Abstract
Sonic hedgehog (Shh) signaling plays a critical role during development and carcinogenesis. While Gli family members govern the transcriptional output of Shh signaling, little is known how Gli-mediated transcriptional activity is regulated. Here we identify the actin-binding protein Missing in Metastasis (MIM) as a new Shh-responsive gene. Together, Gli1 and MIM recapitulate Shh-mediated epidermal proliferation and invasion in regenerated human skin. MIM is part of a Gli/Suppressor of Fused complex and potentiates Gli-dependent transcription using domains distinct from those used for monomeric actin binding. These data define MIM as both a Shh-responsive gene and a new member of the pathway that modulates Gli responses during growth and tumorigenesis.
Keywords: Sonic hedgehog, Gli, MIM, BCC, Actin, transcription
Sonic hedgehog (Shh) signaling controls the proliferation of progenitor cells of many organs (Chuong 1998; Callahan and Oro 2001; Taipale and Beachy 2001; Ruiz i Altaba et al. 2002), and uncontrolled Shh target gene induction promotes a variety of cancers including basal cell carcinomas (BCC) (Callahan and Oro 2001; Pasca di Magliano and Hebrok 2003). Previous studies indicate that most Hedgehog signaling is controlled through the Ci/Gli family of zinc finger transcription factors (Methot and Basler 2001; Bai et al. 2002; Ruiz i Altaba et al. 2002). In vertebrates, three gli genes are present, with Gli1 being predominantly a transcriptional activator and Gli2 and Gli3 acting as both activators and repressors (Matise and Joyner 1999; Ruiz i Altaba et al. 2002). Genetic analysis reveals functional redundancy among these genes, with a combination of activation and repression determining Shh signaling's transcriptional output (Bai et al. 2002; Mill et al. 2003). While the diversity of Shh effects is clear, the mechanisms for controlling Gli transcriptional output in different contexts remain poorly understood.
In flies, regulation of Ci/Gli transcriptional output is in part controlled through associations with cytoplasmic complexes, which include the tumor suppressor Suppressor of Fused (Sufu), Fused, and Costal-2 (Kalderon 2004). In vertebrates, Gli protein subcellular distribution and transcriptional activity are regulated by Sufu (Kogerman et al. 1999; Cheng and Bishop 2002; Dunaeva et al. 2003). Here we identify the novel actin-binding protein MIM as both a Shh-responsive gene and a regulator of Gli transcription.
Results and Discussion
MIM is a Shh-responsive gene
In order to identify new Shh-responsive targets in human skin, we examined cDNA microarray expression profiles from regenerated human epidermis expressing Shh. We have previously shown that tumors arising from such skin share features with basal cell carcinomas (Fig. 1A; Fan et al. 1997). We named the first new Shh-responsive gene characterized from this screen BCC-enriched gene 4 (BEG4). Independently, BEG4 has been previously identified as the actin-binding protein Missing in Metastasis (MIM) (Lee et al. 2002; Mattila et al. 2003; Woodings et al. 2003; Yamagishi et al. 2004), and for simplicity we refer to the gene as MIM for the remainder of this manuscript. MIM is a member of the Wiskott-Aldrich Syndrome family of actin-associated proteins and contains a conserved coiled-coil protein interaction domain and a C-terminal WH2 domain (Supplementary Fig. S1). Previous independent studies have shown that MIM binds monomeric actin through its WH2 domain and bundles F-actin using its N-terminal coiled-coil domain (Lee et al. 2002; Mattila et al. 2003; Woodings et al. 2003; Yamagishi et al. 2004; R. Gonzalez-Quevedo and A. Oro, unpubl.).
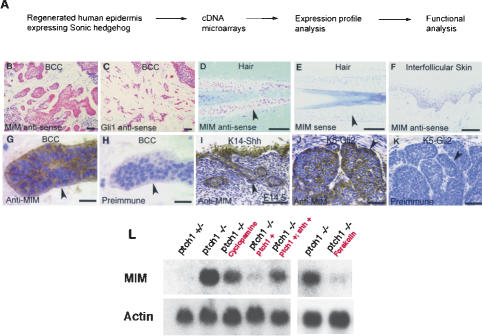
Figure 1.
