Abstract
Mammalian genes frequently present allelic variants that differ in their expression levels and that, in the case of tumor suppressor genes, can be of relevance for cancer susceptibility and aging. We report here the characterization of a novel mouse model with increased activity for the Ink4a and Arf tumor suppressors. We have generated a “super Ink4a/Arf” mouse strain carrying a transgenic copy of the entire Ink4a/Arf locus. Cells derived from super Ink4a/Arf mice have increased resistance to in vitro immortalization and oncogenic transformation. Importantly, super Ink4a/Arf mice manifest higher resistance to cancer compared to normal, nontransgenic, mice. Finally, super Ink4a/Arf mice have normal aging and lifespan. Together, these results indicate that modest increases in the activity of the Ink4a/Arf tumor suppressor result in a beneficial cancer-resistant phenotype without affecting normal viability or aging.
Keywords: Ink4a, Arf, tumor suppression, stress, cancer resistance, aging, senescence
Cancer susceptibility is a quantitative genetic trait that results from complex genetic interactions among a large number of genes (Balmain 2002). It is now well established that for most genetic loci in the human genome there are frequent allelic variants that differ in their expression levels (Yan et al. 2002b; Lo et al. 2003). Based on these and similar observations (Yan and Zhou 2004), it is anticipated that alleles exist for cancer-relevant genes that are expressed at different levels in different individuals and, in this manner, have an impact on cancer susceptibility. For example, an expression allele of the APC tumor suppressor, whose expression is half of the normal allele, is responsible for some cases of familial adenomatous polyposis (Yan et al. 2002a). However, the consequences of modest increases in the activity of tumor suppressor genes remain largely unexplored, not only with regard to cancer susceptibility, but also in relation to aging. Notably, the existence of a reciprocal relation or trade between tumor suppression and aging has been proposed (Tyner et al. 2002; Campisi 2003; Maier et al. 2004). However, this antagonistic relation is based on a limited number of mouse models (see below) and, therefore, new mouse models are still necessary to understand the interplay among tumor suppression, cancer resistance, and aging.
Previously we have generated transgenic mice carrying a single additional copy of an intact p53 gene (Garcia-Cao et al. 2002). “Super p53” mice carrying three functional copies of p53, that is, the two endogenous copies plus the transgenic copy, can be conceptually assimilated to individuals carrying expression allelic variants of p53 that result in a global level of p53 activity that is 1.5-fold higher than normal. Characterization of the super p53 mice indicated that they benefit from a stronger DNA damage checkpoint and increased resistance to cancer without detectable negative effects on aging and lifespan (Garcia-Cao et al. 2002). Two additional mouse models have been reported in which p53 activity has been increased by N-terminal protein truncations that affect the normal properties and regulation of p53 (Tyner et al. 2002; Maier et al. 2004). These two mouse models with increased but aberrantly regulated p53 present accelerated aging and shorter lifespan. A critical difference between the various mouse models with increased p53 activity is that those that do not age prematurely have normally regulated p53 (Garcia-Cao et al. 2002), whereas those that age prematurely express truncated forms of p53 (Tyner et al. 2002; Maier et al. 2004). In this context, we considered the relevance of generating additional mouse models based on tumor suppressors other than p53.
