ABSTRACT
The extent of arsenic contamination in drinking water and its potential threat to human health have resulted in considerable research interest in the microbial species responsible for arsenic reduction. The arsenate reductase gene (arrA), an important component of the microbial arsenate reduction system, has been widely used as a biomarker to study arsenate-reducing microorganisms. A new primer pair was designed and evaluated for quantitative PCR (qPCR) and high-throughput sequencing of the arrA gene, because currently available PCR primers are not suitable for these applications. The primers were evaluated in silico and empirically tested for amplification of arrA genes in clones and for amplification and high-throughput sequencing of arrA genes from soil and groundwater samples. In silico, this primer pair matched (≥90% DNA identity) 86% of arrA gene sequences from GenBank. Empirical evaluation showed successful amplification of arrA gene clones of diverse phylogenetic groups, as well as amplification and high-throughput sequencing of independent soil and groundwater samples without preenrichment, suggesting that these primers are highly specific and can amplify a broad diversity of arrA genes. The arrA gene diversity from soil and groundwater samples from the Cache Valley Basin (CVB) in Utah was greater than anticipated. We observed a significant correlation between arrA gene abundance, quantified through qPCR, and reduced arsenic (AsIII) concentrations in the groundwater samples. Furthermore, we demonstrated that these primers can be useful for studying the diversity of arsenate-reducing microbial communities and the ways in which their relative abundance in groundwater may be associated with different groundwater quality parameters.
IMPORTANCE Arsenic is a major drinking water contaminant that threatens the health of millions of people worldwide. The extent of arsenic contamination and its potential threat to human health have resulted in considerable interest in the study of microbial species responsible for the reduction of arsenic, i.e., the conversion of AsV to AsIII. In this study, we developed a new primer pair to evaluate the diversity and abundance of arsenate-reducing microorganisms in soil and groundwater samples from the CVB in Utah. We observed significant arrA gene diversity in the CVB soil and groundwater samples, and arrA gene abundance was significantly correlated with the reduced arsenic (AsIII) concentrations in the groundwater samples. We think that these primers are useful for studying the ecology of arsenate-reducing microorganisms in different environments.
KEYWORDS: arsenate reductase gene (arrA), high-throughput sequencing, qPCR of arrA gene, arsenic reduction
INTRODUCTION
Arsenic is a major contaminant of drinking water that threatens the health of millions of people worldwide (1–3). Arsenate (AsV) and arsenite (AsIII) are the two dominant forms of naturally occurring inorganic arsenic (2, 4); the latter has been reported to be more toxic and labile in the subsurface, but the relative mobilities of the two oxidation states are dependent on a number of environmental factors, such as pH and mineral composition (5). In nature, although abiotic reduction of AsV to AsIII is possible in the presence of sulfide under strongly acidic conditions (6, 7), microbial reduction of AsV to AsIII is more prevalent and is one of the major causes of arsenic contamination in groundwater (8–17).
The extent of arsenic contamination and its potential threat to human health have resulted in considerable interest in studying microbial species responsible for the reduction of AsV to AsIII. To date, several members of the bacterial phyla Firmicutes and Proteobacteria (Gammaproteobacteria, Deltaproteobacteria, and Epsilonproteobacteria) have shown potential for dissimilatory arsenate reduction (14, 18). These dissimilatory arsenate-respiring bacteria (DARB) have a conserved arsenate reductase gene (arrA) that encodes an arsenate reductase enzyme, an important component of the microbial arsenate reduction system. This gene has been widely used as a biomarker to assess variations in the arsenic-reducing microbial taxa in different environments (11, 17, 19–26).
Monitoring of the arrA gene can provide useful information about the biogeography, biodiversity, and variations of the arsenate-reducing microbial species in different arsenic-contaminated environments, as well as how these microbial species respond to changes in geochemical characteristics. Similarly, studying the relative abundance of the arrA gene in response to changes in geochemical conditions (pH, soil organic matter content, soil water saturation, and soil phosphorous content) in different land use systems may identify factors that possibly enhance or inhibit microbial arsenic reduction in different systems. Three sets of arrA gene primers are being used to amplify arrA gene fragments of variable sizes, but these available primers are not adequate for high-throughput sequencing and/or quantitation with quantitative PCR (qPCR).
The most widely used PCR primers for the assessment of arrA gene diversity (ASF1, AS2F, and AS1R) were developed by Song et al. (20). The length of the amplified PCR product is 630 bp. These primers have been used extensively (11, 17, 21–25) to generate a useful arrA gene database. However, these primers have three potential limitations to their widespread application in studying the ecology of arsenate-reducing microorganisms. First, arrA gene amplification with these primers is based on a nested PCR approach that requires two independent PCRs for the detection, quantification, or sequencing of arrA genes (20). The additional PCR and potential primer biases in the first PCR may compromise our ability to draw any quantitative conclusions regarding species abundance from the nested PCR-generated amplicons. Second, while the amplicon length (630 bp) can be useful for phylogenetic studies, it is too long for qPCR, which normally requires an amplicon size of less than 400 bp. Third, the amplicon length (630 bp) reported by Song et al. (20) is also too long for the currently available high-throughput DNA-sequencing technologies, which can accurately amplify DNA fragments of approximately 500 to 600 bp. The second most widely used PCR primer set for arrA gene amplification (arrA-F and arrA-R) was developed by Malasarn et al. (27). The primers amplify a 148-bp PCR fragment that could potentially be used for qPCR but is too short to provide accurate information about phylogenetic affiliations. Hassan et al. (26) reported nonspecific amplification issues with this primer pair, drawing into question its potential application for qPCR. The third available primer pair (haarrAD1-F and haarrAG2-R), which is less widely reported, was developed by Kulp et al. (12), using a consensus sequence of the arrA gene from seven different pure culture isolates to generate an amplicon of 500 bp. These primers were modified to reflect the archaeal codon bias of extreme halophiles (Halobacterium) and showed successful amplification of the arrA gene from hypersaline environments of Mono Lake and Searles Lake (12); however, these primers were not successful in amplifying the arrA gene from other environments (20, 28). Similar to the approach of Song et al. (20), Blum et al. (29) used nested PCR with arrA gene primers that were designed from Bacillus selenitireducens strain MLS10 (30, 31). This primer pair may not be suitable to capture diverse arrA genes because (i) the primers were designed from the arrA gene sequence of a single pure culture isolate and (ii) the arrA genes amplified by Blum et al. (29) were from a single phylotype that was closely related to the Bacillus arsenicoselenatis strain.
None of the three available primer sets is sufficient for both qPCR and high-throughput sequencing analyses. In the current study, we have focused on the development and evaluation of PCR primers that are useful for quantification of the arrA gene through a qPCR approach and are appropriate lengthwise for phylogenetic studies using high-throughput sequencing. Growth in arrA gene databases since 2009 has allowed us to design new PCR primers that amplify arrA gene fragments of the optimum size directly from environmental samples. The specificity of these new primers was confirmed by (i) amplification of previously generated arrA gene clones from diverse phylogenetic groups (17) and (ii) direct amplification and high-throughput sequencing of arrA gene amplicons from soil and groundwater from the Cache Valley Basin (CVB) in Utah. These primers were also evaluated for the quantification of arrA genes in soil and groundwater well samples through qPCR. We explored the correlation between arrA gene abundance and arsenic concentrations in the CVB groundwater samples, as well as potential associations of various groundwater quality parameters with arsenic concentrations and arrA gene abundance.
RESULTS AND DISCUSSION
Primer evaluation.
