, and this was followed by the selection of high-affinity B-cells, presumably signifying an adaptive humoral immune response.
Methods
Ethics statement
This study was carried out at the microbiologically-secure CSIRO-Australian Animal Health Laboratory (CSIRO-AAHL). All experiments were approved by the CSIRO-AAHL Animal Ethics Committee and performed according to the relevant guidelines and regulations.
Experimental conditions and virus infection
Samples for this experiment were obtained from a previous study62, which includes histopathological data and detailed information on the experimental model. Briefly, juvenile common carp (mean length 12.1 ± 1.0 cm), Cyprinus carpio, were obtained from wild Australian stocks and acclimatized to laboratory conditions for 8 days. The carp were subjected to a 12 h light/12 h dark cycle and were given a commercial fish feed at 1% of their bodyweight per day. For these experiments, we chose 3 different phases of CyHV-3 infection in the fish: “acute”, “persistent” and “reactivation”. The acute group contained fish in the initial phase of active CyHV-3 infection at a permissive temperature; the persistent group included infected fish held at a low, non-permissive temperature for CyHV-3 replication; and reactivation fish were obtained by returning infected fish from the non-permissive temperature to a permissive temperature, thereby inducing an active infection. To achieve the desired groups, 60 carp were infected with 100 TCID50 ml−1 of an Indonesian CyHV-3 isolate (C0763) by immersion for 2 h at 22 °C. For the acute phase of infection, thirty of the infected fish were kept at 22 °C and individual fish were sacrificed as they became moribund. The remaining thirty infected fish were used to obtain the persistent and reactivation groups. They were initially held at 22 °C for 24 h to allow establishment of the CyHV-3 infection, and the infection was subsequently arrested by decreasing the water temperature to 11 °C over a period of 4 days at a rate of 2 to 3 °C per day. After 28 days at the non-permissive temperature, multiple fish were sacrificed for the persistent group. The virus was reactivated in the remaining fish by increasing the water temperature to 22 °C, again over 4 days, at a rate of 2 to 3 °C per day, and fish were sampled when moribund for the reactivation group. In addition to these groups, sixty mock-infected control carp were subjected to the same temperature regimes as the infected fish. The anterior kidney was dissected from sacrificed fish in each of the groups, and frozen at −20 °C in RNAlater (Ambion). After completion of the experiment, total RNA was extracted from the kidney of each fish with the AllPrep DNA/RNA extraction kit (Qiagen) following the manufacturer’s instructions.
RNA-Seq and qRT-PCR
Three fish in each of the acute, persistent and reactivation phases, plus three randomly chosen mock fish, were selected for RNA sequencing (RNA-Seq). The RNA samples were sent to the Australian Genome Research Facility (AGRF, Melbourne, Australia), where messenger RNA (mRNA) was enriched in each sample by selection of polyA + tailed mRNA, which was then sequenced using two 150 bp single-end HiSeq lanes (Illumina). The raw RNA-Seq reads are available in the NCBI Sequence Read Archive under BioProject accession PRJNA314552. After our initial data analysis, one of the mock replicates was identified as a hybrid between common carp and goldfish (Carassius auratus) and was consequently removed from further analysis.
For validation purposes, the expression of several CyHV-3 open reading frames from the same samples used for RNA-Seq were determined using quantitative reverse transcription-PCR (qRT-PCR; for primers see Table 1 in Sunarto et al.)62. Expression from the open reading frames was normalised to expression of carp 18S rRNA to enable sample comparison. Duplicate reactions for each gene and each sample contained 12.5 μl of TaqMan One-Step RT-PCR Master Mix, 1 μl of RT-PCR Enzyme Mix (Applied Biosystems), 1.25 μl each of probe (5 μM) and forward and reverse primers (2 μM for 18S rRNA primers; 18 μM for viral primers), 2 μl of template RNA (100 ng per reaction), plus 5.75 μl of RNase-free water. The reactions were then amplified using a 7500 Fast Real-time PCR System (Applied Biosystems) with the following conditions: 48 °C for 30 min; 95 °C for 10 min; then 40 cycles of 95 °C for 15 s; 60 °C for 1 min. The results were analysed using 7500 software v.2.3 (Applied Biosystems).
