ABSTRACT
Bacteria predominantly exist as members of surfaced-attached communities known as biofilms. Many bacterial species initiate biofilms and adhere to each other using cell surface adhesins. This is the case for numerous ecologically diverse Alphaprotebacteria, which use polar exopolysaccharide adhesins for cell-cell adhesion and surface attachment. Here, we show that Rhodopseudomonas palustris, a metabolically versatile member of the alphaproteobacterial order Rhizobiales, contains a functional unipolar polysaccharide (UPP) biosynthesis gene cluster. Deletion of genes predicted to be critical for UPP biosynthesis and export abolished UPP production. We also found that R. palustris uses UPP to mediate biofilm formation across diverse photoheterotrophic growth conditions, wherein light and organic substrates are used to support growth. However, UPP was less important for biofilm formation during photoautotrophy, where light and CO2 support growth, and during aerobic respiration with organic compounds. Expanding our analysis beyond R. palustris, we examined the phylogenetic distribution and genomic organization of UPP gene clusters among Rhizobiales species that inhabit diverse niches. Our analysis suggests that UPP is a conserved ancestral trait of the Rhizobiales but that it has been independently lost multiple times during the evolution of this clade, twice coinciding with adaptation to intracellular lifestyles within animal hosts.
IMPORTANCE Bacteria are ubiquitously found as surface-attached communities and cellular aggregates in nature. Here, we address how bacterial adhesion is coordinated in response to diverse environments using two complementary approaches. First, we examined how Rhodopseudomonas palustris, one of the most metabolically versatile organisms ever described, varies its adhesion to surfaces in response to different environmental conditions. We identified critical genes for the production of a unipolar polysaccharide (UPP) and showed that UPP is important for adhesion when light and organic substrates are used for growth. Looking beyond R. palustris, we performed the most comprehensive survey to date on the conservation of UPP biosynthesis genes among a group of closely related bacteria that occupy diverse niches. Our findings suggest that UPP is important for free-living and plant-associated lifestyles but dispensable for animal pathogens. Additionally, we propose guidelines for classifying the adhesins produced by various Alphaprotebacteria, facilitating future functional and comparative studies.
KEYWORDS: Rhodopseudomonas, adhesin, biofilm, holdfast, phylogenetic analysis, unipolar polysaccharide
INTRODUCTION
Diverse bacteria produce cell surface adhesins that facilitate attachment to biotic and abiotic surfaces (1, 2). Some of the earliest observations of bacterial adhesion reported bacterial “stars,” later termed rosettes, in which cells aggregate by attaching to each other at a single pole (3). Similarly, initial observations of bacterial adhesion to abiotic surfaces also noted polar attachment (4). It has since been recognized that the same polar adhesins responsible for rosette formation in many alphaproteobacterial species also mediate irreversible attachment to surfaces and thereby act to initiate the formation of surface-associated communities known as biofilms (1, 5, 6).
Polar surface attachment in Alphaprotebacteria has been most well studied in the freshwater bacterium Caulobacter crescentus (5, 7–10) and more recently in the plant pathogen Agrobacterium tumefaciens (11–13). The polar adhesin of C. crescentus and other members of the order Caulobacterales is called holdfast (1, 14). The polar adhesin of A. tumefaciens has been termed unipolar polysaccharide (UPP) (11). These two unipolar adhesins are distinct but share certain genetic, biochemical, and functional characteristics (1, 11). The synthesis of both adhesins involves a Wzy-dependent polysaccharide synthesis and export pathway. For holdfast, this pathway is encoded by the holdfast synthesis (hfs) gene cluster (8, 15). For UPP, the pathway is partially encoded by the core upp biosynthesis gene cluster, with other components encoded separately in the genome (11). The hfsEFGHCBAD and uppABCDEF gene clusters each have distinct organization and content (i.e., synteny) (Fig. S1). Only hfsD and hfsE have close sequence similarity to uppC and uppE, respectively (Table S1), although other genes likely encode functionally analogous proteins between these two gene clusters. A contrasting feature of these two adhesins is that holdfast-mediated adhesion requires proteins encoded by the holdfast anchor (hfa) operon, which keeps holdfast attached to the cell (16). No apparent homologs of hfa genes are encoded by A. tumefaciens (11) or most other Rhizobiales species (Data Set S1). Holdfast and UPP also exhibit some biochemical similarity, as both contain N-acetylglucosamine (7, 11), allowing the adhesins to be visualized by fluorescence microscopy after staining with the fluorophore-conjugated wheat germ agglutinin (5, 17).
Beyond C. crescentus and A. tumefaciens, polar polysaccharide adhesins are also a common morphological trait across ecologically diverse Alphaprotebacteria (1, 14, 18), especially among Rhizobiales species (19–25). However, the genetic and biochemical diversity of the adhesins across this clade is unclear. Furthermore, the potential environment-specific production and/or function of these adhesins remain largely unexplored. Here, we examine polar adhesin production by the Rhizobiales member Rhodopseudomonas palustris. This purple nonsulfur bacterium was first reported to produce a polar adhesin almost 50 years ago (26), but the genes involved in its biosynthesis were never characterized. Additionally, R. palustris is renowned for its metabolic versatility (27), a feature that allowed us to investigate if adhesin production is coordinated by different metabolic modules. We show that the putative R. palustris uppE (RPA2750) and uppC (RPA4833) orthologs are required for synthesis of a UPP adhesin. UPP is differentially required for R. palustris biofilm formation under various conditions but is particularly influential under photoheterotrophic conditions, in which light energy and organic substrates are used to support growth. Moving beyond R. palustris, we also explored whether UPP is associated with different bacterial lifestyles by performing a comparative genomic analysis across diverse Rhizobiales species. Our results indicate that UPP is a conserved ancestral trait of the Rhizobiales, and that upp genes have been independently lost multiple times during the evolution of the Rhizobiales clade. Based on our analysis, we propose that genetic synteny of adhesion biosynthesis genes is a valid criterion on which to designate the polar adhesins of various Rhizobiales members as UPP.
RESULTS AND DISCUSSION
Genomic organization of the putative R. palustris CGA009 core upp gene cluster.
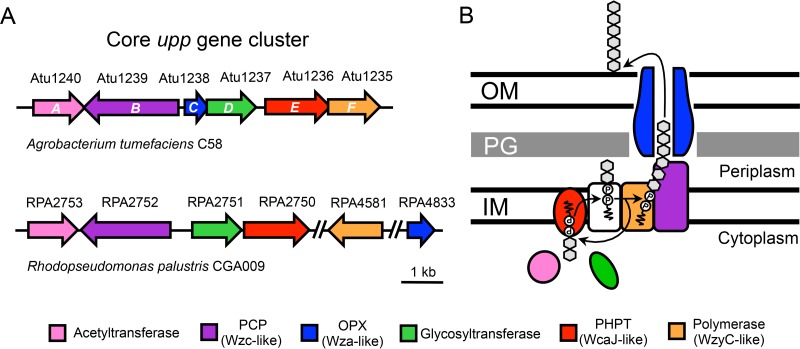
R. palustris has long been known to form rosettes (26, 28); however, the genetic loci responsible for polar adhesin biosynthesis remain uncharacterized. Recently, bioinformatic analysis revealed that R. palustris contains a putative upp gene cluster (23). Such clusters have been shown to function in UPP production in other Rhizobiales species (12, 24). We confirmed that R. palustris CGA009 encodes a putative upp gene cluster using a TBLASTN reciprocal best-hit approach with the A. tumefaciens C58 UppABCDEF proteins as query sequences. We identified four adjacent genes in R. palustris with close identity to A. tumefaciens uppABDE (Fig. 1A and Table S2). The candidate orthologs for both uppC (RPA4833) and uppF (RPA4581) are outside the putative R. palustris uppABDE cluster (RPA2753 to RPA2750) (Fig. 1A). As expected based on species relatedness, the synteny of the putative R. palustris upp gene cluster is more similar to that of the A. tumefaciens upp gene cluster than to the C. crescentus hfs gene cluster (Fig. S1). We did not identify any candidate hfa homologs in R. palustris (Data Set S1), which are required for holdfast anchoring in C. crescentus and which are similarly absent in A. tumefaciens (11).