MIM is a Shh-responsive gene. (A) Strategy to isolate novel Shh-responsive genes. (B,C) In situ hybridizations with anti-sense probes demonstrate that similarly to Gli1 (C), MIM (B) transcripts accumulate in human BCC tumor epithelium (transcript staining is red with toluidine blue counterstain). (D) MIM anti-sense probes show that MIM transcripts also accumulate in the outer root sheath of human anagen hair follicles (red staining highlighted with arrowhead). No staining is seen with control MIM sense probes (lack of red staining highlighted by arrowhead in E). (F) MIM transcripts are not detected within the interfollicular epithelium in human scalp. (G) An affinity-purified, polyclonal, anti-MIM antibody shows MIM protein expression within the epithelium of human BCCs (brown signal highlighted by arrowhead). (H) Tumor epithelium (arrowhead) is only highlighted by the Mayer's hematoxylin counterstain when treated with control preimmune serum from the same animal. MIM immunoreactivity is also seen within the ectopic epithelial skin proliferations formed in embryonic K14-Shh transgenic mice (arrowhead in I) and the BCC-like skin tumors arising in K5-Gli2 transgenic mice (arrowhead in J). (K) No MIM protein is detected in K5-Gli2 transgenic skin tumors stained with preimmune serum. Bars: B–F, 50 μm; G,H, 15 μm; I, 100 μm; J,K, 50 μm. (L) Cultured ptch1 mutant MEFs demonstrate constitutive pathway activation, and accumulate high levels of MIM transcripts. In contrast, in cells in which Shh target genes are suppressed, such as ptch1+/– MEFs or ptch1-null MEFs rescued by retroviral gene transfer with the wild-type ptch1 gene, there are dramatically lower levels of MIM RNA. Similarly to known Shh targets (Taipale et al. 2000), treatment of ptch1 mutant MEFs for 72 h with 8 μM cyclopamine, an inhibitor of smoothened (Chen et al. 2002), or for 24 h with 40 μM forskolin, a negative regulator of Ci/Gli (Wang et al. 1999), reduces MIM accumulation. Reactivating Shh target genes in cultured ptch1–/–; ptch1+ cells by infecting with a Shh retrovirus (ptch–/–; ptch1+; shh+) increases MIM transcripts.
MIM is a Shh-responsive gene. In vivo, MIM transcripts are detected in the outer root sheath of anagen hair follicles, where Shh targets are normally expressed (Oro and Higgins 2003), but are not seen in the interfollicular epithelium (Fig. 1D–F). Similarly to known Shh target genes, such as Gli1 (Fig. 1C), MIM RNA and protein also accumulate at sites of inappropriately active Shh signaling, such as the tumor epithelium of human BCCs (Fig. 1B,G,H). Additionally, MIM protein is seen in the abnormal follicular growths and tumors that arise in transgenic mice ectopically expressing Shh (Fig. 1I) or Gli2 (Fig. 1J,K). In vitro, MIM transcripts are induced using conditions that stimulate both Shh signaling and target gene expression in ptch1 mouse embryo fibroblasts (Fig. 1L; Taipale et al. 2000) and in the ptch1 epithelial line A431 (Koike et al. 2002; data not shown). Taken together, these data show that MIM behaves as a Shh-responsive gene both in vivo and in vitro.
MIM synergizes with Gli to recapitulate Shh effects in skin
The association of MIM expression with tumor growth, and the fact that many Shh target genes have been shown to affect pathway signaling (McMahon 2000; Taipale and Beachy 2001), led us to examine the effects of ectopic MIM expression in regenerated human skin grafts. As previously shown (Fan et al. 1997), grafts expressing Shh demonstrate robust keratinocyte proliferation and deep epithelial ingrowths into the underlying dermis, both features of the anagen hair follicle and BCCs (Fig. 2B,F,H,K). Unlike the tumorigenic phenotypes produced in transgenic mice overexpressing Gli1 or Gli2 (see Fig. 1J,K; Grachtchouk et al. 2000; Nilsson et al. 2000; Oro and Higgins 2003), regenerated human skin grafts expressing only Gli1 demonstrate proliferative rates and architectural features similar to those of control grafts (Fig. 2C,F,K). In addition, ectopic expression of MIM alone also fails to yield a phenotype distinct from controls (Fig. 2D,F,K), indicating that neither gene alone is sufficient to promote the epithelial proliferation and invasion seen in Shh grafts.
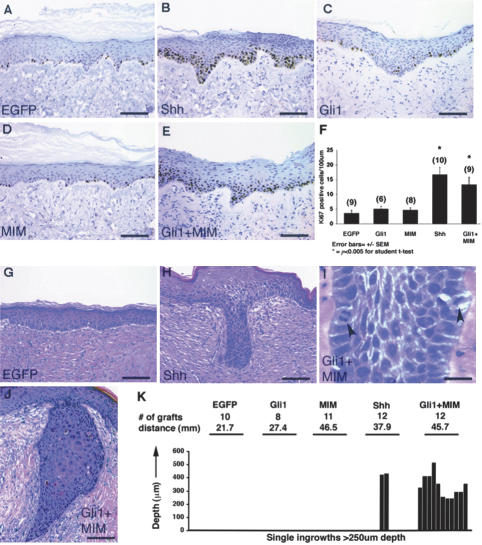
Figure 2.