Data analysis
RNA-Seq reads were cleaned by removing Illumina adapters, trimming sequences with a quality score below 20 and removing reads with fewer than 75 bp with Trimmomatic v.0.3364. Any remaining ribosomal RNA was then removed from the reads using SortMeRNA v.2.065 against the SILVA v.119 ribosomal database66. The high quality mRNA reads were mapped to the Cyprinus carpio genome v.1.030 (available here: www.carpbase.org) and the CyHV-3 genome9 (GenBank accession DQ657948.1) using TopHat v.2.1.067. Successfully mapped reads were assembled into separate transcriptomes for each individual sample using Cufflinks v.2.2.1, before being merged into a master assembly with Cuffmerge v.1.0.0, as recommended68. Counts for each gene were extracted from the mapping files using HTSeq v.0.6.069 with the “union” mode for handling overlapping reads. The raw counts were used for differential expression testing in DESeq2 v.1.6.3, using a significance value of 0.05 and Benjamini & Hochberg corrections for multiple comparisons, as recommended70. Reads per kilobase of transcript per million mapped reads (RPKM) values were calculated using the counts from HTSeq v.0.6.069 in Python v.2.7.11, and normalised log transformations of count data were conducted using DESeq2 v.1.6.370. CummeRbund v.2.8.271 was used to create dendrogram and PCA plots of gene expression similarities between replicates, and matplotlib-venn v.0.11.4 and Seaborn v.0.6.072 were used to plot Venn diagrams and bar graphs. Significant pairwise CyHV-3 gene expression differences and the CyHV-3 expression heatmap were plotted using ggplot2 v.2.1.073, and heatmaps of differentially expressed carp immune genes were constructed using pheatmap v.1.0.8. Samtools depth v.1.374 was used on each of the bam alignment files to extract coverage over the CyHV-3 genome and carp interleukin-10 homologues, which was then normalised to the total number of mapped reads per million in Python v.2.7.11 and plotted using Gviz v.1.14.275. Duplicated carp genes were detected by clustering high quality proteins from the carp genome (greater than 10 amino acids in length and less than 20% stop codons) using orthoMCL76, and extracting immune-related homologues with reciprocal best match results using BLASTp v.2.2.3177 with an e-value of 10−5.
Detection of immunoglobulin repertoire
Immunoglobulin (Ig) diversity is generated through somatic recombination and hypermutation of variable regions, which means that Ig diversity cannot be adequately obtained by mapping RNA-Seq reads to a sequenced genome53,56. This can be overcome by independently assembling RNA-Seq reads into transcriptomes and identifying Ig transcripts in the resulting assembly53. To achieve this, the cleaned mRNA reads from above were re-assembled into separate transcriptomes in a genome-independent manner using Trinity v.2.0.678, with a minimum contig length of 200 and a minimum k-mer coverage of 2. Transcripts with homology to Ig sequences were identified in the assemblies using BLASTn v.2.2.3177 with an e-value of 10−3 against the LIGM-DB database v.1.2.179, which includes Cyprinus carpio sequences. IgBLAST v.1.4.080 was then used on the matching transcripts to identify variable (V), joining (J) and diversity (D) regions, and to determine if they were in-frame and functional. The results were loaded into a Change-O v.0.3.281 database and functional transcripts were assigned into clonal groups (defined as having the same V assignment, J assignment and J length) using DefineClones.py with the “m1n” substitution model and a distance of 7 for calculating distances between the sequences. The model and parameters were selected after visualisation of distance to nearest neighbour plots81. Testing for significant differences in Ig diversity between the infection phases was conducted using testDiversity in Alakazam v.0.2.381 with 1000 bootstrap replicates. The selection pressure on Ig transcripts was estimated using observed and expected mutations and the Bayesian estimation of Antigen-driven SELectIoN (BASELINe) algorithm57. To examine positions of diversity within the V region, a global alignment of Ig transcripts was performed using clustalW82, and the Shannon entropy of each position was calculated using Python v.2.7.12 and plotted with Pandas v.0.17.183.
Amplification and sequencing of IL-10 homologues
PCR was used to confirm the presence of two interleukin-10 (IL-10) homologues in the carp genome. The first homologue (IL-10b; cycg006102) was amplified using the primers IL-10F: CGCCAGCATAAAGAACTCGT and IL-10R: TGCCAAATACTGCTCGATGT13. For the second homologue (IL-10a; cycg035130), the primers IL-10_H2F: CTAGGAGAGCAGCAAACCAGT and IL-10_H2R: TGCCAAATACTGCTCAATGT were designed based on an alignment of homologues in the carp genome assembly30. For each primer set, PCRs were compiled with 12.5 μl of HotStarTaq master mix (Qiagen, Hilden, Germany), 0.5 μl of the forward and reverse primer (18 μM), 9.5 μl of water and 2 μl of template DNA. The conditions for both PCRs were 95 °C for 15 min; then 40 cycles of 95 °C for 30 sec, 50 °C for 30 sec, 72 °C for 30 sec; and a final elongation step of 72 °C for 7 min. Amplified products were purified using the Wizard SV Gel and PCR Clean-Up Kit (Promega, Madison, USA) and sequenced in both directions using a Thermofisher 3500xl Genetic Analyser.