The newly developed primer pair amplifies an arrA gene fragment of about 330 bp that is suitable for both phylogenetically based assessments and quantification of the arrA gene through qPCR. In silico, these primers matched (≥90% DNA identity) 86% of the total arrA gene sequences from GenBank, and an NCBI BLAST search revealed high arrA gene specificity for this primer pair. The forward primer (arrA-CVF1) exactly matched (100% DNA identity) the arrA gene sequences from Geobacter OR-1 (GenBank accession no. AB769875), Geobacter uraniireducens arrA52 (GenBank accession no. JF827128), and Desulfuromonas WB3 (GenBank accession no. KM452746) isolates and showed about 90% DNA identity with the arrA gene sequence from a Chrysiogenes arsenatis strain (GenBank accession no. AY660883) (see Fig. S2A and B in the supplemental material). Similarly, the reverse primer showed high levels of identity with the arrA gene sequences from these isolates (Fig. S2A and B). The protein sequences translated from the forward primer were highly similar to those of the pure culture isolates (Fig. S3).
Although we did not evaluate direct amplification of the arrA genes from pure culture isolates because the isolates were not available in our laboratory, we performed empirical evaluation of the primers through successful amplification of all 54 arrA gene clones of diverse phylogenetic clusters (Fig. S1) from the study by Mirza et al. (17), as well as amplification from soil and groundwater samples collected from the CVB (Fig. S4). A single band of appropriate size (330 bp) was observed on 1% agarose gels for all samples (Fig. S4). Amplicon sequencing from the soil and groundwater samples, based on high-throughput sequencing, revealed that all amplified sequences were related to the arrA gene sequences in GenBank. None of the amplified arrA gene sequences was related to other genes, inasmuch as (i) all amplified sequences from soil and groundwater samples showed high levels of similarity to the arrA genes from GenBank and (ii) phylogenetic analysis of 100 randomly selected arrA gene sequences along with other arsenite oxidase and Mo-protein-related sequences indicated different phylogenetic clustering (Fig. S5). The 100 sequences were randomly selected using a Python script and were translated into amino acids prior to phylogenetic analysis. These findings suggest that this primer pair is highly specific for arrA and can be used for high-throughput sequencing of arrA genes from soil and groundwater samples.
The newly developed PCR primers allowed direct amplification of the arrA gene from native soil and groundwater samples without any preenrichment or incubation of samples under laboratory conditions. Most of the previous studies that focused on the assessment of arrA gene diversity across different environments, including arsenic-contaminated Cambodian sediments (32), aquifer sediments from West Bengal, India (33), estuarine sediments from the Chesapeake Bay (20), streambed sediments from the Inner Coastal Plain in New Jersey (21, 34), water and sediments from Mono Lake and Searles Lake in California (35, 36), sediments from Rifle, Colorado (37), and sediments from the CVB (17), used a preenrichment step before the evaluation of arrA gene diversity. Some studies (12, 25, 38) were able to amplify arrA gene sequences directly from environmental samples using existing primers, without preenrichment, as recently highlighted by Ying et al. (25). Such preenrichment steps are known to reduce both overall microbial diversity and functional microbial diversity and can cause significant shifts in microbial communities toward fewer phylotypes that are better at competing for the available resources under existing growth conditions (39, 40); this results in significant reductions in overall microbial diversity and communities that do not represent the actual populations of the arsenate reductase communities in the natural environment, as suggested by Ying et al. (25). In addition, these enrichment-based microcosm studies did not provide information about the actual abundance of arrA genes in different soil and groundwater environments.
Broader arrA gene diversity detected with the newly developed primers.
Using this primer pair, we observed that amplified arrA gene sequences belonged to eight major phylogenetic clusters at 85% DNA identity (Fig. 1). Only two of the eight clusters (clusters III and IV) contained pure isolates. Cluster III was composed of arrA gene sequences from two Geobacter strains (Geobacter lovleyi SZ and Geobacter uraniireducens) and contained about 16% of the arrA gene sequences (9,774 sequences) in this study (Fig. 1). A large abundance of Geobacter-related phylotypes was observed previously in sediments from sites in West Bengal, India (41), and Rifle, Colorado (37).
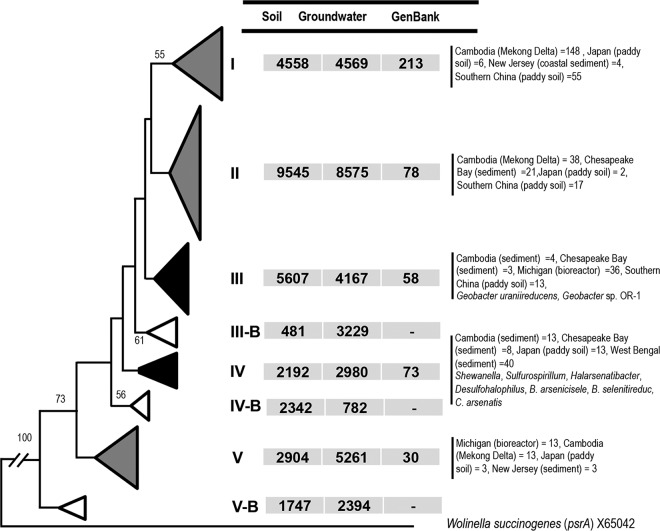
FIG 1.
Maximum likelihood phylogenetic tree based on partial sequences (∼330 bp) of the 61,333 total arsenate reductase (arrA) genes sequenced, representing 253 phylotypes (each containing >5 sequences), and of 762 arrA gene sequences, from cultured or uncultured organisms, in GenBank. The sequences were collapsed into major phylogenetic clusters at 85% DNA identity, and relative distributions of arrA gene sequences determined in the current study or obtained from GenBank are reported. Black triangles, clusters containing sequences from pure culture isolates; gray triangles, clusters containing sequences from uncultured organisms from GenBank; white triangles, clusters unique to this study. Numbers at the branch nodes reflect the maximum likelihood bootstrap support values. The psrA gene from Wolinella succinogenes (GenBank accession no. X65042) was used as an outgroup. Detailed distributions of arrA gene sequences from GenBank are presented in Tables S3 and S4.
The arrA gene cluster IV, represented by arrA genes from several pure culture isolates, including Shewanella, Sulfurospirillum, Halarsenatibacter, Desulfohalophilus, B. selenitireducens, B. arsenicoselenatis, and Chrysiogenes arsenatis (Fig. 1) isolates, contained only 8% of the arrA gene sequences retrieved from the CVB soil and groundwater samples. The other six major clusters, represented by 46,387 sequences, lacked any pure culture representatives. This indicates an exceptionally broad arrA gene diversity (Fig. 1) that extends far beyond the range of the few known species of dissimilatory arsenate-reducing microorganisms. Similar to our results, previous studies observed broad variations in arrA gene sequences from different regions of the world (12, 17, 20, 21, 23–26), despite the limited number of arrA gene sequences analyzed.
Three of eight clusters (clusters IIIB, IVB, and VB) (Fig. 1), represented by 10,975 sequences, did not contain any arrA gene sequences from any cultured or uncultured organism in the GenBank database. This indicates the presence of arrA genes of potential novel lineage that could be unique to the CVB sites. Arsenic-contaminated sites throughout the world, including the CVB, are characterized as shallow Holocene Age alluvial, lacustrine, or deltaic deposits (2). The CVB, unlike the most intensely studied arsenic-contaminated areas of the world, has a semiarid climate and abundant sedimentary, limestone, and dolomite geology (42, 43). The geological sources of As are volcanic deposits that are exposed in the surrounding mountains and the margins of the valley.