MIM synergizes with Gli to recapitulate Shh effects in skin. Representative sections of EGFP (A), Shh (B), Gli1 (C), MIM (D), and Gli1 + MIM (E) regenerated human skin stained with anti-Ki67 antibody, a marker of cell proliferation. Keratinocyte proliferative activity is measured by the number of keratinocytes with brown nuclear staining. (F) Quantification of data in which each graft condition is plotted on the X-axis, and the Y-axis represents the average number of Ki-67 positive keratinocytes per 100 μm of graft tissue. Digits placed over error bars show the number of grafts examined per condition. p values are calculated versus EGFP control grafts. (G–J) H&E-stained sections showing the deeply invasive ingrowths identified in both Shh (H) and Gli1 + MIM (I,J) skin. (K) The number and depth of the invasive ingrowths identified in each graft condition. At top, the histogram lists the number of skin grafts and total graft distance (in millimeters) examined per graft condition. Below, each histogram bar represents one invasive epithelial ingrowth measuring at least 250 μm in depth (e.g., two deep ingrowths were seen in Shh grafts). The height of each bar shows that ingrowth's maximum depth of invasion (Y-axis). Bars: A–E,G,H,J, 100 μm; I, 25 μm.
In contrast, Gli and MIM act synergistically to induce epidermal growth. In grafts coexpressing both Gli1 and MIM, keratinocyte proliferation rates rival those of Shh grafts (Fig. 2E,F). In addition, these grafts show an abundance of deep epithelial ingrowths that are architecturally and cytologically similar to those seen in Shh grafts (Fig. 2I–K). Together, these results suggest that the robust keratinocyte proliferation and invasion seen in Shh grafts are mediated by the combined activities of both Gli transcription factors and MIM.
MIM potentiates Gli-dependent transcription
To test whether MIM affects the transcriptional output of Gli targets, we examined the promoter activity of the Keratin 17 (K17) gene. K17 is one of the earliest markers of the hair follicle lineage and is expressed, similarly to other Shh target genes, throughout BCC tumor epithelium (Fig. 3A,B; McGowan and Coulombe 1998; Kurzen et al. 2001). In mosaic, Gli1-infected skin grafts, cells producing abundant Gli protein also express high levels of endogenous K17, indicating that Gli expression is sufficient to induce K17 (Fig. 3C,D).
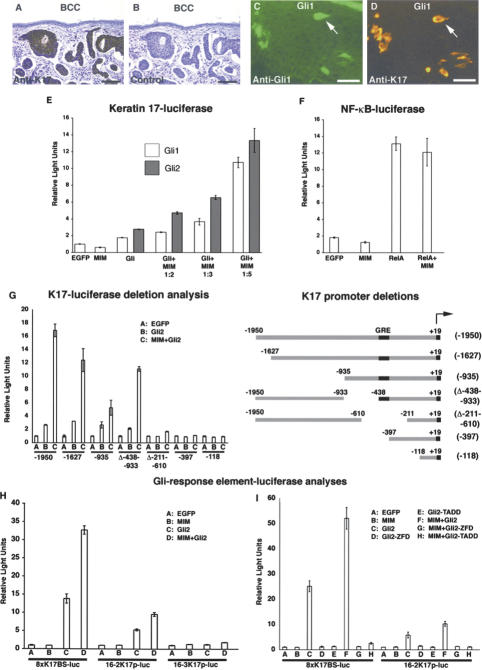
Figure 3.
MIM potentiates Gli-dependent transcription. Polyclonal K17 antibody (A) but not secondary antibody alone (B) reacts with tumor epithelium. Coincident expression of Gli1 (C) and K17 (D) in a skin mosaic demonstrates that Gli activates endogenous K17. Bars: A,B, 100 μm; C,D, 25 μm. (E) Luciferase activities in lysates from primary keratinocytes showing Gli1 (white bars) and Gli2 (gray bars) activate the K17 promoter, and MIM potentiates Gli-dependent transcription in a dosage-sensitive manner. (F) Luciferase activity showing MIM does not potentiate NF-κB transcriptional activity in keratinocytes. (G) Luciferase activity with K17 promoter deletions identifies a 41-bp region required for both Gli responsiveness and MIM potentiation in keratinocytes. (H) MIM potentiates Gli-mediated transcription from multimerized Gli response elements in keratinocytes. Luciferase activities were measured from reporter constructs carrying eight directly repeated copies of wild-type Gli response sequences from the mouse K17 promoter (8xK17BS-luc) (Supplementary Fig. S2), the mouse FOXA2 promoter (16-2K17p-luc), or a mutated element from the mouse FOXA2 promoter (16-3K17p-luc) (Sasaki et al. 1997). (I) Luciferase activities showing that MIM does not potentiate Gli-mediated transcription from multimerized Gli response elements when cotransfected with a Gli2 DNA-binding mutant (Gli2-ZFD) or a Gli2 transactivation mutant (Gli2-TADD). (E–I) Error bars, ±SEM.
We assayed for transcriptional synergy between MIM and Gli using a 2-kb K17 promoter fragment (N. Bianchi, K. McGowan, and P. Coulombe, unpubl.) in primary human keratinocytes. Gli1 or Gli2 expression alone induces K17-luciferase activity two- to threefold, whereas expressing only MIM has no effect on promoter activity. In contrast, expression of Gli1 or Gli2 with MIM results in a dose-dependent potentiation of promoter activity, yielding a >10-fold induction versus controls (Fig. 3E). MIM potentiation appears specific to Gli transcriptional activity, as MIM did not potentiate RelA-dependent transcription from an NF-κB-luciferase reporter (Fig. 3F).