Additional Information
How to cite this article: Neave, M. J. et al. Transcriptomic analysis of common carp anterior kidney during Cyprinid herpesvirus 3 infection: Immunoglobulin repertoire and homologue functional divergence. Sci. Rep. 7, 41531; doi: 10.1038/srep41531 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank Kylie Hall (Fisheries Victoria) and Keith Bell (K & C Global Fisheries, Victoria) for supplying the carp used in this experiment. Funding was provided by the Invasive Animals Cooperative Research Centre of Australia (Project 3W1).
Footnotes
The authors declare no competing financial interests.
Author Contributions M.J.N. analysed the data and wrote the manuscript. A.S. carried out the challenge experiments. A.S. and K.A.M. designed and conceived the experiments. All authors contributed to interpreting the data and writing the manuscript.
References
- Davison A. J. et al. Comparative Genomics of Carp Herpesviruses. J. Virol. 87, 2908–2922 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretzinger A., Fischer-Scherl T., Oumouna M., Hoffmann R. & Truyen U. Mass mortalities in Koi carp, Cyprinus carpio, associated with gill and skin disease. Bull. Eur. Assoc. Fish Pathol. 19, 182–185 (1999). [Google Scholar]
- Hedrick R. P. et al. A Herpesvirus Associated with Mass Mortality of Juvenile and Adult Koi, a Strain of Common Carp. J. Aquat. Anim. Health 12, 44–57 (2000). [DOI] [PubMed] [Google Scholar]
- Uchii K., Matsui K., Iida T. & Kawabata Z. Distribution of the introduced cyprinid herpesvirus 3 in a wild population of common carp, Cyprinus carpio L. J. Fish Dis. 32, 857–864 (2009). [DOI] [PubMed] [Google Scholar]
- Sunarto A., Rukyani A. & Itami T. Indonesian experience on the outbreak of Koi Herpesvirus in Koi and Carp (Cyprinus carpio). Bull. Fish. Res. Agency 15–21 (2005). [Google Scholar]
- FAO. The state of world fisheries and aquaculture (2010).
- Negenborn J., van der Marel M. C., Ganter M. & Steinhagen D. Cyprinid herpesvirus-3 (CyHV-3) disturbs osmotic balance in carp (Cyprinus carpio L.)-A potential cause of mortality. Vet. Microbiol. 177, 280–288 (2015). [DOI] [PubMed] [Google Scholar]
- Rakus K. et al. Cyprinid herpesvirus 3: An interesting virus for applied and fundamental research. Vet. Res. 44, 1–16 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki T. et al. Genome sequences of three koi herpesvirus isolates representing the expanding distribution of an emerging disease threatening koi and common carp worldwide. J. Virol. 81, 5058–65 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilouze M., Dishon A. & Kotler M. Coordinated and sequential transcription of the cyprinid herpesvirus-3 annotated genes. Virus Res. 169, 98–106 (2012). [DOI] [PubMed] [Google Scholar]
- Harkness J. M., Kader M. & DeLuca N. A. Transcription of the herpes simplex virus 1 genome during productive and quiescent infection of neuronal and nonneuronal cells. J. Virol. 88, 6847–61 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunarto A. et al. Koi Herpesvirus Encodes and Expresses a Functional Interleukin-10. J. Virol. 86, 11512–11520 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunarto A. & McColl K. A. Expression of immune-related genes of common carp during cyprinid herpesvirus 3 infection. Dis. Aquat. Organ. 113, 127–135 (2015). [DOI] [PubMed] [Google Scholar]
- Piazzon M. C. et al. Cyprinid Herpesvirus 3 Il10 Inhibits Inflammatory Activities of Carp Macrophages and Promotes Proliferation of Igm + B Cells and Memory T Cells in a Manner Similar to Carp Il10. J. Immunol. 195, 3694–3704 (2015). [DOI] [PubMed] [Google Scholar]
- Yi Y. et al. Functional characterization of viral tumor necrosis factor receptors encoded by cyprinid herpesvirus 3 (CyHV3) genome. Fish Shellfish Immunol. 45, 757–770 (2015). [DOI] [PubMed] [Google Scholar]
- Kattlun J., Menanteau-Ledouble S. & El-Matbouli M. Non-structural protein pORF 12 of cyprinid herpesvirus 3 is recognized by the immune system of the common carp Cyprinus carpio. Dis. Aquat. Organ. 111, 269–273 (2014). [DOI] [PubMed] [Google Scholar]