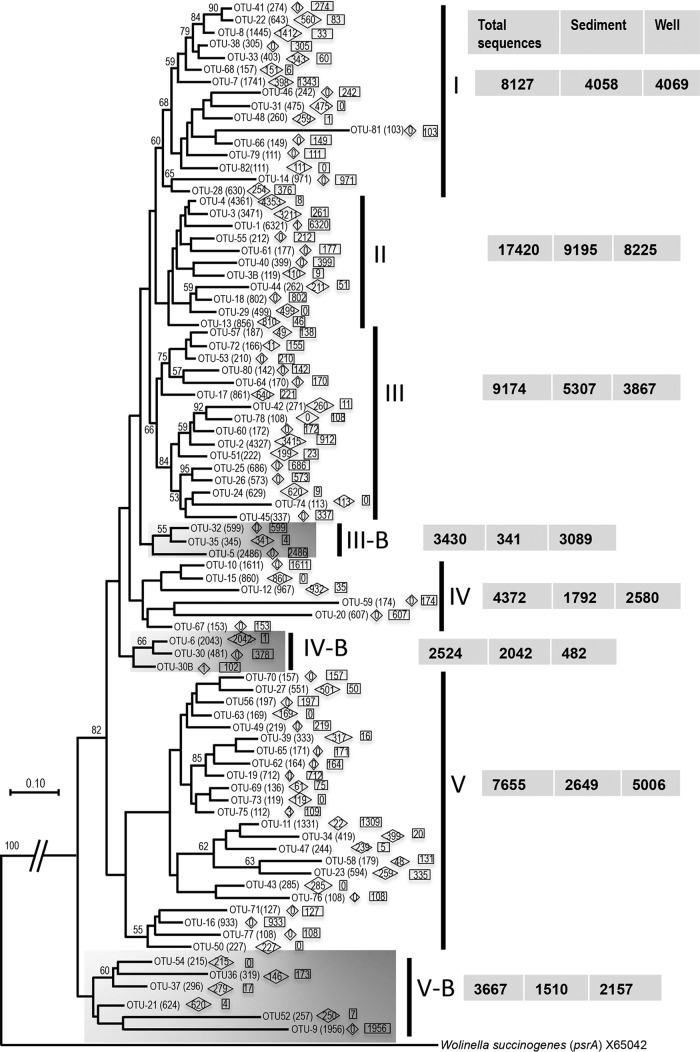
The arrA gene diversity was much greater at the 90% DNA identity level; we detected 253 operational taxonomic units (OTUs) represented by more than five sequences. Eighty-two phylotypes, containing 91% of the total sequences, were represented by more than 100 sequences (Fig. 2). This high level of variation in the amplified arrA gene sequences suggests that this primer pair can be used effectively to capture a wide diversity of arrA genes. Our earlier work on subsurface sediments from the CVB (17) found only 14 such phylotypes. The use of the new, directly applied primers described here, in combination with much deeper high-throughput sequencing, has significantly increased the number of known phylotypes from this valley. We observed an approximately 18-fold increase in the phylotypes with 90% DNA identity that contained more than five arrA gene sequences. The relatively small number of phylotypes detected in the previous study could be due to the sequencing depth; only 290 arrA gene clones were sequenced, compared to the 62,056 sequences retrieved in this study. In addition, as mentioned above, the cloning-based study was conducted using glucose- and groundwater-amended enrichments, which might have reduced the diversity of arrA genes to a few dominant phylotypes. The observed number of arrA gene phylotypes with 90% DNA identity containing more than five sequences in this study was 5-fold greater than the number of phylotypes (containing more than one sequence) reported for the Mekong Delta (25) using direct arrA gene amplification without any enrichment. The relative distributions of arrA gene sequences from soil and groundwater samples were similar within five of the major clusters (clusters I to III, IV, and V) (Fig. 1). Distributions varied within three of those clusters (clusters IIIB, IVB, and V) (Fig. 1). The differences in the distributions of arrA sequences from soil and groundwater samples from the CVB sites observed in Fig. 1 were more prominent at the 90% DNA identity level (Fig. 2). Approximately 46% of OTUs (38 of 82 OTUs) with more than 100 sequences were found in either soil or groundwater samples but not both (Fig. 2). The As-reducing microbial community detected in the groundwater samples was significantly different from that found in the soil samples (P < 0.01) (Fig. S6). This finding suggests that there could be unique populations of arsenate-reducing microbes in either the planktonic or attached (biofilm) communities of the aquifer and that very few populations inhabit both environments, likely because of the differences in the biogeochemical conditions of these two systems and/or the propensity of some organisms to participate in biofilm ecosystems.
FIG 2.
Maximum likelihood phylogenetic tree based on partial arrA gene sequences (∼330 bp) from five soil samples and four groundwater samples from the Cache Valley Basin, amplified with the newly developed PCR primers and sequenced using high-throughput sequencing. Sequences belonging to the same OTU (at 90% identity), as assigned by mothur, were clustered together, and branches within clusters were collapsed to show the overall relationships of these clusters to one another. The major grouping (groups I to V) is consistent with the phylogenetic tree shown in Fig. 1, and three clusters (highlighted in gray) are unique. Numbers at the nodes reflect the maximum likelihood bootstrap support values. A total of 82 OTUs containing more than 100 arrA gene sequences from soil and groundwater samples were used in this analysis. The distributions of arrA gene sequences among the soil and groundwater samples are represented in diamonds and squares, respectively. Numbers at the right correspond to the overall distribution of arrA gene sequences in the major groups and their relative distribution in sediment and groundwater samples. The psrA gene from Wolinella succinogenes (GenBank accession no. X65042) was used as an outgroup.
It is important to mention that, while high levels of arrA gene diversity suggest wide genetic variations of arsenate-reducing microorganisms and possibly relatively high rates of arrA gene evolution, greater gene diversity does not provide any information about the organisms' actual functional capabilities or rates of As reduction in an ecosystem. It is possible that some of the arrA genes are nonfunctional homologs, i.e., highly similar to the arrA gene but not capable of As reduction. Exploring this further will require pure culture isolates containing diverse arrA genes and evaluation of their As reduction rates under controlled conditions.
Quantitation of arrA gene abundance in soil and groundwater samples.
The development of this primer combination permitted the quantitation of dissimilatory arsenate-reducing bacteria in environmental samples. The regression line for the standard curve generated through serial dilutions of arrA gene clones showed a coefficient of determination of 0.996 to 0.997 and a PCR amplification efficiency of 93 to 102%. The specificity of the amplicon was confirmed by melting curve analysis, which indicated the presence of a single peak. We assayed DNA preparations from 29 soil and groundwater samples from the CVB for quantification of the arrA gene. Low levels of variation in arrA gene quantification from three independent qPCRs were observed using the same DNA extracts from soil and groundwater samples. However, the numbers of arrA gene copies varied among different soil and groundwater samples (Fig. 3).
FIG 3.
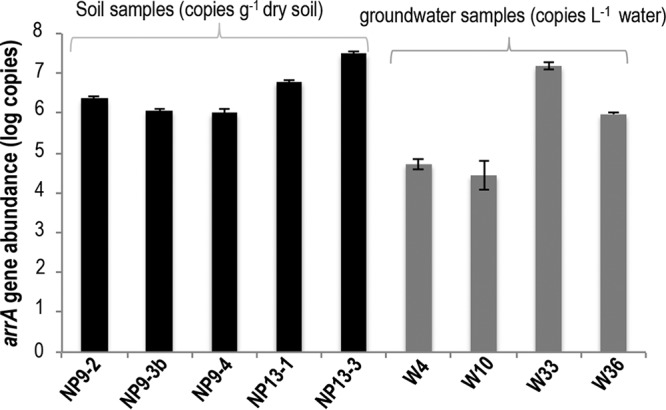
qPCR quantification, using the newly developed arrA gene primers, of arrA gene copies from soil and groundwater samples from the Cache Valley Basin. The gene copy numbers are given on a log scale. Error bars represent standard errors.