MIM potentiation of Gli targets could occur either through the Gli response element or another portion of the K17 promoter/enhancer. Deletion analysis of the K17 promoter shows that constructs retaining 41 nt, 398–438 bp upstream of the 5′-end of the K17 gene, demonstrate both Gli2-responsiveness and MIM-potentiation. Deletions that remove this region lose both activities (Fig. 3G). Within this region are several Gli-like DNA-binding sequences (Supplementary Fig. S2), and to test its sufficiency to respond to Gli and MIM, we constructed a luciferase reporter containing eight directly repeating copies of this element (8xK17BS-luc) (Supplementary Fig. S2). As predicted, the 8xK17BS-luc reporter responded to Gli2 transcriptional activation and resulted in greater than twofold potentiation by MIM (Fig. 3H). These data support the idea that MIM potentiates Gli through its response element. Additionally, differences in MIM potentiation between the K17 response element multimer and the full-length K17 promoter may indicate a requirement for a particular promoter orientation or the necessity of additional sequences outside the core Gli response element for maximum MIM transcriptional potentiation (Fig. 3H).
To test whether MIM can potentiate Gli activity on other known Gli response elements, we examined the mouse FoxA2 floor plate enhancer. Previously, wild-type copies (16-2) of this DNA response element were shown to bind Gli and mediate transcription, while a mutated element (16-3) was unresponsive (Sasaki et al. 1997). Similar to the K17 response element, MIM potentiates Gli2-mediated transcription from the wild-type FOXA2 enhancer in keratinocytes and fails to affect activity of the mutated response element (Fig. 3H). In addition, expression of MIM potentiates Gli-dependent transcription on the FOXA2 promoter in 3T3 cells, demonstrating that MIM transcriptional potentiation can occur on several Gli-responsive promoters and in distinct cell types (Supplementary Fig. S3A). Furthermore, MIM potentiation is dependent on Gli DNA binding and transactivation. In keratinocytes, Gli2 mutants lacking either the zinc finger DNA-binding domain (Gli2-ZFD) or the putative transactivation domain (Gli2-TADD) (Yoon et al. 1998) fail to show MIM-dependent potentiation from the K17 or FOXA2 Gli-response elements (Fig. 3I). Taken together, these data indicate that MIM potentiates the transcriptional activity of Gli.
MIM associates with Gli and Sufu
Previous work has shown that Sufu, Costal-2, and Fused form a complex with Ci/Gli to regulate its activity (Kogerman et al. 1999; Murone et al. 2000; Kalderon 2004). To determine how MIM regulates the transcriptional activity of Gli, we assayed for physical interactions between MIM- and Gli-containing complexes. Using GSTMIM affinity columns, we found that both Gli1 and Gli2 are retained by full-length MIM whereas columns containing GST alone or an N-terminal deletion of MIM, GST-MIMΔN399, do not bind (Fig. 4A,B). In addition to Gli, we discovered that endogenous Sufu is also specifically retained by full-length MIM (Fig. 4A), suggesting that the three proteins form a complex in vitro. In further support of a MIM association, coimmunoprecipitation studies showed that MIM pellets with Gli or Sufu using antibodies to either MIM or the other complex component (Fig. 4C,E). Sufu appears to associate with MIM in the absence of Gli, as epitope-tagged Sufu is retained on the MIM column in the absence of transfected Gli protein (Fig. 4D). Supporting the idea that both proteins make direct, independent contacts with MIM, we found that in vitro translated Gli1 and Sufu bind to GST-MIM, but not GST-MIMΔN399 or GST columns (Fig. 4F). Indicative of the importance of the MIM/Gli complex interactions, the MIMΔN399 mutant that fails to interact with Gli and Sufu showed a markedly reduced capacity to potentiate Gli-dependent transcription (Fig. 4G). Although these results indicate that a MIM/Gli/Sufu complex is important for MIM-mediated transcriptional potentiation, additional MIM mutant analysis is required to better understand the functional relationships between the three proteins.
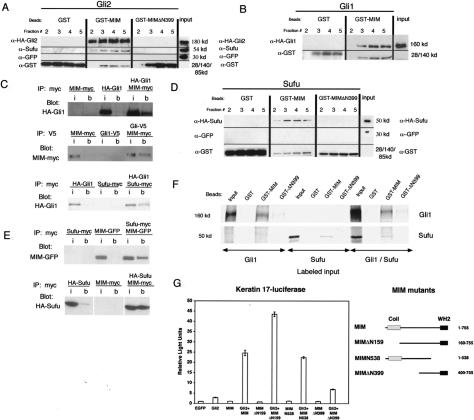
Figure 4.