The Ink4a/Arf locus encodes two tumor suppressor genes, p16Ink4a and p19Arf, which together with p53 are regarded as the most relevant tumor suppressors based on their frequent inactivation in human cancer (Ruas and Peters 1998; Malumbres and Barbacid 2001; Lowe and Sherr 2003). Adjacent to the Ink4a/Arf locus is a paralog of Ink4a, namely, the Ink4b gene that encodes p15Ink4b. Ink4b has a role in tumor suppression, although its relevance is more restricted than in the case of Ink4a (Ruas and Peters 1998; Malumbres and Barbacid 2001; Lowe and Sherr 2003). Both p16Ink4a and p15Ink4b inhibit the activity of the CDK4,6/cycD kinases, thus contributing to the maintenance of the active, growth-suppressive form of the retinoblastoma family of proteins (Lowe and Sherr 2003). On the other hand, the tumor suppressor p19Arf contributes to the stability of p53 by inhibiting the p53-degrading activity of MDM2 (Lowe and Sherr 2003). Through the activation of Rb and p53, the Ink4a/Arf locus is able to induce cell senescence and cell death. There is a large amount of evidence indicating that the Ink4a/Arf locus is a sensor of oncogenic stress, its expression being up-regulated upon the detection of aberrant oncogenic signals (Serrano et al. 1997; Palmero et al. 1998; Serrano and Blasco 2001). In addition, other types of stress have been found to up-regulate the expression of the Ink4a/Arf locus, such as prolonged cultivation in the absence of an appropriate extracellular matrix (Ramirez et al. 2001) or telomere dysfunction (Smogorzewska and de Lange 2002). In mice, it has been well established that the tumor suppression potency of Ink4a and Arf is quantitatively similar when each of them is individually considered (Sharpless et al. 2003, 2004).
In the present work, we generated mice that carry a modest increase in the activity of the Ink4a/Arf locus. This modest increase has a significant impact on cancer resistance and, at the same time, does not impinge on organismal viability or aging.
Results
Generation of transgenic Ink4a/Arf mice
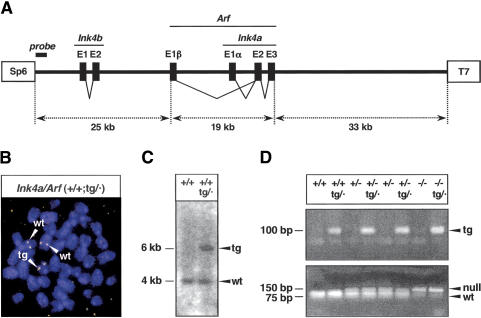
Transgenic Ink4a/Arf mice were obtained by injecting fertilized oocytes with a murine genomic segment of 77 kb that contains all the exons of p15Ink4b, p19Arf, and p16Ink4a (Fig. 1A). One transgenic line was obtained that transmitted the transgene and that appeared complete after analysis of its integrity by PCR and Southern blot (see Materials and Methods). Fluorescence in situ hybridization (FISH) of metaphases from mouse embryo fibroblasts (MEFs) carrying the transgene indicated the presence of a single integration site (Fig. 1B). Furthermore, analysis of transgenic-derived genomic DNA with a probe from a terminal region of the transgene detected the expected band produced by the endogenous alleles plus a single additional band corresponding to the insertion site of the transgene (Fig. 1C). These data strongly suggest that this line of mice contains a single copy of the transgene. In all of our subsequent functional analyses of the transgene, we focused on the functionality of Ink4a and Arf because these genes are known to be more important than Ink4b with regard to tumor suppression in mice (Latres et al. 2000; Sharpless et al. 2003, 2004). For this reason, and also for simplicity, we will refer to the transgenic allele as the Ink4a/Arf transgene, abbreviated as tg. Previous to the characterization of mice carrying the transgenic allele in a wild-type context, that is (+/+;tg/·), we performed the appropriate crosses to put the transgene in a genetic background that is null for the endogenous Ink4a/Arf locus (–/–;tg/·) (Fig. 1D) and, thus, analyzed the functionality of the Ink4a/Arf transgene in the absence of the endogenous genes.
Figure 1.