FIG 1.
Synteny of A. tumefaciens C58 and R. palustris CGA009 core upp gene clusters and proposed protein functions. (A) Genes (arrows) are colored based on functional prediction and sequence similarity (>50% query cover, >25% identity, >40% positives, and an E value <1 × 10−20). Double slashes represent large (>100 kb) genomic regions not shown in the figure. (B) Model of the proposed Wzy-dependent synthesis and export pathway for UPP based on references 15, 29, and 30). Rhizobiales core upp gene clusters lack an important Wzx-like flippase (white), which is contained elsewhere in the genome. Gray hexagons represent repeat saccharide units of the UPP. IM, inner membrane; PG, peptidoglycan; OM, outer membrane; PCP, polysaccharide copolymerase; OPX, outer membrane polysaccharide export; PHPT, polyisoprenyl-phosphate hexose-1-phosphate transferase.
The putative R. palustris uppABDE, uppC, and uppF genes are predicted to encode a partial Wzy-dependent polysaccharide export pathway (Fig. 1B). Wzy-dependent pathways are broadly distributed across Gram-negative bacteria (29) and have been most well characterized in lipopolysaccharide and capsular polysaccharide biosynthesis and export in E. coli (30). We propose a Wzy-dependent model for UPP synthesis and export based on the current understanding of Wzy-dependent pathways (Fig. 1B), similar to what has been proposed for Wzy-dependent holdfast production (15). Briefly, an iterative multienzyme process assembles repeat saccharide units on the inner membrane (IM)-associated lipid carrier undecaprenyl phosphate (und-PP). The assembly is then translocated across the IM and into the periplasm, where the repeat saccharide units are transferred from und-PP to add to the growing polysaccharide chain on another und-PP carrier. Ultimately, the polysaccharide chain is exported onto the cell surface (Fig. 1B). It should be noted that for UPP, certain enzymes thought to be required for synthesis are encoded outside the core upp cluster, such as a flippase (Fig. 1B, white), which is responsible for translocation across the IM. This genetic arrangement is distinct from that of C. crescentus and most other Caulobacterales species, which encode putative Wzx-like flippases (HfsF) in their hfs gene clusters (Fig. S1) (15, 17, 31).
Visualization of R. palustris unipolar adhesin.
To facilitate genetic and phenotypic characterization of the R. palustris adhesin, we first tested if we could visualize adhesin on wild-type (WT) R. palustris cells using the fluorophore-conjugated lectin WGA-488. Adhesins produced by diverse Alphaprotebacteria have been shown to bind WGA (5, 7, 32), which itself binds N-acetylglucosamine residues. When we stained R. palustris with WGA-488, we observed fluorescence at single poles of some individual cells and at the center of every rosette (Fig. 2A). WGA-488 can potentially stain N-acetylglucosamine present in peptidoglycan if the outer membrane is compromised, but we seldom observed nonpolar cell body staining under standard photoheterotrophic conditions. From this, we conclude that we are indeed staining the unipolar adhesin produced by R. palustris, which contains N-acetylglucosamine, similar to the UPP of other Rhizobiales species (11, 23, 24), as well as Caulobacterales holdfast (7, 17).
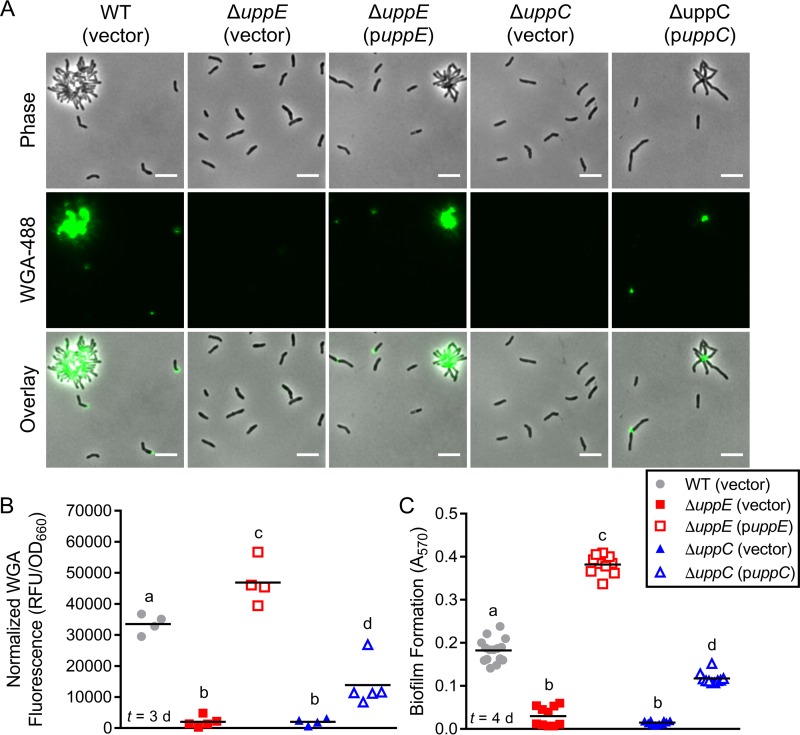
FIG 2.
uppE and uppC are required for UPP biosynthesis, cell-cell adhesion, and biofilm formation. (A) Epifluorescence microscopy of cells stained with WGA-488 after 2 days of growth under standard photoheterotrophic conditions. Scale bars, 5 μm. (B) Normalized total WGA-488 fluorescence from batch UPP quantification following 3 days of growth under standard photoheterotrophic conditions. Different letters indicate significant differences between strains (P < 0.05; one-way analysis of variance [ANOVA] followed by Tukey's multiple-comparison test; n = 4 to 5). (C) Biofilm formation levels (A570) were quantified by CV staining of adherent biomass following 4 days of growth in microtiter wells under standard photoheterotrophic conditions. All strains grew equivalently, so A570 values were not normalized. Different letters indicate significant differences between strains (P < 0.0001; one-way ANOVA followed by Tukey's multiple-comparison test; n = 10 or 15, pooled from three independent experiments). (B and C) Symbols indicate biological replicates and lines indicate the means. Time (t) of sampling following inoculation is indicated in lower left corner.
UppE and UppC are required for R. palustris UPP biosynthesis, cell-cell adhesion, and biofilm formation.
We next addressed the genetic requirements underlying polar adhesin production in R. palustris. In A. tumefaciens, uppE (12, 13) and uppC (C. Fuqua, personal communication) are essential for UPP biosynthesis. Similarly, the uppE ortholog (gmsA) of the root-nodulating symbiont Rhizobium leguminosarum is necessary for root hair attachment (20). In C. crescentus, the putative uppC homolog hfsD is required for holdfast-mediated attachment (8). Thus, we chose the putative uppE and uppC orthologs of R. palustris as targets for in-frame deletions to determine whether they are required for adhesin synthesis.
Deletion of either the putative uppE or uppC ortholog eliminated both rosette formation and WGA-488 binding (Fig. 2A). Complementation of each mutant from a plasmid restored rosette formation as well as unipolar WGA-488 binding to single cells and at the center of rosettes (Fig. 2A). In addition to microscopic visualization of the adhesin on cells, we also developed an assay to quantify adhesin production at the population level by measuring total WGA-488 fluorescence in batch culture samples. Similar to trends observed by microscopy, total WGA-488 fluorescence was significantly lower in the putative ΔuppE or ΔuppC mutant cultures than in the WT and complemented cultures (Fig. 2B). Overall, these results demonstrate an essential role for both of these orthologs in adhesin production in R. palustris. Based on these results, we henceforth refer to these genes as uppE and uppC and to the R. palustris unipolar adhesin as UPP.
Having established that uppE and uppC are critical for R. palustris UPP synthesis and rosette formation, we next assessed if R. palustris UPP contributes to biofilm formation. After 4 days of standard photoheterotrophic growth, the ΔuppE and ΔuppC mutants showed significantly less biofilm formation than the WT and complemented strains (Fig. 2C). Thus, we conclude that UPP is the primary adhesin facilitating biofilm formation under standard photoheterotrophic conditions.
Survey of UPP-mediated biofilm formation across environmental conditions.