MIM associates with Gli and Sufu. (A, top row) A MIM column retains HA-Gli2 and endogenous Sufu as seen by retention in eluate fractions. (Middle row) Coexpressed GFP is not retained. (Lower row) GST fusion proteins. Neither GST nor mutant MIMΔN399 retains Gli2 or Sufu. Lines separate portions of the blot probed with different antibodies. The input represents 1% of total 293 cell lysate loaded on column. (B) MIM column also retains HA-Gli1. The input represents 1% of total 293 cell lysate loaded on column. (C) Immunoprecipitated MIM-myc pellets HA-Gli1 in 293 cell lysates. (i) Input; (b) beads. Immunoprecipitated Gli1-V5 pellets MIM-myc in 293 cell lysates. Immunoprecipitated Sufu-myc also pellets HA-Gli1 in 293 cell lysates. The input in each case represents 0.5% of total input. (D, top row) The MIM column retains HASufu as seen by retention in eluate fractions. (Middle row) Coexpressed GFP is not retained. (Lower lane) GST fusion proteins. Neither GST nor MIMΔN399 retains Sufu. The input represents 1% of total eluate loaded. (E) Immunoprecipitated Sufu-myc pellets MIM-GFP and MIM-myc pellets HA-Sufu when coexpressed in 293 cell lysates. (i) Input; (b) beads. The input represents 0.5% of lysate. No binding of GFP to Sufu-myc was detected (data not shown). (F) In vitro translated Gli1 and Sufu bind to GST-MIM. (Top row) The indicated GST beads were incubated with 35S-labeled protein, and the spun pellet was examined by autoradiography. Note no increase in binding of Sufu or Gli to MIM when both are present. The input represents 50% of reaction used. (G) Luciferase activity of BEG4 mutants missing the monomeric actin-binding domain (MIMN538), the F-actin bundling domain (MIMΔN159), or the N-terminal domain required for association with the Gli complex (MIMΔN399). Error bars, ±SEM.
Analysis of the cytoskeletal-modifying activities of MIM demonstrate that it is a monomeric actin-binding and bundling protein that resides on actin cables (Lee et al. 2002; Mattila et al. 2003; Woodings et al. 2003; Yamagishi et al. 2004; R. Gonzalez-Quevedo and A. Oro, unpubl.). Because actin-binding proteins have been shown to influence transcription (Olave et al. 2002), we tested whether the known actin regulatory activities of MIM are required for Gli-mediated transcriptional potentiation. MIM mutants deficient for either the C-terminal WH2 domain (MIMN538) or the N-terminal actin bundling domain (MIMΔN159) still potentiate Gli-mediated transcription (Fig. 4G). These results show that the known actin regulatory activities of MIM are distinct from its ability to potentiate transcription.
Our data demonstrate that MIM is a Shh-responsive gene that can potentiate Gli transcriptional activity. Unlike Ptch1 and Hip, which control pathway activity through Shh binding (Taipale and Beachy 2001), MIM appears to regulate target gene expression through its association with the Gli complex. MIM's role in regulating the cytoskeleton and transcription indicates, that like β-catenin (Moon et al. 2002), plakoglobin (Maeda et al. 2004), and p120 (Prokhortchouk et al. 2001), it is a member of the growing family of cytoskeletal components that associate with transcription factors to affect nuclear signaling.
We have shown that MIM binds to Gli and Sufu complexes and that an N-terminal region of MIM is required for both complex associations and transcriptional potentiation. Analysis of MIM deletion mutants identifies this Gli-potentiation domain (GliP) domain to lie between amino acids 160 and 399 (Supplementary Fig. S1). The fact that a portion of this region is conserved in sequenced metazoan organisms suggests its transcriptional regulatory properties play an important role in other organisms.
Proteins in the Gli complex are believed to control Gli nuclear localization, protein accumulation, and transcriptional activity/target specificity. Protein stability experiments suggest that MIM does not alter the half-life of Gli protein (C. Callahan and A. Oro, unpubl.). Additionally, while MIM, like Sufu and Gli, contains a putative nuclear export signal used in nucleo-cytoplasmic shuttling (Supplementary Fig. S1), wild-type or potentiating mutants of MIM do not appear to alter the subcellular localization of Gli (Supplementary Fig. S3B). These data point to a role for MIM in regulating Gli transcriptional activity and/or target specificity. In summary, our data demonstrate that MIM is a new member of the Shh signaling pathway and implicate MIM in the regulation of Gli target gene responses during both development and tumorigenesis.
Materials and methods
cDNA microarray analysis
Total RNA from 3-wk Shh grafts (Fan et al. 1997) was compared to that of 1-wk Shh grafts or 1- or 3-wk EGFP control grafts on Human 6000 oligo arrays (Affymetrix). Each graft type was harvested in triplicate and the genes whose expressions were more than threefold greater or lower in 3-wk Shh versus control grafts were subjected to expression analysis using human and mouse Hedgehog-responsive tissues. Initially 20 genes were identified with significantly different expression levels in 3-wk Shh grafts, and two genes from this group were expressed specifically in Shh-responsive tissues.
In situ hybridization
For in situ hybridizations, paraffin sections were processed and hybridized with 35S UTP-labeled probes as previously described (Oro et al. 1997). Probes were generated using human Shh (gift of C. Tabin, Boston, MA), human Gli1 (gift of K. Kinzler, Baltimore, MD), and human MIM 3′-UTR sequences (GenBank AI198284). Toluidine blue (0.2% in water) was used to counterstain tissues.