The plasmid extraction efficiency was approximately 87% ± 5%, compared to nonfiltered transformed Escherichia coli cells. This suggests that, since the total DNA extraction efficiency in actual samples was not taken into account, arrA gene values reported in this study may be underestimates. A comparison of arrA gene abundances in undiluted and diluted (1:10) DNA samples from five randomly selected groundwater wells (wells 5, 8, 12, 16, and 36) showed no difference, suggesting the absence of potential PCR inhibitors in these groundwater samples. This was also evident in the 260-nm/280-nm spectrophotometric absorbance ratios (>1.80), which suggested that the extracted DNA was of good quality.
We did not apply any adjustment factor for the numbers of arrA gene copies per genome, because only a few genomes containing the arrA gene are available to date. Six of eight clusters lacked any pure culture representative (Fig. 1). The isolated strains Parasutterella excrementihominis strain YIT 11859, Geobacter strain OR-1, Desulfitobacterium hafniense strain DCB-2, and Shewanella strain ANA-3 (43–47) contained a single arrA gene copy. Without pure culture isolates from each of the diverse phylogenetic groups, application of any adjustment factor is not justified, and we have reported the results as copies of the arrA gene per gram of soil or per liter of groundwater instead of numbers of arsenate-reducing microorganisms per gram of soil or per liter of groundwater.
Correlation between arrA gene abundance and arsenic concentrations.
We observed a significant correlation between arrA gene abundance and reduced arsenic (AsIII) concentrations in the groundwater samples (Pearson's correlation coefficient = 0.82; P < 0.001) (Fig. 4). Four of the groundwater samples analyzed were associated with the City of Logan landfill. Those samples contained high AsIII concentrations (1.27 to 50 μg/liter [0.017 to 0.67 μM]) and greater arrA gene abundance (5.96 to 8.17 log copies/liter) (Table S1). These landfill samples contributed to the strength of the association between arrA gene abundance and AsIII concentrations. The remaining 20 of 24 groundwater samples were collected from CVB domestic wells. The AsIII concentrations in those samples ranged from 0.1 to 15.6 μg/liter (0.001 to 0.20 μM), and the arrA gene abundance levels ranged from 3.05 to 5.74 log copies/liter of groundwater (Table S1). In some of the samples, such as those from wells 6, 8, 9, and 18, we observed the presence of the arrA gene despite very low AsIII concentrations (∼0.1 μg/liter). This could be due to the fact that some of the microorganisms containing arrA genes possess a wide range of metabolic processes, as observed in the case of most of the currently available pure culture isolates containing arrA genes, including Shewanella strain ANA-3, Geobacter strain OR-1, and Sulfurospirillum. Those organisms have the ability to reduce As but also actively grow by using alternative electron acceptors such as FeIII, NO3−, and SO4−2 and/or by reducing other metals. Another possibility could be that these organisms are present in the environment but are not active. This assumption could be better tested by using arrA gene transcripts as predictors of As reduction in the presence of alternative terminal electron acceptors. Previously, Giloteaux et al. (37) observed a correlation between arrA gene transcripts of Geobacter strains and rates of As reduction. In their study, the abundance of arrA gene transcripts remained high despite decreases in the rates of As reduction, suggesting that other factors may have roles in regulating the expression of the arrA gene.
FIG 4.
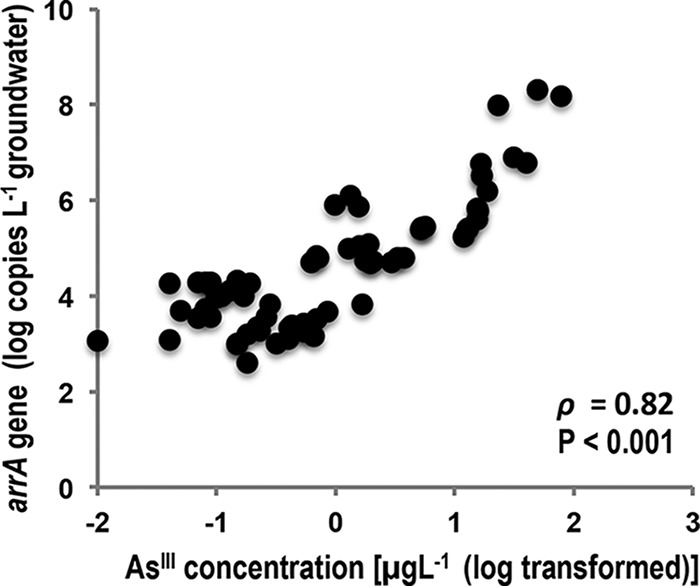
Pearson's correlation analysis of arrA gene abundance and reduced As (AsIII) concentrations. The AsIII concentrations were logarithmically transformed, and the strength of association is represented by Pearson's correlation coefficient (ρ).
We used multivariate analysis to explore relationships among groundwater quality parameters, including arrA gene abundance and total As and AsIII concentrations, in the various samples (Fig. 5). In the nonmetric dimensional scaling (NMDS) plot, the direction and length of the arrows indicate the nature and strength of multivariate associations (Fig. 5). Significant associations existed between arrA gene abundance, As concentrations (total As and AsIII concentrations), and several other water quality parameters, such as dissolved organic carbon (DOC) levels, alkalinity, electrical conductivity (EC), and K+, Na+, Ba2+, Mn, and Cl− levels, in the samples collected from the landfill site. Some factors, such as high As concentrations and DOC levels, may have a direct influence on the abundance of arsenate-reducing microorganisms, while associations with other factors, including K+, Na+, Ba2+, and Cl− concentrations, may be due to the nature of the landfill site and not have any direct relation with the As reduction process. It is important to mention that, when the data for the landfill samples were removed from the analysis and only the data for the samples from CVB domestic wells were compared, the AsIII levels and arrA gene abundance remained correlated, as indicated by the direction of the arrows in Fig. S7, and arrA gene abundance was closely associated with wells 17 and 19, which contained high concentrations of AsIII (12.6 to 15.6 μg/liter [0.17 to 0.21 μM]). Fe and Mn levels were other factors associated with those two wells (Fig. S7), suggesting that the reduction of As might have been coupled with Fe and Mn reduction at those two sites. Wells 8 and 9, containing high concentrations of AsV, were negatively associated with arrA gene abundance (Fig. S7).
FIG 5.
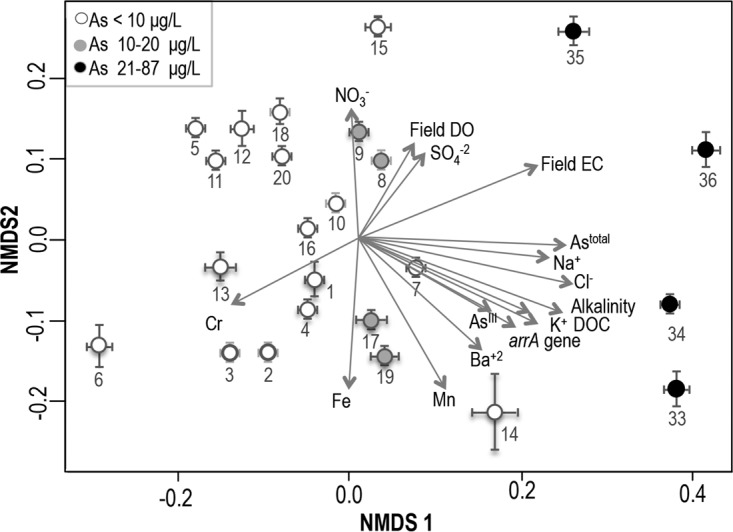
NMDS plots representing different groundwater quality parameters, based on a Euclidean distance matrix. The black circles represent four samples that were associated with the Logan landfill site. The lengths and directions of the arrows indicate the strength and nature, respectively, of the associations of different water quality parameters.