R. palustris is metabolically versatile, allowing it to adopt distinct lifestyles to thrive under diverse conditions. When growing anaerobically in light, R. palustris performs anoxygenic photosynthesis to transform energy (27). During phototrophic growth, R. palustris can obtain carbon by consuming organic substrates (photoheterotrophy) or by fixing CO2 (photoautotrophy) (27). It can also grow by aerobic respiration in the dark (chemoheterotrophy). Additionally, R. palustris is a diazotroph, meaning it can grow with N2 gas as the sole nitrogen source by the process of N2 fixation (33). While R. palustris has almost exclusively been studied under freshwater conditions, it was recently noted that an environmental isolate could grow in salt concentrations of up to 4.5% (34).
The metabolic versatility of R. palustris provided an opportunity to assess whether UPP-mediated surface attachment and biofilm formation are favored by some growth conditions over others. To address this, we examined UPP production and biofilm formation under various growth conditions for both WT R. palustris and the ΔuppE mutant. We proceeded with only the ΔuppE mutant because we did not observe any phenotypic differences between the ΔuppE and ΔuppC mutants (Fig. 2). We chose growth conditions that encompass both the metabolic capabilities of R. palustris (e.g., N2 fixation and photoautotrophy) and abiotic conditions it might normally encounter (e.g., low inorganic phosphate [Pi] and high salinity). Total WGA-488 fluorescence values were not compared across conditions, as they were not always reflective of UPP synthesis. For example, some growth conditions, such as low Pi, resulted in occasional staining at both poles and at what appeared to be cell division septa, suggesting that WGA-488 was staining N-acetylglucosamine moieties in peptidoglycan (Fig. S2).
UPP-assisted biofilm formation is favored by R. palustris in adverse photoheterotrophic environments.
We first examined if biofilm formation was stimulated or inhibited in response to three adverse photoheterotrophic conditions. These conditions are considered to be less favorable for R. palustris growth due to nutrient limitation (low Pi), less-preferred nutrients (N2 fixation), or osmotic stress (high salinity). Thus, we used these conditions to assess whether biofilm formation might function to increase R. palustris survival under suboptimal conditions or to foster persistence in favorable environments (2, 35). We also examined if UPP is utilized by R. palustris across these growth conditions. Two main trends were observed under all three adverse conditions. First, WT R. palustris formed more biofilm under all adverse conditions than standard photoheterotrophic conditions (Fig. 3A), even though standard conditions supported the highest growth rates and highest cell densities (data not shown). Second, UPP contributed to biofilm formation under all photoheterotrophic conditions, as the WT formed more biofilm than the ΔuppE mutant in each case (Fig. 3A). These biofilm trends were consistent with microscopy results, which showed that WT R. palustris exhibited comparable WGA staining patterns under standard and adverse photoheterotrophic conditions (Fig. 3B). Beyond this, there were also condition-specific phenotypes observed.
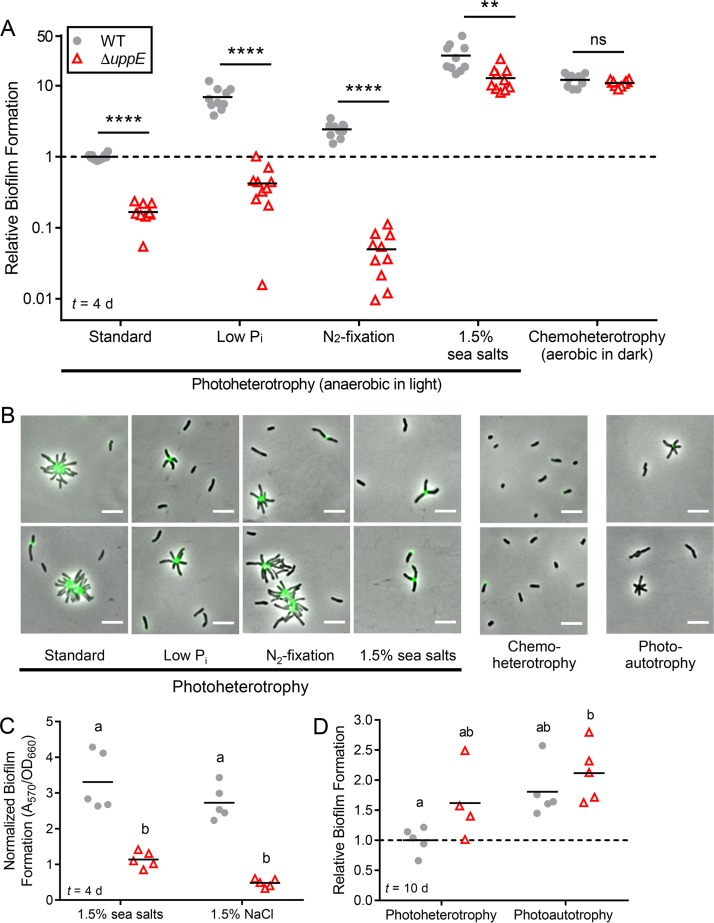
FIG 3.
UPP is important for biofilm formation across photoheterotrophic conditions. (A) Biofilm formation levels were normalized to final planktonic cell density (A570/OD660) and then made relative to normalized WT standard photoheterotrophic values, which was set to 1. **, P < 0.01; ****, P < 0.0001; ns, not significant; based on multiple unpaired, two-tailed t tests without assuming equal variance and followed by Holm-Šídák correction for multiple comparisons; n = 10, pooled from two independent experiments. Significance is only indicated for pairwise comparisons between WT and the ΔuppE mutant under each condition because the assumption of homogeneity of variances was violated in comparisons across conditions. Results from other statistical analyses comparing across conditions are listed in Table S3. (B) Epifluorescence microscopy of cells stained with WGA-488 after 3 days of photoheterotrophic or chemoheterotrophic growth and after 8 days of photoautotrophic growth. Scale bars, 5 μm. (C) Biofilm formation normalized to final planktonic cell density (A570/OD660) following 4 days of photoheterotrophic growth with 1.5% sea salts or 1.5% NaCl. Different letters indicate significant differences between groups (P < 0.05; two-way ANOVA followed by Tukey's multiple-comparison test; n = 5). (D) Relative biofilm formation (A570/OD660) after 10 days of photoheterotrophic or photoautotrophic growth, with WT values under standard photoheterotrophic conditions set to 1. Different letters indicate statistically significant differences between groups (P < 0.05; two-way ANOVA followed by Tukey's multiple-comparison test; n = 4 to 5). (A, C, and D) Symbols indicate biological replicates, and lines indicate the means. Time (t) of sampling following inoculation is indicated in lower left corner.
Under low-Pi conditions, the ΔuppE mutant formed loosely attached lawns at the bottom of microtiter wells. These lawns were easily disrupted and washed away. Such lawns were not formed by the ΔuppE mutant under standard conditions and were unlike all WT photoheterotrophic biofilms, which were firmly attached to the sides and bottom of the wells. The genetic and biochemical basis for these loose biofilms remains to be determined. Stimulation of biofilm formation in response to Pi limitation has also been observed in A. tumefaciens (12, 36). This common observation raises the possibility that increased biofilm formation is a conserved response to Pi limitation across some Rhizobiales species. It has been speculated that low Pi serves as a signal to A. tumefaciens that plant surfaces are nearby, as plants sequester Pi, locally depleting it from the rhizosphere (36). Given that no symbiotic association between R. palustris and plants has been identified, it is possible that biofilm formation serves a different function in this species, such as increasing survival when essential nutrients, such as Pi, are limiting.
We also observed 2-fold higher biofilm levels by WT under N2-fixing conditions than under standard conditions (Fig. 3A). N2 fixation is energetically expensive compared to using other nitrogen sources, such as NH4+, and is therefore tightly regulated (33, 37). We hypothesize that increased aggregation under N2-fixing conditions might function to help retain costly NH4+, which can passively diffuse out of cells as NH3 (38).