Northern blots
Mouse embryonic fibroblasts (MEFs) mutant for ptch1 and infected with Ptch1 (Bailey et al. 2002) or Shh (Fan et al. 1997) retroviruses were cultured as previously described (Bailey et al. 2002). MEFs were treated with 8 μM cyclopamine (gift of P. Beachy, Baltimore, MD) for 72 h or 40 μM forskolin (Sigma) for 24 h where indicated. Blots were probed with a mouse MIM probe (AW323872) or β-actin control (Clontech).
Antibody generation and immunohistochemistry
Rabbit anti-human MIM antiserum was generated by immunizing with bacterially expressed GST-MIM amino acids 1–277. The serum was affinity-purified by application to a 6XHIS-tagged MIM 1–277 affinity column (Amino Link Kit; Pierce), and characterization will be published elsewhere (R. Gonzalez-Quevedo and A.E. Oro, in prep.). Anti-MIM immunohistochemistry was performed with the affinity-purified MIM antibody (1:65) or preimmune serum (1:65). Ki-67 expression was detected by mouse anti-Ki67 at 1:50 (KIF5; DAKO). Anti-K17 immunohistochemistry was performed on paraffin-embedded sections of human BCCs using mouse anti-CK17 at 1:75 (Chemicon International, Inc.). Anti-Gli1 immunohistochemistry was performed on frozen sections of Gli1 skin grafts using goat anti-Gli1 at 1:25 (sc6152; Santa Cruz Biotech). Anti-K17 immunohistochemistry was performed on frozen sections of Gli1 skin grafts using mouse anti-CK17 at 1:25 (DAKO). Anti-myc subcellular localization was performed using rabbit anti-myc (A-14; Santa Cruz Biotech) at 1:250 followed by goat anti-rabbit (605 nm; QuantumDot) at 1:100. Anti-HA subcellular localization was performed using mouse anti-HA (Covance) at 1:1000 followed by goat anti-mouse (525 nm; QuantumDot) at 1:100. Mayer's hematoxylin was used to counterstain tissues.
Regenerated human skin grafts
Regenerated human grafts were made as previously described (Fan et al. 1997). Means and standard errors for different transgene conditions were compared using the unpaired, two-tailed, Student's t-test. For quantification, more than 20 mm of tissue (parallel to skin surface) was examined for each transgene graft type. Care of animals followed Stanford University Animal Care Committee-approved protocols.
MIM affinity assays
Bacteria expressing the indicated GST fusion protein were grown to log phase and induced with 1 mM IPTG (Invitrogen). Cleared lysates were prepared by digesting with lysozyme (Sigma) followed by sonication and Triton X-100 solubilization. Following centrifugation, supernatants were incubated with glutathione-Sepharose 4B beads (Amersham) and washed with PBS, 1% Triton X-100. Cleared lysates for Gli1, Gli2, and Sufu binding were made by transfecting 293T cells and lysing in PBS with 1% Igepal, 1 mM sodium orthovanadate and sodium fluoride. After binding to protein-bound glutathione beads, mixtures were loaded onto columns, allowed to settle, and washed with 30 volumes of 1% Triton X-100. GST columns were eluted into seven 0.3-mL fractions with 100 mM Tris (pH 8.8), 20 mM reduced glutathione, 120 mM NaCl, and 1% Triton X-100 with bound protein eluting in fractions 2–5. Blotted protein fractions were visualized with one of the following as indicated: mouse anti-HA 1:1000 (Covance), goat anti-Sufu (M-15) 1:100 (Santa Cruz Biotech), mouse anti-GFP 1:3000 (Roche), or mouse anti-GST 1:10,000 (Novagen). In all affinity column figures, the amount of protein loaded in input lanes was one-hundredth the amount of protein used in the assay. 35S-labeled mouse Sufu, Gli1, or control luciferase protein was made according to the manufacturer's directions (Promega). Protein was incubated with 1% bovine serum albumin and GST beads for 1 h at room temperature. Pelleted beads were washed with 1% Triton X in PBS four times and then resolved by SDS-PAGE. The resultant gel was dried and exposed to film at room temperature.
Coimmunoprecipitation studies
Lysates for immunoprecipitations were made by lysing 293 cells transfected with indicated plasmids in 20 mM HEPES (pH 7.9), 150 mM NaCl, 1% Igepal, 0.5 mM sodium orthovanadate, and 0.25 mM sodium fluoride. Lysates were precleared with beads and then incubated with appropriate antibody for 2 h at 4°C. Addition of Protein A/G beads (Santa Cruz Biotechnology) was followed by serial centrifugation and washing with lysis buffer and 0.2% Igepal. The pellets were separated by PAGE, transferred to nitrocellulose, and interrogated using the indicated antibody by Western blot analysis. MIM-GFP was visualized with mouse anti-GFP 1:3000 (Roche). In each reaction, no immunoprecipitation was seen when precipitating antibody was omitted. In Figure 4E, no immunoprecipitation was seen between Sufu and GFP alone (data not shown). The input lane contained 0.5% of lysate used.