Concluding remarks.
New arrA gene primers have been developed for direct amplification, quantification, and sequencing of the arrA gene from environmental samples. The combined use of these primers and high-throughput sequencing facilitated a nearly 18-fold increase in the number of arrA phylotypes represented by more than five sequences in the CVB in Utah. Although the number of samples sequenced in this study was limited, our findings revealed high levels of diversity and the presence of novel lineages of arrA genes. Larger-scale targeted analyses of the biogeographic distribution of arrA genes, along with isolation and identification of organisms containing such genes, are essential for determining the worldwide biodiversity of the genes and the associated microorganisms.
Overall, arrA gene phylogenetic clustering of the 330-bp fragments sequenced using the newly developed PCR primers corroborated the phylogenetic representativeness of the phylogenetic tree generated from arrA clones and other sequences of larger size (630 bp) from GenBank, because the positions of GenBank arrA gene sequences among the different major phylogenetic groups (groups I to V) were consistent (Fig. 1; also see Fig. S1 and Tables S3 and S4 in the supplemental material). These findings indicated that the 330-bp arrA gene amplicon from these primers could be used for phylogenetically based analysis of the arsenate reductase gene.
We observed a significant correlation between arrA gene abundance quantified through qPCR and AsIII concentrations in the groundwater samples. The successful amplification and quantification of the arrA gene from these groundwater samples, with diverse hydrogeochemical characteristics, suggest that these primers can be used effectively for quantification of the arrA gene in diverse ecological environments of the CVB in Utah.
The development of this new primer pair will facilitate data acquisition from natural arsenate-reducing populations and better description of the biodiversity of such populations. These primers could also be very useful for studying the dynamics of arsenate-reducing communities from different phylogenetic groups in the context of arsenic contamination of groundwater. The use of these primers in qPCR can help in the study of variations in the relative abundance of arsenate-reducing microbial species, both seasonally and in response to different biogeochemical treatments, to identify the geochemical factors responsible for increased arsenic concentrations in groundwater.
MATERIALS AND METHODS
Development and evaluation of arrA primers.
We designed PCR primers to target the arrA gene by using the GenBank database (July 2015). A total of 762 arrA gene sequences of appropriate length (600 bp) from GenBank, including sequences from cultured and uncultured organisms, were aligned using Muscle (48). This alignment was used in HYDEN software (49) to generate one forward primer and two reverse primers for different conserved regions unique to the arrA gene. The HYDEN software identifies conserved regions in the DNA, uses variants of simple approximation algorithms, and runs a “greedy hill-climbing” algorithm procedure to improve the primer coverage. The standard parameters used for each primer were as follows: number of degenerate positions, 2; maximum number of mismatches, 2; primer length, 20 bp; number of base primers for contraction/expansion algorithms, 50. The primers were further evaluated visually by checking the alignment with Clustal X (50) and Sequencher 5.1 (Gene Codes Corp., Ann Arbor, MI). The primer specificity was confirmed with a NCBI BLAST search.
Among the primers, arrA-CVF1 (5′-CACAGCGCCATCTGCGCCGA-3′) and arrA-CVR1 (5′-CCGACGAACTCCYTGYTCCA-3′) matched (≥90% DNA identity) about 86% of total arrA gene sequences. The amplicon size (∼330 bp) generated by these primers was optimal both for the phylogenetically based assessments and for qPCR. This primer pair was tested for the amplification of 54 clones containing arrA genes from diverse phylogenetic groups. The clones were generated previously (17) from CVB sediments using the nested PCR primer sets developed by Song et al. (20). Pure culture isolates containing the arrA gene have been very limited and confined to one or two major phylogenetic groups (subgroups III and IV in Fig. 1). In contrast, the arrA gene clones from CVB sediments are highly diverse and distributed across different major phylogenetic groups (17). The newly developed PCR primers were evaluated by using these arrA gene clones to show that the primers can successfully amplify arrA genes from diverse phylogenetic groups and also that the target region for these primers is conserved across different phylogenetic clusters. The specificity of the new primer pair was also evaluated for direct amplification of arrA genes from soil and groundwater samples from the CVB. The specificity of the PCR amplicons generated from these soil and groundwater samples was confirmed using high-throughput sequencing.
Soil and groundwater sample collection and chemical analyses.
Soil cores were collected from two sites (NP9 and NP13) upgradient of the City of Logan municipal solid waste landfill. Geoprobe direct-push coring was used to collect the sediments in plastic sleeves (diameter, 2.54 cm; length, 152 cm) at a maximum depth of 93.5 cm. Samples from different layers were sectioned in a vinyl anaerobic chamber (Coy Laboratory Products, Grass Lake, MI) in 100% N2 and were immediately stored in a −70°C freezer for DNA extraction. Further details on sample collection and sectioning were reported previously (42). Three layers from NP9 and two layers from NP13 (see Table S1 in the supplemental material) were used to evaluate the primers.
Groundwater samples were collected from 24 wells located in the CVB (the locations are described in Table S1). Twenty of the samples were from domestic wells, used for irrigation and potentially for household use. The sampling locations were random across the basin, but selection was limited by access to the well. The other four-well samples were from monitoring wells located downgradient of the City of Logan municipal landfill and were potentially influenced by landfill leachate.
The wells were pumped until the EC and dissolved oxygen (DO) readings were stable. Field-portable probes were used to analyze the equilibrated water samples for EC, DO, pH, and temperature. Water samples were collected in triplicate. Arsenic species were separated using Dowex 1X8 anion exchange resin (60/100 mesh) (51). With acidification of the sample to pH 4, the neutral AsIII oxyanion passes through the column while the negatively charged AsV adsorbs to the resin. Samples for total trace element analysis were acidified in the field, whereas samples for anion and DOC analysis were filtered in the field (0.2 μm) and stored on ice. Samples were returned to the laboratory for analysis within 6 h after collection. Total trace element analysis was performed by inductively coupled plasma-mass spectrometry (ICP-MS) (Agilent 7700x), after sample digestion with nitric acid. Anions were analyzed using a Dionex ion chromatograph (IC-3000) (52), with KOH gradient elution. DOC was analyzed using a combustion Teledyne TOC analyzer (Apollo 900) (53). Arsenic concentrations in the groundwater ranged from 0.5 to 87 μg/liter (0.01 to 1.17 μM) (Table S1). The groundwater across the study area is alkaline; the samples downgradient from the landfill were characterized by high EC and DOC levels.
The soils were extracted using a saturation paste with deionized water. The pore water extracts were analyzed using the methods described for groundwater. Pore water arsenic levels ranged from 10.5 to 158 μg/liter (0.14 to 2.11 μM) (Table S1), and other pore water properties are reported in Table S1. The geology of this site and the geochemistry of As in this area were described previously (42).
DNA extraction and arrA gene amplification from soil and groundwater samples.
Genomic DNA from the five soil samples in the CVB (Table S1) was extracted using the PowerSoil DNA isolation kit (Mo Bio, Carlsbad, CA). All extraction steps were performed according to the manufacturer's instructions, and DNA samples were stored at −20°C until analysis.
DNA was also extracted from 24 groundwater well samples collected from the CVB (Table S1). One liter of groundwater per well was filtered through 0.22-μm Sterivex filters (Millipore Corp., Billerica, MA) using a peristaltic pump (Masterflex; Cole-Parmer Co, Vernon Hills, IL). The filters were broken and the membranes were removed, cut into small pieces using sterile scissors, and then placed in 1.5-ml microcentrifuge tubes for DNA extraction as described above.