In contrast to all other photoheterotrophic conditions, ΔuppE mutant biofilm levels were 13-fold higher under 1.5% sea salt conditions than WT cells under standard conditions, despite lacking UPP (Fig. 3A). Similar trends were seen with 1.5% NaCl, confirming that the enhanced biofilm formation of both the WT and the ΔuppE mutant was due to high salinity and not another component of the sea salt supplement (Fig. 3C). The high ΔuppE mutant biofilm levels under high-salinity conditions suggest that additional factors besides UPP contribute to this response. Thus, while UPP-mediated surface attachment contributes to robust biofilm formation by R. palustris during photoheterotrophic growth, UPP is less crucial under high-salinity conditions.
UPP-independent biofilm formation is stimulated by nonphotoheterotrophic conditions.
We also examined UPP production and biofilm formation under chemoheterotrophic and photoautotrophic conditions. Under chemoheterotrophic conditions, UPP was not necessary for biofilm formation, as WT and the ΔuppE mutant formed similar levels of biofilm. We were surprised by this result, as it suggested that biofilm formation was entirely UPP independent. Aerobically grown bacteria typically adhere near the air-liquid interface (39). However, the adherent biomass of both the WT and the ΔuppE aerobic biofilms was at the bottom of the microtiter well, suggesting that R. palustris might preferentially form biofilms at microaerobic or anaerobic zones. In support of this, the adherent biomass was pigmented, indicating the production of bacteriochlorophyll and carotenoids, which is stimulated in response to low O2 (40). Additionally, chemoheterotrophic conditions seem to favor biofilm formation, as WT and ΔuppE biofilm levels were approximately 12-fold higher than those of the WT under standard photoheterotrophic conditions. (Fig. 3A). Separately, although WGA-488 staining was observed on some single cells, we did not observe any rosettes under chemoheterotrophic conditions (Fig. 3B). It is therefore possible that UPP is produced but is dispensable for chemoheterotrophic biofilm formation.
During photoautotrophy with sodium bicarbonate as the carbon source and thiosulfate as an electron donor, R. palustris has a specific growth rate approximately one-fourth that of during photoheterotrophic growth (41, 42). Because of the slower growth, we extended photoautotrophic incubations from 3 days for epifluorescence microscopy and 4 days for biofilm assay to 8 days and 10 days, respectively, in order to allow cultures to reach final densities similar to those observed after 3 to 4 days of photoheterotrophic growth. After 8 days of photoautotrophic growth, we observed WT rosettes that stained very little or not at all with WGA-488, suggesting that less UPP is produced or that UPP composition is different under these conditions (Fig. 3B). Under photoautotrophic conditions, the WT and the ΔuppE mutant showed similar levels of biofilm formation (Fig. 3D), suggesting that biofilm formation was UPP independent. Similar trends were seen in parallel control cultures for which we allowed for 10 days of photoheterotrophic growth (Fig. 3D), unlike results from the 4-day photoheterotrophic experiments, where the ΔuppE mutant formed less biofilm than WT (Fig. 3A). There are multiple nonmutually exclusive explanations for why the difference in biofilm formation between the WT and the ΔuppE mutant after 4 days was not also observed after 10 days. Because UPP is thought to mediate the initial irreversible surface attachment of cells (5), prolonging the incubation period may have led to some degradation of UPP and/or might have allowed sufficient time for as-of-yet unknown adhesins or other factors, such as DNA release following cell lysis, to facilitate attachment. Such factors could also contribute to the increased biofilm formation observed across the different conditions tested herein.
Overall, our survey of R. palustris biofilm formation across growth conditions can be summarized as follows. UPP mediates biofilm formation under photoheterotrophic conditions, especially those photoheterotrophic conditions that are less favorable to growth. Under certain photoheterotrophic conditions, such as high salinity, biofilm formation involves additional factors that are independent of UPP. Finally, chemoheterotrophic and photoautotrophic conditions also stimulate biofilm formation but in a manner that appears to be independent of UPP.
Conservation of core upp biosynthesis genes across Rhizobiales species.
Beyond C. crescentus, R. leguminosarum, A. tumefaciens, and now R. palustris, the characterization of polar adhesins in other Alphaprotebacteria has been cursory. Historically, all polar adhesins were referred to as holdfast. However, designation of alphaproteobacterial adhesins has been complicated by functional differences. For example, the polar glucomannan adhesin of R. leguminosarum plays a unique role in root hair attachment but is not required for attachment to abiotic surfaces (19, 20). The R. leguminosarum glucomannan biosynthesis gene cluster is orthologous to the A. tumefaciens uppABCDEF cluster, which A. tumefaciens uses to attach to both biotic and abiotic surfaces (5, 11, 12). Thus, R. leguminosarum polar glucomannan and A. tumefaciens UPP are homologous adhesins with functional differences. Also contributing to the ambiguity in classifying previously identified Rhizobiales polar adhesins is the compositional diversity (1, 12, 20–22). For example, A. tumefaciens UPP contains N-acetylgalactosamine in addition to N-acetylglucosamine (12), the R. leguminosarum glucomannan adhesin contains primarily glucose and mannose (19), the Bradyrhizobium japonicum polar adhesin contains galactose and lactose (22), and the Hyphomicrobium polar adhesin likely contains galactose and mannose (21). We therefore propose that alphaproteobacterial adhesins be classified according to genetic synteny. Based on the synteny (Fig. 1) and functional requirement of upp orthologs for adhesin production (Fig. 2), we conclude that R. palustris produces UPP.
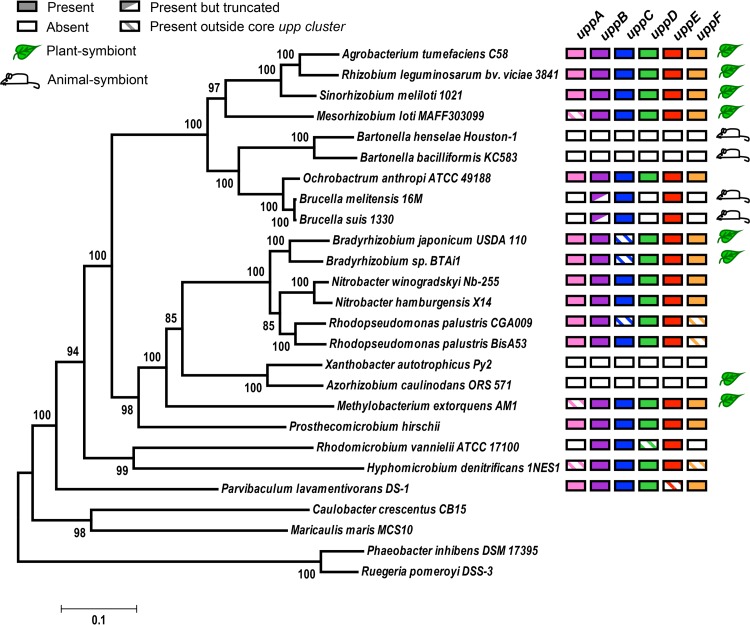
With the criterion of genetic synteny in mind, we explored the phylogenetic distribution and genomic organization of the core uppABCDEF orthologs across 22 Rhizobiales species, representing the lifestyle diversity of this clade (Fig. 4). The topology of this tree is largely consistent with the alphaproteobacterial phylogeny inferred from a concatenation of 104 protein alignments (43). Our analysis revealed broad conservation of putative upp gene clusters, indicating that UPP is an ancestral trait of the Rhizobiales clade. Almost all of the Rhizobiales plant symbionts, including the plant pathogen A. tumefaciens, the root-nodulating diazotrophs R. leguminosarum, S. meliloti, Mesorhizobium loti, and B. japonicum, the stem-nodulating photosynthetic diazotroph Bradyrhizobium sp. strain BTAi, and the leaf epiphyte Methylobacterium extorquens, contain complete or near-complete upp gene clusters (Fig. 4). The exception to this trend is the root-nodulating diazotroph Azorhizobium caulinodans (44), which does not contain a upp cluster (Fig. 4 and Data Set S2). We were also unable to identify a upp cluster in Xanthobacter autotrophicus, a free-living diazotroph closely related to A. caulinodans (Fig. 4). This absence suggests that the upp cluster was lost before these lineages split. Despite the absence of a upp cluster in A. caulinodans, it still appears to produce a polar adhesin and form rosettes (25). Upon closer examination of the A. caulinodans ORS571 genome, we identified a putative Wzy-like polysaccharide biosynthesis gene cluster with high similarity to the Vibrio fischeri symbiosis polysaccharide (syp) locus (Data Set S3) (45). These putative syp homologs seem to have been acquired horizontally and might have been coopted for polar polysaccharide synthesis in A. caulinodans.