- McColl K. A., Cooke B. D. & Sunarto A. Viral biocontrol of invasive vertebrates: Lessons from the past applied to cyprinid herpesvirus-3 and carp (Cyprinus carpio) control in Australia. Biol. Control 72, 109–117 (2014). [Google Scholar]
- Weber M. J. & Brown M. L. Effects of Common Carp on Aquatic Ecosystems 80 Years after ‘Carp as a Dominant’: Ecological Insights for Fisheries Management. Rev. Fish. Sci. 17, 524–537 (2009). [Google Scholar]
- Koehn J. D. Carp (Cyprinus carpio) as a powerful invader in Australian waterways. Freshw. Biol. 49, 882–894 (2004). [Google Scholar]
- Perelberg A., Ilouze M., Kotler M. & Steinitz M. Antibody response and resistance of Cyprinus carpio immunized with cyprinid herpes virus 3 (CyHV-3). Vaccine 26, 3750–3756 (2008). [DOI] [PubMed] [Google Scholar]
- Adkison M. A., Gilad O. & Hedrick R. P. An enzyme linked immunosorbent assay (ELISA) for detection of antibodies to the Koi herpes virus (KHV) in the serum of Koi Cyprinus carpio. Fish Pathol 40, 53–62 (2005). [Google Scholar]
- Kongchum P. et al. Association between IL-10a single nucleotide polymorphisms and resistance to cyprinid herpesvirus-3 infection in common carp (Cyprinus carpio). Aquaculture 315, 417–421 (2011). [Google Scholar]
- Rakus K. et al. Major histocompatibility (MH) class II B gene polymorphism influences disease resistance of common carp (Cyprinus carpio L.). Aquaculture 288, 44–50 (2009). [DOI] [PubMed] [Google Scholar]
- Rakus K. L. et al. Gene expression analysis of common carp (Cyprinus carpio L.) lines during Cyprinid herpesvirus 3 infection yields insights into differential immune responses. Dev. Comp. Immunol. 37, 65–76 (2012). [DOI] [PubMed] [Google Scholar]
- Forlenza M. et al. Transcription of signal-3 cytokines, IL-12 and IFNαβ, coincides with the timing of CD8αβ up-regulation during viral infection of common carp (Cyprinus carpio L.). Molecular Immunology 45, 1531–1547 (2008). [DOI] [PubMed] [Google Scholar]
- Torrent F. et al. The amino-terminal domain of ORF149 of koi herpesvirus is preferentially targeted by IgM from carp populations surviving infection. Arch. Virol. doi: 10.1007/s00705-016-2934-4 (2016). [DOI] [PubMed] [Google Scholar]
- Magadan S., Sunyer O. J. & Boudinot P. In Pathogen-Host Interactions: Antigenic Variation v. Somatic Adaptations (eds. Hsu E. & Du Pasquier L.) 235–264, doi: 10.1007/978-3-319-20819-0_10 (Springer International Publishing, 2015). [DOI] [Google Scholar]
- Savan R., Igawa D. & Sakai M. Cloning, characterization and expression analysis of interleukin-10 from the common carp, Cyprinus carpio L. Eur. J. Biochem. 270, 4647–4654 (2003). [DOI] [PubMed] [Google Scholar]
- Piazzon M. C., Wentzel A. S., Wiegertjes G. F. & Forlenza M. Carp Il10a and Il10b exert identical biological activities in vitro, but are differentially regulated in vivo. Dev. Comp. Immunol. doi: 10.1016/j.dci.2016.08.016 (2016). [DOI] [PubMed] [Google Scholar]
- Xu P. et al. Genome sequence and genetic diversity of the common carp, Cyprinus carpio. Nat. Publ. Gr. 46, 1212–1219 (2014). [DOI] [PubMed] [Google Scholar]
- Waters J. P., Pober J. S. & Bradley J. R. Tumour necrosis factor in infectious disease. J. Pathol. 230, 132–147 (2013). [DOI] [PubMed] [Google Scholar]
- Smith C. A. et al. T2 open reading frame from the shope fibroma virus encodes a soluble form of the TNF receptor. Biochem. Biophys. Res. Commun. 176, 335–342 (1991). [DOI] [PubMed] [Google Scholar]
- Ouyang P. et al. The IL-10 homologue encoded by cyprinid herpesvirus 3 is essential neither for viral replication in vitro nor for virulence in vivo. Vet. Res. 44, 1–18 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenkranz D. et al. Identification of envelope protein pORF81 of koi herpesvirus. J. Gen. Virol. 89, 896–900 (2008). [DOI] [PubMed] [Google Scholar]
- Zhou J. et al. Vaccination of plasmid DNA encoding ORF81 gene of CJ strains of KHV provides protection to immunized carp. Vitr. Cell. Dev. Biol. - Anim. 50, 489–495 (2014). [DOI] [PubMed] [Google Scholar]