DNA extracts from previously generated arrA gene clones (17), as well as DNA extracted from the five soil samples and four randomly selected groundwater samples (wells 4, 10, 33, and 36), were amplified using the newly developed primers to evaluate amplicon specificity. All PCR amplifications were performed in 50-μl reaction volumes containing 1× PCR buffer, with 2 mM MgCl2, 0.2 μM each primer, 1.8 mM MgCl2, 200 μM deoxynucleotide triphosphates, 20 ng of template, and 1 μl of FastStart high-fidelity PCR system enzyme (Roche Applied Sciences). The PCR conditions were as follows: 3 min at 95°C, 35 cycles of denaturation at 94°C for 45 s, primer annealing at 60°C for 45 s, and extension at 72°C for 1 min, and a final extension for 7 min at 72°C.
For metagenomic sequencing, DNA from soil (five samples) and groundwater (four samples) samples was amplified with the newly developed primers linked to adapter and unique barcode sequences (Table S2). A 5-μl aliquot of each amplified arrA gene product was used to evaluate successful PCR amplification and to verify the appropriate amplicon size by electrophoresis on a 1% agarose gel. The remaining 45 μl of amplified PCR product was purified with Agencourt AMPure beads (Beckman Coulter, Brea, CA). Purified PCR products from each of the nine samples were pooled in equimolar concentrations and were sequenced with an Ion Torrent PGM system (Life Technologies) at the Utah State University Center for Integrated Biosystems.
High-throughput sequence analysis.
Amplified sequences were screened for quality control, including minimum read lengths (300 bp), maximum homopolymers of >8 bases, sequences with unidentified bases (N), and sequences with more than one inexact match with the unique barcode identifier and perfect primer matches, by using the Ribosomal Database Project (RDP) pipeline. All high-quality sequences (62,056 sequences) were confirmed as arrA gene sequences by using the NCBI standalone local BLAST search application, and subsequent phylogenetic analysis showed their close relatedness to the previously described arrA gene sequences. None of the amplified arrA gene sequences was related to arsenite oxidase genes (aioA and arxA) or other Mo-containing proteins. To assess variation or diversity, the amplified arrA gene sequences were aligned with Muscle. These aligned arrA gene DNA sequences were used to create a distance matrix, followed by clustering into OTUs at 85% and 90% DNA identity, as described previously (17, 25), using mothur (54).
Phylogenetic analyses.
For phylogenetic analyses, we used two arrA gene data sets, i.e., (i) 630-bp arrA gene sequences generated previously from the CVB in Utah (17), using primers described by Song et al. (20), to determine the phylogenetic clustering based on larger fragments and (ii) 330-bp arrA gene sequences generated in the current study, along with trimmed arrA gene clone sequences from our previous study, to compare the phylogenetic clustering based on smaller arrA gene products. Overall, three maximum likelihood (ML) phylogenetic trees were constructed using FastTree 2.0.0 (55). The arrA gene sequences were aligned using Muscle, and tree branches were collapsed into subgroups based on OTUs (sequences belonging to the same OTU) using MEGA version 5.2 (56). The first phylogenetic tree contained the 54 arrA gene clones (630 bp) that were reported previously (17), using the primers developed by Song et al. (20), along with the arrA gene sequences from GenBank (Fig. S1). These clones were initially used for evaluation of the new primers. This phylogenetic tree was constructed to determine the distribution of the tested arrA gene clones among diverse phylogenetic groups and to evaluate the overall phylogenetic grouping of longer arrA gene fragments (630 bp).
The second ML phylogenetic tree was constructed using representative arrA gene sequences from the OTUs containing more than five sequence (253 OTUs) for the soil and groundwater samples (Fig. 1). These sequences represented 61,333 arrA gene sequences, accounting for about 99% of the total arrA gene sequences that were generated using high-throughput sequencing. We used OTUs containing more than five sequences as a conservative approach to avoid any potential biases that might be caused by sequencing artifacts. The arrA gene sequences from GenBank that fully overlapped the arrA gene region amplified with our primers and that were used for construction of the first phylogenetic tree (arrA clones and sequences from GenBank) were also included in this analysis. All arrA gene sequences were trimmed to an equal length (∼330 bp). This tree was constructed to evaluate the relative positions of previously described arrA genes from GenBank and the arrA gene sequences amplified in this study using high-throughput sequencing. The complete phylogenetic tree was too large to publish in a convenient format; hence, we collapsed branches to show major groupings at 85% DNA identity and we presented the abundance of our sequences relative to other sequences from GenBank (Fig. 1). The detailed distribution of the arrA gene sequences from GenBank, among different phylogenetic groups associated with this phylogenetic tree, are presented in Tables S3 and S4. The third phylogenetic tree contained representative arrA gene sequences from different OTUs at 90% DNA identity, a previously used cutoff level for this gene in sediment from the CVB (17) and from the Mekong Delta in Cambodia (25). For this phylogenetic tree, we selected sequences from the OTUs represented by >100 sequences, and the number of representative sequences selected for each OTU was proportional to the number of arrA gene sequences of the OTU (Fig. 2).
Quantitative PCR.
The arrA gene abundance was determined using qPCR, which was carried out in 25-μl volumes containing 12.50 μl of iTaq Fast SYBR Green Supermix with ROX (Bio-Rad, Hercules, CA), 100 nM primers (arrA-CVF1 and arrA-CV1R), and 10 ng of template DNA. PCR conditions were as follows: 94°C for 5 min and then 40 cycles of denaturation at 94°C for 45 s, annealing at 60°C for 1 min, and extension at 72°C for 1 min. PCR-grade water was used as a negative control. The specificity of the qPCR products was confirmed by melting curve analysis. A standard curve was generated from serial dilutions (100 to 10−9) of plasmid DNA containing a fragment of the arrA gene. The qPCR efficiency (E) was calculated according to the equation E = 10(−1/slope). Three independent qPCRs were performed using the same DNA extracts from CVB soil and groundwater samples.
We evaluated the approximate DNA extraction efficiency by inoculating 1 ml of E. coli cells (in triplicate) containing arrA gene plasmids into 1 liter of groundwater, followed by filtration and extraction of DNA from the filters. We compared the arrA gene copies retrieved from uninoculated groundwater samples, groundwater samples inoculated with 1 ml of arrA-containing E. coli cells, and samples extracted directly without filtration, to estimate the loss of plasmid copies during the filtration step. We also compared arrA gene abundance in undiluted and diluted (1:10) DNA samples from five randomly selected groundwater wells (wells 5, 8, 12, 16, and 36), to evaluate the presence of PCR inhibitors in the groundwater samples.
Statistical analyses.
Overall, differences in the arsenate-reducing community structures in different soil and groundwater samples were calculated from Bray-Curtis dissimilarity at 97% DNA identity and were visualized by NMDS. The significance of the differences was assessed through analysis of similarity (ANOSIM), as described previously (17).
Correlations between arrA gene abundance and AsIII concentrations in groundwater were evaluated using the Pearson product-moment correlation coefficient, and the significance of the differences was explored with two-tailed t tests using R software (www.R-project.org). The AsIII concentrations were logarithmically transformed to normalize the data distribution. The associations of various groundwater quality parameters with arsenic concentrations (both total As and AsIII concentrations) and arrA gene abundance for different groundwater wells were explored through multivariate analysis using a Euclidean distance matrix, and results were visualized with NMDS plots in the Vegan Community Ecology Package of R software. All data were standardized or scaled before the multivariate analyses.
Accession number(s).
The arrA gene sequences from both soil and groundwater samples were deposited in the NCBI Sequence Read Archive (SRA) under accession number SRP067270.
Supplementary Material
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02725-16.