FIG 4.
Conservation of core UPP biosynthesis genes among Rhizobiales species. A maximum likelihood phylogeny was inferred based on a concatenated alignment of 6 conserved housekeeping proteins using an LG+G+I substitution model (58) with four discrete gamma categories and invariable sites in MEGA6 (56). The tree with the highest log likelihood is shown. Node values indicate branch support from 100 bootstrap replicates. Scale bar represents the number of substitutions per site along branches. Leaf and mouse symbols indicate known plant and animal symbiotic relationships, respectively.
While UPP is well conserved in plant-associating Rhizobiales species, the opposite is true for animal pathogens. This trend was first noted upon the initial discovery of the upp gene cluster in R. leguminosarum, which noted that this cluster is absent in the Rhizobiales intracellular mammalian pathogen Brucella melitensis (20). Rather than being entirely absent (20), our data corroborate more recent bioinformatic evidence (23) that Brucella spp. contain a cluster of 3 putative upp orthologs (uppBCE) (Fig. 4 and S1, and Data Set S2). It is not known whether this partial upp cluster is involved in the synthesis of a functional UPP. In the closely related intracellular animal pathogens of the genera Bartonella, upp orthologs are completely absent (Fig. 4 and Data Set S2). In contrast, the soil-dwelling opportunistic human pathogen Ochrobactrum anthropi (46), which is more closely related to Brucella than Bartonella, contains a complete uppABCDEF gene cluster (Fig. 4). Ochrobactrum spp. are thought to be rhizosphere community members but are capable of infecting animal hosts (46, 47). We hypothesize that the entire upp cluster was first lost in the Bartonella lineage during adaptation to an intracellular lifestyle after diverging from Brucella/Ochrobactrum. More recently, the Brucella lineage has similarly lost multiple upp orthologs during its transition to becoming intracellular pathogens. The independent loss of upp orthologs in both Bartonella and Brucella would suggest convergent evolution upon adaptation to intracellular niches within animal hosts, supporting the hypothesis that UPP is not important for such lifestyles. Conversely, the conservation of upp orthologs in plant symbionts and free-living species suggests that UPP is beneficial in other diverse environments. Considering this, we hypothesize that Ochrobactrum anthropi has retained the complete upp cluster because it is typically free-living in the soil and thus benefits from producing UPP.
Unipolar adhesins are also used by Alphaprotebacteria outside the Rhizobiales. In the order Caulobacterales, hfs and hfa gene clusters for holdfast synthesis are well conserved (17, 31). Despite the differences in synteny between the upp and hfs gene clusters (Fig. S1), both encode Wzy-dependent pathways for polar polysaccharide synthesis and export, and uppC and uppE show sequence similarity to hfsD and hfsE, respectively (Table S1). Because of these similarities, we hypothesize that holdfast and UPP share an evolutionary origin and that the upp and hfs loci diversified in genomic organization following divergence of the Rhizobiales and Caulobacterales clades.
Other alphaproteobacterial species of the marine Roseobacter clade within the order Rhodobacterales also produce polar adhesins and form rosettes but do not contain either upp or hfs or hfa homologs (18) (Data Sets S1 and S2). The polar polysaccharide adhesin of the Roseobacter species Phaeobacter inhibens contains N-acetylglucosamine based on WGA binding, indicating that the biochemical composition is at least somewhat similar to that of UPP and holdfast (32). In this case, polar adhesion synthesis is encoded on a plasmid, since plasmid curing prevented P. inhibens rosette formation and diminished attachment to abiotic surfaces and algal cells (6). Furthermore, genetic disruption of the plasmid-encoded putative rhamnose operon lowered biofilm formation (48). Plasmids encoding putative rhamnose operons are widely distributed among other Roseobacter species (48), suggesting that polar polysaccharide synthesis and export in this clade are genetically distinct from those of UPP and holdfast. It is not clear whether acquisition of these plasmids led to the loss of gene clusters similar to either upp or hfs loci.
While polar polysaccharide adhesins are a common morphological trait across ecologically diverse Alphaprotebacteria, there is considerable genetic, compositional, and functional variation, which likely reflects adaptation to different niches. We propose here that genetic synteny of biosynthetic loci is a suitable criterion on which to base classification of polar adhesins. This criterion bypasses uncertainty arising from compositional differences while highlighting the shared underlying biosynthetic pathway. As such, holdfast and UPP are distinct adhesins despite facile similarities. Likewise, the A. caulinodans adhesin and the Roseobacter rhamnose adhesins should each receive their own designation, as they are genetically distinct from both holdfast and UPP, as well as from each other. Adoption of a unified classification scheme will facilitate both the comparison of adhesins and the exploration of functional differences within and between adhesin types.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All R. palustris strains were derived from CGA009 (27) and are listed in Table 1. Unless otherwise indicated, R. palustris was grown statically in 10 ml of defined photosynthetic medium (PM) (49) in sealed 16-ml anaerobic tubes with argon gas in the headspace. All R. palustris cultures were incubated at 30°C. All phototrophic cultures were illuminated with a 60-W light bulb. For all heterotrophic conditions, PM was supplemented with succinate as the sole carbon source (15 mM in liquid cultures or 10 mM in agar). Incubation in PM with 15 mM succinate and light are here referred to as standard photoheterotrophic conditions. For low-inorganic phosphate (Pi) conditions, PM was modified by replacing Na2HPO4 and KH2PO4 (12.5 mM each) with equimolar concentrations of Na2SO4 and K2SO4. A 1:1 molar mixture of Na2HPO4 and KH2PO4 was added for a final Pi concentration of 30 μM. For N2-fixing conditions, (NH4)2SO4 was omitted from PM, and argon was replaced with N2. For high-salinity conditions, PM was supplemented with 1.5% (wt/vol) sea salts (Sigma) or NaCl. For chemoheterotrophic conditions, cultures were grown in 10 ml of aerobic PM supplemented with 0.05% yeast extract in addition to 15 mM succinate in 50-ml Erlenmeyer flasks shaken at 225 rpm in darkness. For photoautotrophic conditions, anaerobic PM was supplemented with 60 mM NaHCO3 as the inorganic carbon source and 30 mM Na2S2O3 as an inorganic electron donor. Plasmid-harboring R. palustris strains were grown with 50 μg/ml gentamicin in liquid culture and 100 μg/ml gentamicin on agar plates. The Escherichia coli strains used for cloning (DH5-α and S17-1) were grown aerobically in Luria-Bertani medium (BD) supplemented with 15 μg/ml gentamicin when necessary.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and/or descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| R. palustris | ||
| CGA009 | Wild-type strain | 27 |
| CGA4000 | CGA009 derivative; ΔuppE (ΔRPA2750) mutant | This study |
| CGA4022 | CGA009 derivative; ΔuppC (ΔRPA4833) mutant | This study |
| E. coli | ||
| S17-1 | thi pro hsdR hsdM+ recA; chromosomal insertion of RP4-2 (Tc::Mu Km::Tn7) | 51 |
| DH5-α | F− λ− recA1 Δ(lacZYA-argF)U169 hsdR17 thi-1 gyrA96 supE44 endA1 relA1 φ80lacZΔM15 | Thermo Fisher Scientific |
| Plasmids | ||
| pJQ200KS | Gmr; sacB; R. palustris suicide vector | 50 |
| pJQ-RPA2750 | Gmr; sacB; derived from pJQ200KS; deletion vector for uppE (RPA2750) | 59 |
| pJQ-RPA4833 | Gmr; sacB; derived from pJQ200KS; deletion vector for uppC (RPA4833) | This study |
| pGEM | High-copy-no. cloning vector for insertion of PCR products | Promega |
| pBBPgdh | Gmr; broad-host-range cloning vector with constitutive R. palustris gapdh promoter | 53 |
| pBBP-RPA2750 | Gmr; derived from pBBPgdh; complementation vector for ΔuppE (ΔRPA2750) | This study |
| pBBP-RPA4833 | Gmr; derived from pBBPgdh; complementation vector for ΔuppC (ΔRPA4833) | This study |
Gmr, gentamicin resistant.