- Adamek M. et al. Cyprinid herpesvirus 3 infection disrupts the skin barrier of common carp (Cyprinus carpio L.). Vet. Microbiol. 162, 456–470 (2013). [DOI] [PubMed] [Google Scholar]
- Lee X. et al. Transcriptomic analysis of koi (Cyprinus carpio) spleen tissue upon cyprinid herpesvirus 3 (CyHV3) infection using next generation sequencing. Fish Shellfish Immunol. 3 (2015). [DOI] [PubMed] [Google Scholar]
- Miest J. J. et al. Differential effects of alloherpesvirus CyHV-3 and rhabdovirus SVCV on apoptosis in fish cells. Vet. Microbiol. 176, 19–31 (2015). [DOI] [PubMed] [Google Scholar]
- Saura M. et al. An antiviral mechanism of nitric oxide: Inhibition of a viral protease. Immunity 10, 21–28 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu D. et al. Microfibrillar-associated protein 4 (MFAP4) genes in catfish play a novel role in innate immune responses. Dev. Comp. Immunol. 35, 568–579 (2011). [DOI] [PubMed] [Google Scholar]
- Pijanowski L. et al. Carp neutrophilic granulocytes form extracellular traps via ROS-dependent and independent pathways. Fish Shellfish Immunol. 34, 1244–1252 (2013). [DOI] [PubMed] [Google Scholar]
- Latina A. et al. Np63 targets cytoglobin to inhibit oxidative stress-induced apoptosis in keratinocytes and lung cancer. Oncogene 35, 1493–1503 (2016). [DOI] [PubMed] [Google Scholar]
- Sharma R., Yang Y., Sharma A., Awasthi S. & Awasthi Y. C. Antioxidant Role of Glutathione S-Transferases: Protection Against Oxidant Toxicity and Regulation of Stress-Mediated Apoptosis. Antioxid. Redox Signal. 6, 289–300 (2004). [DOI] [PubMed] [Google Scholar]
- Wu Y. et al. Human glutathione S-transferase P1-1 interacts with TRAF2 and regulates TRAF2-ASK1 signals. Oncogene 25, 5787–5800 (2006). [DOI] [PubMed] [Google Scholar]
- Duell B. L. et al. Recent insights into microbial triggers of interleukin-10 production in the host and the impact on infectious disease pathogenesis. FEMS Immunology and Medical Microbiology 64, 295–313 (2012). [DOI] [PubMed] [Google Scholar]
- Merle N. S., Noe R., Halbwachs-Mecarelli L., Fremeaux-Bacchi V. & Roumenina L. T. Complement system part II: Role in immunity. Frontiers in Immunology 6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Beurden S. J. et al. The alloherpesviral counterparts of interleukin 10 in European eel and common carp. Fish and Shellfish Immunology 31, 1211–1217 (2011). [DOI] [PubMed] [Google Scholar]
- David L., Blum S., Feldman M. W., Lavi U. & Hillel J. Recent Duplication of the Common Carp (Cyprinus carpio L.) Genome as Revealed by Analyses of Microsatellite Loci. Mol. Biol. Evol. 20, 1425–1434 (2003). [DOI] [PubMed] [Google Scholar]
- Li J.-T. et al. The fate of recent duplicated genes following a fourth-round whole genome duplication in a tetraploid fish, common carp (Cyprinus carpio). Sci. Rep. 5, 8199 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.-T., Li J.-T., Zhang X.-F. & Sun X.-W. Transcriptome analysis reveals the time of the fourth round of genome duplication in common carp (Cyprinus carpio). BMC Genomics 13, 96 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedicenti O., Collins C., Wang T., McCarthy U. & Secombes C. J. Which Th pathway is involved during late stage amoebic gill disease? Fish Shellfish Immunol. 46, 417–425 (2015). [DOI] [PubMed] [Google Scholar]
- Moore K. W., de Waal Malefyt R., Coffman R. L. & O’Garra A. Interleukin-10 and the interleukin-10 receptor. Annu. Rev. Immunol. 19, 683–765 (2001). [DOI] [PubMed] [Google Scholar]
- Blachly J. S. et al. Immunoglobulin transcript sequence and somatic hypermutation computation from unselected RNA-seq reads in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 112, 4322–7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein J. A., Jiang N., White R. A., Fisher D. S. & Quake S. R. High-Throughput Sequencing of the Zebrafish Antibody Repertoire. Science 324, 807–810 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes G. D. et al. Cryptic hybridization and introgression between invasive Cyprinid species Cyprinus carpio and Carassius auratus in Australia: Implications for invasive species management. Anim. Conserv. 15, 83–94 (2012). [Google Scholar]