See Supplemental Material for luciferase assays and subcellular localization experiments, plus details of expression and reporter constructs.
Acknowledgments
We thank the Oro lab, M.P. Scott, G. Barsh, M. Cleary, G. Crabtree, and P. Khavari for critical input; N. Patil for Affymetrix microarrays, C.C. Hui for mouse Sufu; S. Cheng for Sufu; A. Dlugosz for Gli2 tumors; and P. Khavari for LZRS-RelA and NF-κB-luc. This work was supported by grants to C.A.C. (NIH K08 AR047803 and the Walter V. and Idun Y. Berry Fellowship), P.C. (NIH R01 AR44232), and A.E.O. (NIH R01 AR46786-02 and P01AR44012).
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1221804.
References
- Bai C.B., Auerbach, W., Lee, J.S., Stephen, D., and Joyner, A.L. 2002. Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development 129: 4753-4761. [DOI] [PubMed] [Google Scholar]
- Bailey E.C., Milenkovic, L., Scott, M.P., Collawn, J.F., and Johnson, R.L. 2002. Several PATCHED1 missense mutations display activity in patched1-deficient fibroblasts. J. Biol. Chem. 277: 33632-33640. [DOI] [PubMed] [Google Scholar]
- Callahan C.A. and Oro, A.E. 2001. Monstrous attempts at adnexogenesis: Regulating hair follicle progenitors through Sonic hedgehog signaling. Curr. Opin. Genet. Dev. 11: 541-546. [DOI] [PubMed] [Google Scholar]
- Chen J.K., Taipale, J., Cooper, M.K., and Beachy, P.A. 2002. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes & Dev. 16: 2743-2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S.Y. and Bishop, J.M. 2002. Suppressor of Fused represses Gli-mediated transcription by recruiting the SAP18–mSin3 corepressor complex. Proc. Natl. Acad. Sci. 99: 5442-5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong C.M. 1998. Molecular basis of epithelial appendage morphogenesis. R.G. Landes, Austin, TX.
- Dunaeva M., Michelson, P., Kogerman, P., and Toftgard, R. 2003. Characterization of the physical interaction of Gli proteins with SUFU proteins. J. Biol. Chem. 278: 5116-5122. [DOI] [PubMed] [Google Scholar]
- Fan H., Oro, A.E., Scott, M.P., and Khavari, P.A. 1997. Induction of basal cell carcinoma features in transgenic human skin expressing Sonic hedgehog. Nat. Med. 3: 788-792. [DOI] [PubMed] [Google Scholar]
- Grachtchouk M., Mo, R., Yu, S., Zhang, X., Sasaki, H., Hui, C., and Dlugosz, A. 2000. Basal cell carcinomas in mice overexpressing Gli2 in skin. Nat. Genet. 24: 216-217. [DOI] [PubMed] [Google Scholar]
- Kalderon D. 2004. Hedgehog signaling: Costal-2 bridges the transduction gap. Curr. Biol. 14: R67-R69. [DOI] [PubMed] [Google Scholar]
- Kogerman P., Grimm, T., Kogerman, L., Krause, D., Undén, A.B., Sandstedt, B., Toftgård, R., and Zaphiropoulos, P.G. 1999. Mammalian suppressor-of-fused modulates nuclear-cytoplasmic shuttling of Gli-1. Nat. Cell Biol. 1: 312-319. [DOI] [PubMed] [Google Scholar]
- Koike C., Mizutani, T., Ito, T., Shimizu, Y., Yamamichi, N., Kameda, T., Michimukai, E., Kitamura, N., Okamoto, T., and Iba, H. 2002. Introduction of wild-type patched gene suppresses the oncogenic potential of human squamous cell carcinoma cell lines including A431. Oncogene 21: 2670-2678. [DOI] [PubMed] [Google Scholar]
- Kurzen H., Esposito, L., Langbein, L., and Hartschuh, W. 2001. Cytokeratins as markers of follicular differentiation: An immunohistochemical study of trichoblastoma and basal cell carcinoma. Am. J. Dermatopathol. 23: 501-509. [DOI] [PubMed] [Google Scholar]
- Lee Y.G., Macoska, J.A., Korenchuk, S., and Pienta, K.J. 2002. MIM, a potential metastasis suppressor gene in bladder cancer. Neoplasia 4: 291-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda O., Usami, N., Kondo, M., Takahashi, M., Goto, H., Shimokata, K., Kusugami, K., and Sekido, Y. 2004. Plakoglobin (γ-catenin) has TCF/LEF family-dependent transcriptional activity in β-catenin-deficient cell line. Oncogene 23: 964-972. [DOI] [PubMed] [Google Scholar]
- Matise M.P. and Joyner, A.L. 1999. Gli genes in development and cancer. Oncogene 18: 7852-7859. [DOI] [PubMed] [Google Scholar]
- Mattila P.K., Salminen, M., Yamashiro, T., and Lappalainen, P. 2003. Mouse MIM, a tissue-specific regulator of cytoskeletal dynamics, interacts with ATP-actin monomers through its C-terminal WH2 domain. J. Biol. Chem. 278: 8452-8459. [DOI] [PubMed] [Google Scholar]