R. palustris strain construction.
All plasmids and primers are listed in Tables 1 and 2, respectively. Deletion vectors for uppC (RPA4833) and uppE (RPA2750) were generated by PCR amplification of the genomic regions flanking the gene to be deleted, as described previously (41). PCR product pairs were fused by overlap extension PCR and cloned into pJQ200SK (50). Vectors were introduced into R. palustris by conjugation with E. coli S17-1 (51) or by electroporation (52). Complementation vectors for uppC and uppE were generated by PCR amplification of each gene along with the putative ribosomal binding site. PCR products were cloned into pBBPgdh (53), and complementation and empty pBBPgdh vectors were introduced into R. palustris by conjugation with E. coli S17-1.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′→3′)a | Description (restriction site) |
|---|---|---|
| UuppE-XbaI | CGCGGTGGCGGCCGCTCTAGAAAGCATCACGGATCTGTTCGTCTG | uppE (RPA2750) upstream flanking region (XbaI) |
| UuppE-delR | GCGAACGCCTCAGTAGGTACCGCTGATCGGCTCCATCTGTTCATG | uppE (RPA2750) upstream in-frame deletion reverse |
| DuppE-delF | ATGGAGCCGATCAGCGGTACCTACTGAGGCGTTCGCTCTTCAACA | uppE (RPA2750) downstream in-frame deletion forward |
| DuppE-BamHI | TTCCTGCAGCCCGGGGGATCCAGAGCAACAACAACCAAAGGGAGC | uppE (RPA2750) downstream flanking region (BamHI) |
| uppE-compF-BamHI | CTGATCTAGAAGCACGGTGGATATGGATTCC | uppE (RPA2750) complementation forward (BamHI) |
| uppE-compR-XbaI | GACTGGATCCCCGGACGACAAAGTCGTG | uppE (RPA2750) complementation reverse (XbaI) |
| UuppC-XbaI | GACTTCTAGAACCCATTTCGTGAGTGGCAACC | uppC (RPA4833) upstream flanking region (XbaI) |
| UuppC-delR | AGAACCAGCGTTCGATGATCATCGATACTTGAAACGCGC | uppC (RPA4833) upstream in-frame deletion reverse |
| DuppC-delF | GATGATCATCGAACGCTGGTTCTGAACCGG | uppC (RPA4833) downstream in-frame deletion forward |
| DuppC-BamHI | GACTTCTAGACGGTTTCGAACTCGGGGGTTAT | uppC (RPA4833) downstream flanking region (BamHI) |
| uppC-compF-BamHI | ACAGCGGGATCCCGTGGCGAGGGATGGC | uppC (RPA4833) complementation forward (BamHI) |
| uppC-compR-XbaI | ACAGCGTCTAGATCAGAACCAGCGTTCGCCGA | uppC (RPA4833) complementation reverse (XbaI) |
Restriction sites are underlined.
Epifluorescence microscopy and image analysis.
Unless stated otherwise, R. palustris cultures used for microscopy were grown in liquid without agitation for 2 to 3 days, except for photoautotrophic cultures, which were grown for 8 days. Culture samples were centrifuged, and the cell pellet was resuspended in 0.1 mM phosphate-buffered saline (PBS) (Fisher Scientific) to an optical density at 660 nm (OD660) between 0.6 and 0.9. Wheat germ agglutinin Alexa Fluor 488 conjugate (WGA-488) (Molecular Probes) was added to cells suspended in PBS at a final concentration of 2 μg/ml and incubated in darkness at room temperature for 15 min. Cells were washed with PBS three times to remove unbound dye and then resuspended in PBS. Cells were imaged on agarose pads using a Nikon Eclipse 90i light microscope equipped with a 100× oil immersion objective and a Photometrics Cascade 1K electron-multiplying charge-coupled-device (EMCCD) camera and processed using the Nikon NIS-Elements software. Images were subsequently analyzed using the ImageJ distribution Fiji (54).
Batch UPP quantification via total WGA fluorescence.
R. palustris cultures were grown under standard photoheterotrophic conditions for 3 days to early stationary phase. A 400-μl sample of each culture was centrifuged, and the cell pellet was resuspended in 400 μl of PBS. A 100-μl aliquot of each cell suspension was set aside for use as the unstained control. WGA-488 was added to the remaining 300 μl of resuspended cells to a final concentration of 1.5 μg/ml and incubated in darkness at room temperature for 15 min. WGA-488-stained cells were washed three times with PBS and then resuspended in 120 μl of PBS to account for cells lost during washes. A 100-μl aliquot of the stained cells and the reserved unstained samples were each transferred to empty wells of a black polystyrene 96-well μClear flat-bottom microtiter plate (Greiner Bio-One). Fluorescence (top-120, excitation 485/20; emission 528/20) and the OD660 were measured using a Synergy H1 microplate reader (BioTek). Fluorescence readings were normalized to cell densities (relative fluorescent units [RFU]/OD660), and background fluorescence was removed by subtracting RFU/OD660 values of unstained samples from the WGA-488-stained samples.
Crystal violet microtiter plate biofilm assay.
Biofilm formation was quantified using a modified version a crystal violet microtiter plate assay (39). Briefly, starter cultures were grown under standard photoheterotrophic conditions supplemented with 0.1% yeast extract. A 1.5-μl volume of stationary-phase culture was used to inoculate the wells of a lidded untreated polystyrene 24-well plate (Corning) containing 1.5 ml of the specified sterile medium. All plates were incubated statically at 30°C. For anaerobic phototrophic growth conditions, plates were incubated in a BD GasPak EZ container with two EZ anaerobe container system sachets (BD) and illuminated by two 60-W light bulbs, one on either side of the container. For chemoheterotrophic growth, plates were incubated aerobically in darkness. For all heterotrophic growth conditions, plates were incubated for 4 days. For photoautotrophic growth conditions (and paired heterotrophic controls), plates were incubated for 10 days. After incubation, plates were shaken at 150 rpm for 3 min on a flat-bed rotary shaker to disrupt loosely attached cells. A 400-μl aliquot of culture was removed for quantifying cell density (OD660) for normalization. Then, 400 μl of 0.1% (wt/vol) crystal violet (CV) was added to each well, and plates were incubated statically at room temperature for 15 min. Wells were then washed 3 times with 2 ml of deionized water to remove unbound CV. Next, 750 μl of 10% (vol/vol) acetic acid was added to each well, followed by shaking at 150 rpm for 3 min to solubilize bound CV. A 150-μl sample of solubilized CV was transferred to a 96-well plate, and absorbance at 570 nm (A570) was measured. Uninoculated wells containing sterile medium were treated the same way as described above to determine background A570, which was subsequently subtracted from all A570 measurements.
Identification of orthologous core upp gene clusters and phylogenetic analysis.
The putative orthologs of the core UPP biosynthesis genes in R. palustris CGA009 (GenBank accession no. BX571963.1) were initially identified by reciprocal best-hit analysis using the UppABCDEF proteins of A. tumefaciens C58 (GenBank accession no. AE007869.2) as the query sequences for a TBLASTN search against the translated nucleotide database of R. palustris CGA009. The best hits in R. palustris CGA009 were subsequently used as query sequences for a BLASTX search against the proteome of A. tumefaciens C58. All putative R. palustris orthologs showed >50% query cover and an E value of <1 × 10−20 (Table S2). Previous studies noted that the core uppABCDEF biosynthesis gene cluster is conserved among several Rhizobiales species (20, 23), which was confirmed by using BLASTP with A. tumefaciens C58 UppABCDEF proteins as query sequences (Data Set S2). Several additional species that contain complete or near-complete upp gene clusters were also identified using BLASTP (minimum threshold for homology of query cover, >50%; E value, <1 × 10−10) (Data Set S2).