Generation of a transgenic Ink4a/Arf allele. (A) Structure of the transgene. The transgene contains the complete exon structure of three genes, Ink4b, Arf, and Ink4a. Boxes labeled SP6 and T7 represent short terminal sequences (of 20 bp) used for the identification of the transgene by PCR (see Materials and Methods). A thick bar near the SP6 terminus indicates the location of the probe used for the Southern blot shown in C. (B) Fluorescent in situ hybridization of the transgene in a representative MEF metaphase. The fluorescent probe consists of the complete transgene. The transgene is located in a chromosome that is smaller than the two homologs containing the endogenous alleles. (C) Southern blot to detect the SP6-terminal region of the transgene. Genomic DNA digested with BamHI produced a band of ∼4 kb, present in both wild-type and transgenic mice, and a single additional band of ∼6 kb specific to transgenic mice. The probe used consisted of a PCR fragment of ∼380 bp (schematically shown in A) that was generated by primers SP6-PAC and SP6-tg (see Materials and Methods). (D) PCR genotyping of the various Ink4a/Arf alleles. (Top) Detection of the transgene by a PCR reaction based on the T7-terminal sequence of the transgene. (Bottom) The endogenous Ink4a/Arf allele was indirectly genotyped through a linked polymorphism in the Tyrp1 (tyrosinase-related protein 1) locus, which is located at a genetic distance of ∼4.7 cM. The Ink4a/Arf-wild-type (wt) allele is linked to the wild-type Tyrp1 allele, whereas the Ink4a/Arf-null allele is linked to the mutant Tyrp1(brown) allele (Sviderskaya et al. 2003). The two Tyrp1 alleles can be distinguished by a diagnostic TaqI site (Zdarsky et al. 1990) present in Tyrp1-wt (linked to Ink4a/Arf-wt), but absent in Tyrp1(brown) (linked to Ink4a/Arf-null). A PCR product that contains the reported TaqI polymorphism was digested with TaqI to determine the presence or absence of the TaqI site.
The Ink4a/Arf transgene is functional in MEFs
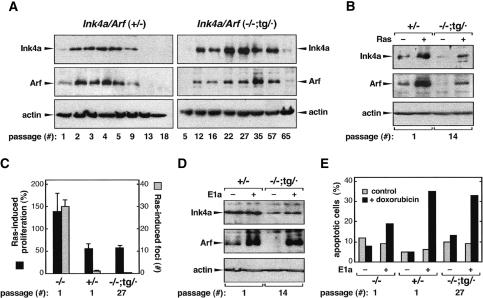
We began by analyzing the functionality of the transgenic allele in MEFs using well established in vitro assays. Under standard cultivation conditions, MEFs experience stress that results in the progressive accumulation of p16Ink4a and p19Arf (Palmero et al. 1997; Zindy et al. 1997, 1998). Typically, the endogenous Ink4a/Arf locus becomes active during the very first passages (3T3 serial passage protocol), reaching maximal levels before passage 5 (Fig. 2A, left panel). We observed that the transgene was activated by in vitro culture stress in Ink4a/Arf (–/–;tg/·) MEFs although, in contrast to the endogenous locus, its activity became noticeable after passage 5, reaching its maximum between passages 30 and 40 (Fig. 2A, right panel; see also Supplementary Fig. S1). It is interesting to note that the kinetics of activation of Ink4a and Arf were parallel within a given culture, indicating that, as in the case of the endogenous locus, the transgenic allele retains coordinated activation of the two genes in response to culture stress. Further recapitulating the behavior of Ink4/Arf (+/–) MEFs, continued passage of Ink4a/Arf (–/–;tg/·) cultures beyond the point of maximal accumulation of p16Ink4a and p19Arf favored the selection of cells that did not express these proteins (Fig. 2A; Supplementary Fig. S1). In fact, high-passage Ink4a/Arf (–/–;tg/·) cultures (>60 passages) invariably lost the transgene, as determined by Southern blot (data not shown). The observed selection against cells that express transgenic-derived p16Ink4a or p19Arf reflects the antiproliferative activity of the transgenic Ink4a/Arf allele.
Figure 2.