- McGowan K.M. and Coulombe, P.A. 1998. Onset of keratin 17 expression coincides with the definition of major epithelial lineages during skin development. J. Cell Biol. 143: 469-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon A.P. 2000. More surprises in the Hedgehog signaling pathway. Cell 100: 185-188. [DOI] [PubMed] [Google Scholar]
- Methot N. and Basler, K. 2001. An absolute requirement for Cubitus interruptus in Hedgehog signaling. Development 128: 733-742. [DOI] [PubMed] [Google Scholar]
- Mill P., Mo, R., Fu, H., Grachtchouk, M., Kim, P.C., Dlugosz, A.A., and Hui, C.C. 2003. Sonic hedgehog-dependent activation of Gli2 is essential for embryonic hair follicle development. Genes & Dev. 17: 282-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon R.T., Bowerman, B., Boutros, M., and Perrimon, N. 2002. The promise and perils of Wnt signaling through β-catenin. Science 296: 1644-1646. [DOI] [PubMed] [Google Scholar]
- Murone M., Luoh, S.M., Stone, D., Li, W., Gurney, A., Armanini, M., Grey, C., Rosenthal, A., and deSauvage, F.J. 2000. Gli regulation by the opposing activities of fused and suppressor of fused. Nat. Cell Biol. 2: 310-312. [DOI] [PubMed] [Google Scholar]
- Nilsson M., Unden, A., Krause, D., Malmqwist, U., Raza, K., Zaphiropoulos, P., and Toftgard, R. 2000. Induction of basal cell carcinomas and trichoepitheliomas in mice overexpressing Gli-1. Proc. Natl. Acad. Sci. 97: 3438-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olave I.A., Reck-Peterson, S.L., and Crabtree, G.R. 2002. Nuclear actin and actin-related proteins in chromatin-remodeling. Annu. Rev. Biochem. 71: 755-781. [DOI] [PubMed] [Google Scholar]
- Oro A.E. and Higgins, K.M. 2003. Hair cycle regulation of Hedgehog signal reception. Dev. Biol. 255: 238-248. [DOI] [PubMed] [Google Scholar]
- Oro A.E., Higgins, K.M., Hu, Z., Bonifas, J.M., Epstein Jr., E.H., and Scott, M.P. 1997. Basal cell carcinomas in mice overexpressing Sonic hedgehog. Science 276: 817-821. [DOI] [PubMed] [Google Scholar]
- Pasca di Magliano M. and Hebrok, M. 2003. Hedgehog signalling in cancer formation and maintenance. Nat. Rev. Cancer 3: 903-911. [DOI] [PubMed] [Google Scholar]
- Prokhortchouk A., Hendrich, B., Jorgensen, H., Ruzov, A., Wilm, M., Georgiev, G., Bird, A., and Prokhortchouk, E. 2001. The p120 catenin partner Kaiso is a DNA methylation-dependent transcriptional repressor. Genes & Dev. 15: 1613-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz i Altaba A., Sanchez, P., and Dahmane, N. 2002. Gli and hedgehog in cancer: Tumours, embryos and stem cells. Nat. Rev. Cancer 2: 361-372. [DOI] [PubMed] [Google Scholar]
- Sasaki H., Hui, C., Nakafuku, M., and Kondoh, H. 1997. A binding site for Gli proteins is essential for HNF-3β floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development 124: 1313-1322. [DOI] [PubMed] [Google Scholar]
- Taipale J. and Beachy, P.A. 2001. The Hedgehog and Wnt signalling pathways in cancer. Nature 411: 349-354. [DOI] [PubMed] [Google Scholar]
- Taipale J., Chen, J.K., Cooper, M.K., Wang, B., Mann, R.K., Milenkovic, L., Scott, M.P., and Beachy, P.A. 2000. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature 406: 1005-1009. [DOI] [PubMed] [Google Scholar]
- Wang G., Wang, B., and Jiang, J. 1999. Protein Kinase A antagonizes Hedgehog signaling by regulating both the activator and repressor forms of Cubitus interruptus. Genes & Dev. 13: 2828-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodings J.A., Sharp, S.J., and Machesky, L.M. 2003. MIM-B, a putative metastasis suppressor protein, binds to actin and to protein tyrosine phosphatase delta. Biochem. J. 371: 463-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi A., Masuda, M., Ohki, T., Onishi, H., and Mochizuki, N. 2004. A novel actin-bundling/filopodium-forming domain conserved in insulin receptor tyrosine kinase substrate p53 and missing in metastasis protein. J. Biol. Chem. 279: 14929-14936. [DOI] [PubMed] [Google Scholar]
- Yoon J.W., Liu, C.Z., Yang, J.T., Swart, R., Iannaccone, P., and Walter-house, D. 1998. Gli activates transcription through a herpes simplex viral protein 16-like activation domain. J. Biol. Chem. 273: 3496-3501. [DOI] [PubMed] [Google Scholar]