REFERENCES
- 1.Nordstrom DK. 2002. Worldwide occurrences of arsenic in ground water. Science 296:2143–2145. doi: 10.1126/science.1072375. [DOI] [PubMed] [Google Scholar]
- 2.Smedley PL, Kinniburgh DG. 2002. A review of the source, behaviour and distribution of arsenic in natural waters. Appl Geochem 17:517–568. doi: 10.1016/S0883-2927(02)00018-5. [DOI] [Google Scholar]
- 3.Rahman MM, Ng JC, Naidu R. 2009. Chronic exposure of arsenic via drinking water and its adverse health impacts on humans. Environ Geochem Health 31(Suppl 1):189–200. doi: 10.1007/s10653-008-9235-0. [DOI] [PubMed] [Google Scholar]
- 4.Lloyd JR, Oremland RS. 2006. Microbial transformations of arsenic in the environment: from soda lakes to aquifers. Elements 2:85–90. doi: 10.2113/gselements.2.2.85. [DOI] [Google Scholar]
- 5.Dixit S, Hering JG. 2003. Comparison of arsenic(V) and arsenic(III) sorption onto iron oxide minerals: implications for arsenic mobility. Environ Sci Technol 37:4182–4189. doi: 10.1021/es030309t. [DOI] [PubMed] [Google Scholar]
- 6.Cherry JA, Shaikh AU, Tallman DE, Nicholson RV. 1979. Arsenic species as an indicator of redox conditions in groundwater. J Hydrol 43:373–392. doi: 10.1016/0022-1694(79)90182-3. [DOI] [Google Scholar]
- 7.Rochette EA, Bostick BC, Li G, Fendorf S. 2000. Kinetics of arsenate reduction by dissolved sulfide. Environ Sci Technol 34:4714–4720. doi: 10.1021/es000963y. [DOI] [Google Scholar]
- 8.Dowdle PR, Laverman AM, Oremland RS. 1996. Bacterial reduction of arsenic(V) to arsenic(III) in anoxic sediments. Appl Environ Microbiol 62:1664–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rowland HAL, Pederick RL, Polya DA, Pancost RD, Van Dongen BE, Gault AG, Vaughan DJ, Bryant C, Anderson B, Lloyd JR. 2007. The control of organic matter on microbially mediated iron reduction and arsenic release in shallow alluvial aquifers, Cambodia. Geobiology 5:281–292. doi: 10.1111/j.1472-4669.2007.00100.x. [DOI] [Google Scholar]
- 10.Newman DK, Kennedy EK, Coates JD, Ahmann D, Ellis DJ, Lovely DR, Morel FMM. 1997. Dissimilatory arsenate and sulfate reduction in Desulfotomaculum auripigmentum sp. nov. Arch Microbiol 168:380–388. doi: 10.1007/s002030050512. [DOI] [PubMed] [Google Scholar]
- 11.Héry M, Rizoulis A, Sanguin H, Cooke DA, Pancost RD, Polya DA, Lloyd JR. 2015. Microbial ecology of arsenic-mobilizing Cambodian sediments: lithological controls uncovered by stable-isotope probing. Environ Microbiol 17:1857–1869. doi: 10.1111/1462-2920.12412. [DOI] [PubMed] [Google Scholar]
- 12.Kulp TR, Hoeft SE, Miller G, Saltikov C, Murphy JN, Han S, Lanoil B, Oremland RS. 2006. Dissimilatory arsenate and sulfate reduction in sediments of two hypersaline, arsenic-rich soda lakes: Mono and Searles Lakes, California. Appl Environ Microbiol 72:6514–6526. doi: 10.1128/AEM.01066-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oremland RS, Stolz JF. 2003. The ecology of arsenic. Science 300:939–944. doi: 10.1126/science.1081903. [DOI] [PubMed] [Google Scholar]
- 14.Oremland RS, Stolz JF. 2005. Arsenic, microbes and contaminated aquifers. Trends Microbiol 13:45–49. doi: 10.1016/j.tim.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 15.Das S, Liu CC, Jean JS, Liu T. 2016. Dissimilatory arsenate reduction and in situ microbial activities and diversity in arsenic-rich groundwater of Chianan Plain, Southwestern Taiwan. Microb Ecol 71:365–374. doi: 10.1007/s00248-015-0650-3. [DOI] [PubMed] [Google Scholar]
- 16.Das S, Liu CC, Jean JS, Lee CC, Yang HJ. 2016. Effects of microbially induced transformations and shift in bacterial community on arsenic mobility in arsenic-rich deep aquifer sediments. J Hazard Mater 310:11–19. doi: 10.1016/j.jhazmat.2016.02.019. [DOI] [PubMed] [Google Scholar]
- 17.Mirza BS, Muruganandam S, Meng X, Sorensen DL, Dupont RR, McLean JE. 2014. Arsenic(V) reduction in relation to iron(III) transformation and molecular characterization of the structural and functional microbial community in sediments of a basin-fill aquifer in northern Utah. Appl Environ Microbiol 80:3198–3208. doi: 10.1128/AEM.00240-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perez-Jimenez JR, DeFraia C, Young LY. 2005. Arsenate respiratory gene (arrA) for Desulfosporosinus sp. strain Y5. Biochem Biophys Res Commun 338:825–829. doi: 10.1016/j.bbrc.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 19.Messens J, Silver S. 2006. Arsenate reduction: thiol cascade chemistry with convergent evolution. J Mol Biol 362:1–17. doi: 10.1016/j.jmb.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 20.Song B, Chyun E, Jaffé PR, Ward BB. 2009. Molecular methods to detect and monitor dissimilatory arsenate-respiring bacteria (DARB) in sediments. FEMS Microbiol Ecol 68:108–117. doi: 10.1111/j.1574-6941.2009.00657.x. [DOI] [PubMed] [Google Scholar]
- 21.Mumford AC, Barringer JL, Benzel WM, Reilly PA, Young LY. 2012. Microbial transformations of arsenic: mobilization from glauconitic sediments to water. Water Res 46:2859–2868. doi: 10.1016/j.watres.2012.02.044. [DOI] [PubMed] [Google Scholar]
- 22.Ohtsuka T, Yamaguchi N, Makino T, Sakurai K, Kimura K, Kudo K, Homma E, Dong DT, Amachi S. 2013. Arsenic dissolution from Japanese paddy soil by a dissimilatory arsenate-reducing bacterium Geobacter sp. OR-1. Environ Sci Technol 47:6263–6271. [DOI] [PubMed] [Google Scholar]
- 23.Upadhyaya G, Clancy TM, Brown J, Hayes KF, Raskin L. 2012. Optimization of arsenic removal water treatment system through characterization of terminal electron accepting processes. Environ Sci Technol 46:11702–11709. doi: 10.1021/es302145q. [DOI] [PubMed] [Google Scholar]
- 24.Zhang SY, Zhao FJ, Sun GX, Su JQ, Yang XR, Li H, Zhu YG. 2015. Diversity and abundance of arsenic biotransformation genes in paddy soils from southern China. Environ Sci Technol 49:4138–4146. doi: 10.1021/acs.est.5b00028. [DOI] [PubMed] [Google Scholar]
- 25.Ying SC, Damashek J, Fendorf S, Francis CA. 2015. Indigenous arsenic(V)-reducing microbial communities in redox-fluctuating near-surface sediments of the Mekong Delta. Geobiology 13:581–587. doi: 10.1111/gbi.12152. [DOI] [PubMed] [Google Scholar]
- 26.Hassan Z, Sultana M, Van Breukelen BM, Khan SI, Röling WFM. 2015. Diverse arsenic- and iron-cycling microbial communities in arsenic-contaminated aquifers used for drinking water in Bangladesh. FEMS Microbiol Ecol 91:fiv026. doi: 10.1093/femsec/fiv026. [DOI] [PubMed] [Google Scholar]
- 27.Malasarn D, Saltikov CW, Campbell KM, Santini JM, Hering JG, Newman DK. 2004. arrA is a reliable marker for As(V) respiration. Science 306:455. doi: 10.1126/science.1102374. [DOI] [PubMed] [Google Scholar]