Function of the Ink4a/Arf transgene in MEFs. (A) Activation of the transgenic Ink4a/Arf allele in response to in vitro culture stress. MEFs of the indicated genotype were serially passaged according to the 3T3 protocol. In the case of Ink4a/Arf (–/–;tg/·) MEFs (right panels), transgene-produced p16Ink4a and p19Arf reached maximal levels at around passage 35. In the case of Ink4a/Arf (+/–) MEFs (left panels), maximal levels were reached at around passage 4. In both cases, cells expressing p16Ink4a and p19Arf, either from the transgene or from the endogenous allele, were overgrown by cells that had lost expression of p16Ink4a and p19Arf. This overall pattern was reproducibly observed in four independent (–/–;tg/·) MEF cultures, each from a different embryo (see Supplementary Fig. S1). (B) Activation of the transgenic Ink4a/Arf allele in response to Ras-induced oncogenic stress. MEFs of the indicated genotype and passage were retrovirally transduced with an empty vector or with a vector expressing oncogenic H-RasV12, and the levels of p16Ink4a and p19Arf were determined at day 2 post-selection (see Supplemental Material). Immunoblots are representative of three assays with independent MEF cultures. (C) The transgenic Ink4a/Arf allele provides neoplastic resistance to oncogenic Ras. (Black bars) MEFs of the indicated genotype and passage were infected with oncogenic Ras or with empty vector, and their proliferation was measured at day 0 and day 4 post-selection. The ratio between day 4 and day 0 in Ras-infected cells was normalized as the percentage of the corresponding ratio in vector-infected cells. (Gray bars) MEFs (106 cells) were transfected with a plasmid encoding oncogenic Ras, and 3 wk later the total number of neoplastic foci was scored. Data for proliferation and neoplastic foci correspond to the average and standard deviation of three and five assays, respectively, with independent MEF cultures. (D) Activation of the transgenic Ink4a/Arf allele in response to E1a-induced oncogenic stress. MEFs were retrovirally transduced with an empty vector or with a vector expressing E1a, and the levels of p16Ink4a and p19Arf were determined by immunoblotting. Data are representative of three assays with independent MEF cultures. (E) The transgenic Ink4a/Arf allele provides increased sensitivity to E1a-mediated apoptosis. MEFs retrovirally transduced with E1a (+) or empty vector (–) were treated or not with doxorubicin, as indicated, and apoptotic cells were quantified 24 h later by cytometry. Data correspond to a representative assay from a total of three assays with independent MEF cultures.
Oncogenic stress is regarded as the most relevant stimulus sensed by the Ink4a/Arf locus (Serrano and Blasco 2001; Lowe and Sherr 2003). In particular, high levels of oncogenic Ras up-regulate both p16Ink4a and p19Arf, which results in decreased proliferation and resistance to neoplastic transformation (Serrano et al. 1997; Palmero et al. 1998). On the other hand, the viral oncoprotein E1a activates p19Arf, but not p16Ink4a, and results in increased sensitivity to undergo apoptosis (de Stanchina et al. 1998). The transgenic Ink4a/Arf allele retained its ability to sense oncogenic stress and, thus, oncogenic Ras up-regulated both p16Ink4a and p19Arf (Fig. 2B), whereas E1a up-regulated p19Arf (Fig. 2D). The up-regulation of transgene-derived p16Ink4a and p19Arf upon oncogenic stress was observed regardless of the passage number of the cells, but the absolute levels were proportionate to the corresponding basal levels (data not shown). Accordingly, the biological effects of the transgenic allele were evident only in MEFs at high-passage (negative data for low-passage MEFs are not shown). High-passage (–/–;tg/·) MEFs (passage 27) decreased their proliferation after introduction of oncogenic Ras (Fig. 2C, black bars) and were resistant to Ras-driven neoplastic transformation (Fig. 2C, gray bars). This behavior was similar to that of (+/–) MEFs, and opposite to that of (–/–) MEFs (Fig. 2C). In the case of E1a, high-passage (–/–;tg/·) MEFs (passage 27) became sensitized to DNA damage-induced apoptosis in a manner comparable to (+/–) MEFs (Fig. 2E).