For phylogenetic analysis, amino acid sequences for 6 conserved housekeeping proteins, GyrA, GyrB, RpoA, RpoB, FusA, and RecA, from 26 alphaproteobacterial species were individually aligned using MUSCLE (55) with default settings in MEGA6 (56). Gaps and ambiguous sites were removed from alignments using Gblocks (57), with a minimum block length of 10 positions, and gaps were allowed at a position for no more than half of the sequences. The sequences were subsequently concatenated in the order shown above. The final concatenated alignment contained 4,379 amino acid positions (92% of the original positions). Phylogeny was inferred for the concatenated amino acid sequence using the maximum likelihood method based on the Le and Gascuel (LG) model (58), with 4 discrete gamma categories, which allowed for some sites to be evolutionarily invariable, implemented in MEGA6 (56). The LG+G+I model was selected because it was the best-fitting substitution model based on having the lowest Bayesian information criterion score. Initial tree(s) for the heuristic search were obtained by applying the neighbor-joining method to a matrix of pairwise distances estimated using a Jones-Taylor-Thornton model.
Statistical analysis.
All statistical analyses were performed using GraphPad Prism version 6.07. Additional information about statistical analyses is in the figure legends. For Fig. 3A, multiple statistical analyses were performed to reach a consensus for comparisons across growth conditions, which are summarized in Table S3.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yves Brun for use of microscopy facilities and reagents, Clay Fuqua for sharing unpublished data, and members of the Brun and Fuqua labs for thoughtful discussions.
This work was supported in part by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, under award DE-SC0008131, and the Indiana University College of Arts & Sciences.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.03035-16.
REFERENCES
- 1.Berne C, Ducret A, Hardy GG, Brun YV. 2015. Adhesins involved in attachment to abiotic surfaces by Gram-negative bacteria. Microbiol Spectr 3:MB-0018-2015. doi: 10.1128/microbiolspec.MB-0018-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petrova OE, Sauer K. 2012. Sticky situations: key components that control bacterial surface attachment. J Bacteriol 194:2413–2425. doi: 10.1128/JB.00003-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun AC, Elrod RP. 1946. Stages in the life history of Phytomonas tumefaciens. J Bacteriol 52:695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zobell CE. 1943. The effect of solid surfaces upon bacterial activity. J Bacteriol 46:39–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li G, Brown PJB, Tang JX, Xu J, Quardokus EM, Fuqua C, Brun YV. 2012. Surface contact stimulates the just-in-time deployment of bacterial adhesins. Mol Microbiol 83:41–51. doi: 10.1111/j.1365-2958.2011.07909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frank O, Michael V, Päuker O, Boedeker C, Jogler C, Rohde M, Petersen J. 2015. Plasmid curing and the loss of grip–the 65-kb replicon of Phaeobacter inhibens DSM 17395 is required for biofilm formation, motility and the colonization of marine algae. Syst Appl Microbiol 38:120–127. doi: 10.1016/j.syapm.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Merker RI, Smit J. 1988. Characterization of the adhesive holdfast of marine and freshwater caulobacters. Appl Environ Microbiol 54:2078–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith CS, Hinz A, Bodenmiller D, David E, Larson DE, Brun YV. 2003. Identification of genes required for synthesis of the adhesive holdfast in Caulobacter crescentus. J Bacteriol 185:1432–1442. doi: 10.1128/JB.185.4.1432-1442.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bodenmiller D, Toh E, Brun YV. 2004. Development of surface adhesion in Caulobacter crescentus. J Bacteriol 186:1438–1447. doi: 10.1128/JB.186.5.1438-1447.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fiebig A, Herrou J, Fumeaux C, Radhakrishnan SK, Viollier PH, Crosson S. 2014. A cell cycle and nutritional checkpoint controlling bacterial surface adhesion. PLoS Genet 10:e1004101. doi: 10.1371/journal.pgen.1004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomlinson AD, Fuqua C. 2009. Mechanisms and regulation of polar surface attachment in Agrobacterium tumefaciens. Curr Opin Microbiol 12:708–714. doi: 10.1016/j.mib.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu J, Kim J, Danhorn T, Merritt PM, Fuqua C. 2012. Phosphorus limitation increases attachment in Agrobacterium tumefaciens and reveals a conditional functional redundancy in adhesin biosynthesis. Res Microbiol 163:674–684. doi: 10.1016/j.resmic.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu J, Kim J, Koestler BJ, Choi JH, Waters CM, Fuqua C. 2013. Genetic analysis of Agrobacterium tumefaciens unipolar polysaccharide production reveals complex integrated control of the motile-to-sessile switch. Mol Microbiol 89:929–948. doi: 10.1111/mmi.12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poindexter JS. 1964. Biological properties and classification of the Caulobacter group. Bacteriol Rev 28:231–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toh E, Kurtz HD Jr, Brun YV. 2008. Characterization of the Caulobacter crescentus holdfast polysaccharide biosynthesis pathway reveals significant redundancy in the initiating glycosyltransferase and polymerase steps. J Bacteriol 190:7219–7231. doi: 10.1128/JB.01003-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardy GG, Allen RC, Toh E, Long M, Brown PJB, Cole-Tobian JL, Brun YV. 2010. A localized multimeric anchor attaches the Caulobacter holdfast to the cell pole. Mol Microbiol 76:409–427. doi: 10.1111/j.1365-2958.2010.07106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wan Z, Brown PJB, Elliott EN, Brun YV. 2013. The adhesive and cohesive properties of a bacterial polysaccharide adhesin are modulated by a deacetylase. Mol Microbiol 88:486–500. doi: 10.1111/mmi.12199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slightom RN, Buchan A. 2009. Surface colonization by marine roseobacters: integrating genotype and phenotype. Appl Environ Microbiol 75:6027–6037. doi: 10.1128/AEM.01508-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laus MC, Logman TJ, Lamers GE, Van Brussel AAN, Carlson RW, Kijne JW. 2006. A novel polar surface polysaccharide from Rhizobium leguminosarum binds host plant lectin. Mol Microbiol 59:1704–1713. doi: 10.1111/j.1365-2958.2006.05057.x. [DOI] [PubMed] [Google Scholar]
- 20.Williams A, Wilkinson A, Krehenbrink M, Russo DM, Zorreguieta A, Downie JA. 2008. Glucomannan-mediated attachment of Rhizobium leguminosarum to pea root hairs is required for competitive nodule infection. J Bacteriol 190:4706–4715. doi: 10.1128/JB.01694-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moore RL, Marshall KC. 1981. Attachment and rosette formation by hyphomicrobia. Appl Environ Microbiol 42:751–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loh JT, Ho SC, de Feijter AW, Wang JL, Schindler M. 1993. Carbohydrate binding activities of Bradyrhizobium japonicum: unipolar localization of the lectin BJ38 on the bacterial cell surface. Proc Natl Acad Sci U S A 90:3033–3037. doi: 10.1073/pnas.90.7.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams M, Hoffman MD, Daniel JJ, Madren SM, Dhroso A, Korkin D, Givan SA, Jacobson SC, Brown PJB. 2016. Short-stalked Prosthecomicrobium hirschii cells have a Caulobacter-like cell cycle. J Bacteriol 198:1149–1159. doi: 10.1128/JB.00896-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schäper S, Krol E, Skotnicka D, Kaever V, Hilker R, Becker A. 2016. Cyclic di-GMP regulates multiple cellular functions in the symbiotic alphaproteobacterium Sinorhizobium meliloti. J Bacteriol 198:521–535. doi: 10.1128/JB.00795-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu C, Lee K, Wang Y, Peng M, Lee K, Suzuki S, Suzuki T, Oyaizu H. 2011. Involvement of the azorhizobial chromosome partition gene (parA) in the onset of bacteroid differentiation during Sesbania rostrata stem nodule development. Appl Environ Microbiol 77:4371–4382. doi: 10.1128/AEM.02327-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whittenbury R, McLee AG. 1967. Rhodopseudomonas palustris and Rh. viridis–photosynthetic budding bacteria. Arch Mikrobiol 334:324–334. [DOI] [PubMed] [Google Scholar]