- 28.Escudero LV, Casamayor EO, Chong G, Pedrós Alió C, Demergasso C. 2013. Distribution of microbial arsenic reduction, oxidation, and extrusion genes along a wide range of environmental arsenic concentrations. PLoS One 8:e78890. doi: 10.1371/journal.pone.0078890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blum JS, McCann SH, Bennett S, Miller LG, Stolz JF, Stoneburner B, Saltikov C, Oremland RS. 2016. A microbial arsenic cycle in sediments of an acidic mine impoundment: Herman Pit, Clear Lake, California. Geomicrobiol J 33:677–689. doi: 10.1080/01490451.2015.1080323. [DOI] [Google Scholar]
- 30.Ranganathan M. 2005. Arr genes from arsenate-respiring low G+C Gram positive bacteria Bacillus selenitireducens strain MLS10 and Clostridium sp. strain OhILAs. Master's thesis Duquesne University, Pittsburgh, PA. [Google Scholar]
- 31.Stolz JF, Berekaa MM, Fischer E, Polshyna G, Thangavelu M, Dheer R, Ranganathan M, Garci Moyano A, El Assar S, Basu P. 2011. Methods for detection of arsenate respiring bacteria: advances, cautions, and caveats, p 283–296. In Stolz JF, Oremland RS (ed), Microbial metabolism of metals and metalloids: advances and applications. American Society for Microbiology, Washington, DC. [Google Scholar]
- 32.Lear G, Song B, Gault A, Polya DA, Lloyd JR. 2007. Molecular analysis of arsenate-reducing bacteria within Cambodian sediments following amendment with acetate. Appl Environ Microbiol 73:1041–1048. doi: 10.1128/AEM.01654-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Héry M, Gault AG, Rowland HAL, Lear G, Polya DA, Lloyd JR. 2008. Molecular and cultivation-dependent analysis of metal-reducing bacteria implicated in arsenic mobilisation in south-east Asian aquifers. Appl Geochem 23:3215–3223. doi: 10.1016/j.apgeochem.2008.07.003. [DOI] [Google Scholar]
- 34.Barringer JL, Mumford A, Young LY, Reilly PA, Bonin JL, Rosman R. 2010. Pathways for arsenic from sediments to groundwater to streams: biogeochemical processes in the Inner Coastal Plain, New Jersey, USA. Water Res 44:5532–5544. doi: 10.1016/j.watres.2010.05.047. [DOI] [PubMed] [Google Scholar]
- 35.Kulp TR, Han S, Saltikov CW, Lanoil BD, Zargar K, Oremland RS. 2007. Effects of imposed salinity gradients on dissimilatory arsenate reduction, sulfate reduction, and other microbial processes in sediments from two California Soda Lakes. Appl Environ Microbiol 73:5130–5137. doi: 10.1128/AEM.00771-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoeft SE, Kulp TR, Han S, Lanoil B, Oremland RS. 2010. Coupled arsenotrophy in a hot spring photosynthetic biofilm at Mono Lake, California. Appl Environ Microbiol 76:4633–4639. doi: 10.1128/AEM.00545-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giloteaux L, Holmes DE, Williams KH, Wrighton KC, Wilkins MJ, Montgomery AP, Smith JA, Orellana R, Thompson CA, Roper TJ, Long PE, Lovley DR. 2013. Characterization and transcription of arsenic respiration and resistance genes during in situ uranium bioremediation. ISME J 7:370–383. doi: 10.1038/ismej.2012.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hollibaugh JT, Budinoff C, Hollibaugh RA, Ransom B, Bano N. 2006. Sulfide oxidation coupled to arsenate reduction by a diverse microbial community in a soda lake. Appl Environ Microbiol 72:2043–2049. doi: 10.1128/AEM.72.3.2043-2049.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mirza BS, Rodrigues JLM. 2012. Development of a direct isolation procedure for free-living diazotrophs under controlled hypoxic conditions. Appl Environ Microbiol 78:5542–5549. doi: 10.1128/AEM.00714-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tilman D. 1982. Resource competition and community structure. Princeton University Press, Princeton, NJ. [PubMed] [Google Scholar]
- 41.Héry M, Van Dongen BE, Gill F, Mondal D, Vaughan DJ, Pancost RD, Polya DA, Lloyd JR. 2010. Arsenic release and attenuation in low organic carbon aquifer sediments from West Bengal. Geobiology 8:155–168. doi: 10.1111/j.1472-4669.2010.00233.x. [DOI] [PubMed] [Google Scholar]
- 42.Meng X, Dupont RR, Sorensen DL, Jacobson AR, McLean JE. 2015. Mineralogy and geochemistry affecting arsenic solubility in sediment profiles from the shallow basin-fill aquifer of Cache Valley Basin, Utah. Appl Geochem doi: 10.1016/j.apgeochem.2015.12.011. [DOI] [Google Scholar]
- 43.Meng X, Dupont RR, Sorensen DL, Jacobson AR, McLean JE. 2016. Arsenic solubilization and redistribution under anoxic conditions in three aquifer sediments from a basin-fill aquifer in northern Utah: the role of natural organic carbon and carbonate minerals. Appl Geochem 66:250–263. doi: 10.1016/j.apgeochem.2016.01.004. [DOI] [Google Scholar]
- 44.Li X, Zhang L, Wang G. 2014. Genomic evidence reveals the extreme diversity and wide distribution of the arsenic-related genes in Burkholderiales. PLoS One 9:e92236. doi: 10.1371/journal.pone.0092236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ehara A, Suzuki H, Amachi S. 2015. Draft genome sequence of Geobacter sp. strain OR-1, an arsenate-respiring bacterium isolated from Japanese paddy soil. Genome Announc 3(1):e01478-14. doi: 10.1128/genomeA.01478-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saltikov CW, Newman DK. 2003. Genetic identification of a respiratory arsenate reductase. Proc Natl Acad Sci U S A 100:10983–10988. doi: 10.1073/pnas.1834303100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim SH, Harzman C, Davis JK, Hutcheson R, Broderick JB, Marsh TL, Tiedje JM. 2012. Genome sequence of Desulfitobacterium hafniense DCB-2, a Gram-positive anaerobe capable of dehalogenation and metal reduction. BMC Microbiol 12:21. doi: 10.1186/1471-2180-12-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Linhart C, Shamir R. 2007. Degenerate primer design: theoretical analysis and the HYDEN program. Methods Mol Biol 402:221–244. [DOI] [PubMed] [Google Scholar]
- 50.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilkie JA, Hering JG. 1998. Rapid oxidation of geothermal arsenic(III) in stream waters of the eastern Sierra Nevada. Environ Sci Technol 32:657–662. doi: 10.1021/es970637r. [DOI] [Google Scholar]
- 52.Dionex Corp. 2006. Determination of inorganic anions and organic acids in fermentation broths. Application note 123. Dionex Corp., Sunnyvale, CA. [Google Scholar]
- 53.American Public Health Association (APHA). 2012. Standard methods for the examination of water and wastewater, p 21–22. In Rice EW, Baird RB, Eaton AD, Clesceri LS (ed), Method 5310B High temperature combustion method, 22nd ed, vol 5 American Public Health Association, Washington DC. [Google Scholar]
- 54.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2: approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.