- Yaari G. & Kleinstein S. H. Practical guidelines for B-cell receptor repertoire sequencing analysis. Genome Med. 7, 121 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaari G., Uduman M. & Kleinstein S. H. Quantifying selection in high-throughput Immunoglobulin sequencing data sets. Nucleic Acids Res. 40, 10–12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Waldbieser G. C. & Lobb C. J. The Nucleotide Targets of Somatic Mutation and the Role of Selection in Immunoglobulin Heavy Chains of a Teleost Fish. J. Immunol. 176, 1655–1667 (2006). [DOI] [PubMed] [Google Scholar]
- Magadán-Mompó S., Zimmerman A. M., Sánchez-Espinel C. & Gambón-Deza F. Immunoglobulin light chains in medaka (Oryzias latipes). Immunogenetics 65, 387–396 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marianes A. E. & Zimmerman A. M. Targets of somatic hypermutation within immunoglobulin light chain genes in zebrafish. Immunology 132, 240–255 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiraporn P., Sasimanas U. & Prapansak S. Diversity analysis of the immunoglobulin M heavy chain gene in Nile tilapia, Oreochromis niloticus (Linnaeus). African J. Biotechnol. 14, 2282–2299 (2015). [Google Scholar]
- Sunarto A. et al. Characteristics of cyprinid herpesvirus 3 in different phases of infection: implications for disease transmission and control. Virus Res. 188, 45–53 (2014). [DOI] [PubMed] [Google Scholar]
- Sunarto A. et al. Isolation and characterization of koi herpesvirus (KHV) from Indonesia: Identification of a new genetic lineage. J. Fish Dis. 34, 87–101 (2011). [DOI] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M. & Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopylova E., Noe L. & Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28, 3211–3217 (2012). [DOI] [PubMed] [Google Scholar]
- Quast C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–78 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P. T. & Huber W. HTSeq A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W. & Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff L., Trapnell C. & Kelley D. cummeRbund: Analysis, exploration, manipulation, and visualization of Cufflinks high-throughput sequencing data. Bioconductor. 2013 (2013). [Google Scholar]
- Waskom M. et al. seaborn: v0.6.0 (June 2015), doi: 10.5281/zenodo.19108 (2015). [DOI]
- Wickham H. ggplot2: elegant graphics for data analysis at http://had.co.nz/ggplot2/book (Springer: New York, 2009). [Google Scholar]
- Li H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahne F. et al. Gviz: Plotting data and annotation information along genomic coordinates. Bioconductor. 2015 (2015). [Google Scholar]
- Li L., Stoeckert C. J. & Roos D. S. OrthoMCL: Identification of Ortholog Groups for Eukaryotic Genomes. Genome Res. 13, 2178–2189 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W. & Lipman D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- Haas B. J. et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudicelli V. et al. IMGT/LIGM-DB, the IMGT® comprehensive database of immunoglobulin and T cell receptor nucleotide sequences. Nucleic Acids Res. 34, D781–D784 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Ma N., Madden T. L. & Ostell J. M. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 41, W34–W40 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N. T. et al. Change-O: A toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics 31, 3356–3358 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007). [DOI] [PubMed] [Google Scholar]
- McKinney W. Data Structures for Statistical Computing in Python. In Proceedings of the 9th Python in Science Conference (eds. van der Walt S. & Millman J.) 51–56 (2010). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.