- 27.Larimer FW, Chain P, Hauser L, Lamerdin J, Malfatti S, Do L, Land ML, Pelletier DA, Beatty JT, Lang AS, Tabita FR, Gibson JL, Hanson TE, Bobst C, Torres JLTY, Peres C, Harrison FH, Gibson J, Harwood CS. 2004. Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris. Nat Biotechnol 22:55–61. doi: 10.1038/nbt923. [DOI] [PubMed] [Google Scholar]
- 28.Westmacott D, Primrose SB. 1976. Synchronous growth of Rhodopseudomonas palustris from the swarmer phase. J Gen Microbiol 94:117–125. doi: 10.1099/00221287-94-1-117. [DOI] [PubMed] [Google Scholar]
- 29.Cuthbertson L, Mainprize IL, Naismith JH, Whitfield C. 2009. Pivotal roles of the outer membrane polysaccharide export and polysaccharide copolymerase protein families in export of extracellular polysaccharides in Gram-negative bacteria. Microbiol Mol Biol Rev 73:155–177. doi: 10.1128/MMBR.00024-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whitfield C. 2006. Biosynthesis and assembly of capsular polysaccharides. Annu Rev Biochem 75:39–68. doi: 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
- 31.Brown PJB, Hardy GG, Trimble MJ, Brun YV. 2008. Complex regulatory pathways coordinate cell-cycle progression and development in Caulobacter crescentus. Adv Microb Physiol 54:1–101. doi: 10.1016/S0065-2911(08)00001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Segev E, Tellez A, Vlamakis H, Kolter R. 2015. Morphological heterogeneity and attachment of Phaeobacter inhibens. PLoS One 10:e0141300. doi: 10.1371/journal.pone.0141300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oda Y, Samanta SK, Rey FE, Wu L, Liu X, Yan T, Zhou J, Harwood CS. 2005. Functional genomic analysis of three nitrogenase isozymes in the photosynthetic bacterium Rhodopseudomonas palustris. J Bacteriol 187:7784–7794. doi: 10.1128/JB.187.22.7784-7794.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adessi A, Concato M, Sanchini A, Rossi F, de Philippis R. 2016. Hydrogen production under salt stress conditions by a freshwater Rhodopseudomonas palustris strain. Appl Microbiol Biotechnol 100:2917–2926. doi: 10.1007/s00253-016-7291-4. [DOI] [PubMed] [Google Scholar]
- 35.Jefferson KK. 2004. What drives bacteria to produce a biofilm? FEMS Microbiol Lett 236:163–173. doi: 10.1111/j.1574-6968.2004.tb09643.x. [DOI] [PubMed] [Google Scholar]
- 36.Danhorn T, Hentzer M, Givskov M, Parsek MR, Fuqua C. 2004. Phosphorus limitation enhances biofilm formation of the plant pathogen Agrobacterium tumefaciens through the PhoR-PhoB regulatory system. J Bacteriol 186:4492–4501. doi: 10.1128/JB.186.14.4492-4501.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heiniger EK, Oda Y, Samanta SK, Harwood CS. 2012. How posttranslational modification of nitrogenase is circumvented in Rhodopseudomonas palustris strains that produce hydrogen gas constitutively. Appl Environ Microbiol 78:1023–1032. doi: 10.1128/AEM.07254-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim M, Zhang Z, Okano H, Yan D, Groisman A, Hwa T. 2012. Need-based activation of ammonium uptake in Escherichia coli. Mol Syst Biol 8:616. doi: 10.1038/msb.2012.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Toole GA, Pratt LA, Watnick PI, Newman DK, Weaver VB, Kolter R. 1999. Genetic approaches to study biofilms. Methods Enzymol 310:91–109. doi: 10.1016/S0076-6879(99)10008-9. [DOI] [PubMed] [Google Scholar]
- 40.Rey FE, Harwood CS. 2010. FixK, a global regulator of microaerobic growth, controls photosynthesis in Rhodopseudomonas palustris. Mol Microbiol 75:1007–1020. doi: 10.1111/j.1365-2958.2009.07037.x. [DOI] [PubMed] [Google Scholar]
- 41.Rey FE, Oda Y, Harwood CS. 2006. Regulation of uptake hydrogenase and effects of hydrogen utilization on gene expression in Rhodopseudomonas palustris. J Bacteriol 188:6143–6152. doi: 10.1128/JB.00381-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang JJ, Heiniger EK, McKinlay JB, Harwood CS. 2010. Production of hydrogen gas from light and the inorganic electron donor thiosulfate by Rhodopseudomonas palustris. Appl Environ Microbiol 76:7717–7722. doi: 10.1128/AEM.01143-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams KP, Sobral BW, Dickerman AW. 2007. A robust species tree for the Alphaproteobacteria. J Bacteriol 189:4578–4586. doi: 10.1128/JB.00269-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee K, De Backer P, Aono T, Liu C, Suzuki S, Suzuki T, Kaneko T, Yamada M, Tabata S, Kupfer DM, Najar FZ, Wiley GB, Roe B, Binnewies TT, Ussery DW, D'Haeze W, Herder J. Den, Gevers D, Vereecke D, Holsters M, Oyaizu H. 2008. The genome of the versatile nitrogen fixer Azorhizobium caulinodans ORS571. BMC Genomics 9:271. doi: 10.1186/1471-2164-9-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shibata S, Yip ES, Quirke KP, Ondrey JM, Visick KL. 2012. Roles of the structural symbiosis polysaccharide (syp) genes in host colonization, biofilm formation, and polysaccharide biosynthesis in Vibrio fischeri. J Bacteriol 194:6736–6747. doi: 10.1128/JB.00707-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chain PSG, Lang DM, Comerci DJ, Malfatti SA, Vergez LM, Shin M, Ugalde RA, Garcia E, Tolmasky ME. 2011. Genome of Ochrobactrum anthropi ATCC 49188T, a versatile opportunistic pathogen and symbiont of several eukaryotic hosts. J Bacteriol 194:4274–4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barquero-Calvo E, Conde-Alvarez R, Chacón-Díaz C, Quesada-Lobo L, Martirosyan A, Guzmán-Verri C, Iriarte M, Mancek-Keber M, Jerala R, Gorvel JP, Moriyon I, Moreno E, Chaves-Olarte E. 2009. The differential interaction of Brucella and Ochrobactrum with innate immunity reveals traits related to the evolution of stealthy pathogens. PLoS One 4:e5893. doi: 10.1371/journal.pone.0005893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Michael V, Frank O, Bartling P, Scheuner C, Göker M, Brinkmann H, Petersen J. 2016. Biofilm plasmids with a rhamnose operon are widely distributed determinants of the “swim-or-stick” lifestyle in roseobacters. ISME J 10:2498–2513. doi: 10.1038/ismej.2016.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim M, Harwood C. 1991. Regulation of benzoate-CoA ligase in Rhodopseudomonas palustris. FEMS Microbiol Lett 83:199–203. doi: 10.1111/j.1574-6968.1991.tb04440.x-i1. [DOI] [Google Scholar]
- 50.Quandt J, Hynes MF. 1993. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene 127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 51.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Nat Biotechnol 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 52.Pelletier DA, Hurst GB, Foote LJ, Lankford PK, McKeown CK, Lu T, Schmoyer DD, Shah MB, Hervey WJ Jr, McDonald WH, Hooker BS, Cannon WR, Daly DS, Gilmore JM, Wiley HS, Auberry ODL, Wang Y, Larimer FW, Kennel SJ, Doktycz MJ, Morrell-Falvey JL, Owens ET, Buchanan MV. 2008. A general system for studying protein-protein interactions in Gram-negative bacteria research articles. J Proteome Res 7:3319–3328. doi: 10.1021/pr8001832. [DOI] [PubMed] [Google Scholar]
- 53.McKinlay JB, Harwood CS. 2010. Carbon dioxide fixation as a central redox cofactor recycling mechanism in bacteria. Proc Natl Acad Sci U S A 107:11669–11675. doi: 10.1073/pnas.1006175107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Castresana J. 2000. Selection of conserved block from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 58.Le SQ, Gascuel O. 2008. An improved general amino acid replacement matrix. Mol Biol Evol 25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 59.LaSarre B, McCully AL, Lennon JT, McKinlay JB. 2016. Microbial mutualism dynamics governed by dose-dependent toxicity of cross-fed nutrients. ISME J; doi: 10.1038/ismej.2016.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.