These observations indicate that the transgenic Ink4a/Arf allele is partially attenuated compared to the endogenous allele but retains its main attributes as a sensor of oncogenic stress, with the ability to prevent neoplastic transformation in vitro.
The Ink4a/Arf transgene is functional in mice
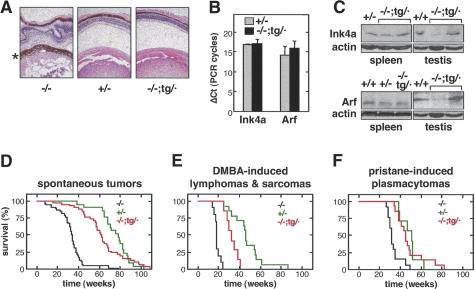
Absence of functional Ink4a or Arf has well characterized effects in mice. We began by analyzing a developmental phenotype that is proprietary of Arf deficiency, namely, the aberrant presence of vascular embryonic remnants in the adult eye that results in severe ocular aberrations (McKeller et al. 2002). As reported, we observed retrolental tissue and morphological aberrations in the retina and lens of Arf (–/–) mice (data not shown), as well as in Ink4a/Arf (–/–) mice (Fig. 3A, left). In contrast, the eyes of Ink4a/Arf (–/–;tg/·) and (+/–) mice were normal and had no evidence of retrolental tissue (Fig. 3A, center and right). These results indicate that the transgene is functional regarding the activity of p19Arf in the regression of the embryonic hyaline vasculature of the eye.
Figure 3.
Function of the Ink4a/Arf transgene in vivo. (A) The transgenic Ink4a/Arf allele rescues the aberrant eye phenotype of Arf-deficient mice. The pictures show representative sections of eyes from adult mice (8–12 mo old) of the indicated genotypes stained with hematoxylin and eosin. In agreement with the previously reported phenotype of Arf-null mice (McKeller et al. 2002), the eyes of Ink4a/Arf (–/–) mice are severely distorted due to an aberrant layer of tissue derived from the embryonic hyaline vasculature (marked with an asterisk). The eyes of Ink4a/Arf (–/–;tg/·) mice were normal and indistinguishable from the eyes of Ink4a/Arf (+/–) mice. (B) The transgenic Ink4a/Arf allele directs the expression of Ink4a-specific and Arf-specific mRNA. Ink4a and Arf transcripts were detected by real-time quantitative RT–PCR using primers that specifically distinguish both transcripts. Values indicate the average and standard deviation of the difference in PCR cycles (ΔCt) with respect to β-actin (it should be noted that the higher the value, the lower the expression). The observed differences were not statistically significant. Total RNA was obtained from testis of mice (∼1.5 yr old) of the indicated genotype. Each measure is derived from at least two independent assays, each of them with triplicate PCRs. (C) Transgenic Ink4a/Arf mice express transgene-derived p16Ink4a and p19Arf proteins. Protein extracts were obtained from spleen and testis of mice (∼1.5 yr old) of the indicated genotype. The samples from testis in the Ink4a and Arf immunoblots correspond to the same mice. The two lanes labeled (–/–;tg/·) correspond to different mice. (D) The transgenic Ink4a/Arf allele delays spontaneous tumorigenesis in the absence of endogenous Ink4a/Arf alleles. Survival curve of mice of the indicated genotype. The size of each group of mice was as follows: (–/–), n = 32; (+/–), n = 14; (–/–;tg/·), n = 36. Table 1 shows the incidence of spontaneous neoplasias in these mice. (E) The transgenic Ink4a/Arf allele delays DMBA-induced tumorigenesis in the absence of endogenous Ink4a/Arf alleles. Survival curve of mice treated with DMBA to induce lymphomas and fibrosarcomas (see Table 1). The size of each group of mice was as follows: (–/–), n = 14; (+/–), n = 14; (–/–;tg/·), n = 13. (F) The transgenic Ink4a/Arf allele delays pristane-induced tumorigenesis in the absence of endogenous Ink4a/Arf alleles. Survival curve of mice treated with pristane to induce plasmacytomas and peritoneal tumors. The size of each group of mice was as follows: (–/–), n = 13; (+/–), n = 7; and (–/–;tg/·), n = 14.
In mice, the Ink4a/Arf locus is expressed at very low levels, and it is generally detectable only in adult or old mice (Palmero et al. 1997; Zindy et al. 1997, 1998). By quantitative RT–PCR, we were able to detect the mRNA coding for p16Ink4a and p19Arf in testis from (–/–;tg/·) mice, and their respective levels were not significantly different from those in (+/–) mice (Fig. 3B). Also, we detected transgene-derived proteins p16Ink4a and p19Arf in spleen and testis from (–/–;tg/·) mice and, again, at levels generally similar to those of (+/–) mice (Fig. 3C).
The tumor suppressor activity of the transgenic Ink4a/Arf allele was evaluated by monitoring the appearance of spontaneous and chemically induced tumors in groups of mice of the three relevant genotypes, namely (+/–), (–/–;tg/·), and (–/–). The viability of Ink4a/Arf (–/–;tg/·) mice was significantly higher than the viability of Ink4a/Arf (–/–) mice, although not identical to that of Ink4a/Arf (+/–) mice (Fig. 3D). The tumor spectrum of Ink4a/Arf (–/–;tg/·) mice was characterized by a high prevalence of histiocytic sarcomas and a modest incidence of lymphomas and fibrosarcomas, a pattern of tumors broadly similar to that of (+/–) mice (Table 1). In contrast, Ink4a/Arf (–/–) mice had a high incidence of both histiocytic sarcomas and lymphomas (Table 1). The tumor suppressor activity of the transgenic allele was further evaluated using chemical carcinogenesis. Treatment of neonates with DMBA induced a variety of tumor types (Table 1), and Ink4a/Arf (–/–;tg/·) mice showed a level of cancer resistance that was significantly above that of Ink4a/Arf (–/–) mice (Fig. 3E). Moreover, in the case of pristane-induced plasmacytomas, Ink4a/Arf (–/–;tg/·) mice had a level of susceptibility that was indistinguishable from that of Ink4a/Arf (+/–) mice (Fig. 3F).
Table 1.
Tumor spectrum in Ink4a/Arf (-/-;tg/·) mice
|
Ink4a/Arfa
|
|||
|---|---|---|---|
| (-/-) | (+/-) | (-/-;tg/·) | |
| Spontaneous tumors | |||
| histiocytic sarcomas | 16/31 (52%) | 9/13 (69%) | 16/29 (55%) |
| lymphomas | 14/31 (45%) | 3/13 (23%) | 5/29 (17%) |
| fibrosarcomas | 6/31 (19%) | 1/13 (8%) | 5/29 (17%)b |
| otherc | 2/31 (6%) | 0/13 (0%) | 3/29 (10%) |
| DMBA-induced tumors | |||
| histiocytic sarcomas | 1/13 (8%) | 4/10 (40%) | 0/11 (0%) |
| lymphomas | 3/13 (23%) | 1/10 (10%) | 3/11 (27%) |
| fibrosarcomas | 11/13 (85%) | 1/10 (10%) | 5/11 (45%) |
| otherd | 0/13 (0%) | 3/10 (30%) | 4/11 (36%) |
Number of affected mice followed by the total number of mice (the corresponding percentage is given in parentheses). Some mice developed more than one tumor
Two myosarcomas have been included in this group
In the (-/-) mice, the two tumors correspond to hemangiosarcomas; in the (-/-;tg/·) mice the tumors were one hemangiosarcoma, one liver carcinoma, and one lung adenocarcinoma
In the (+/-) mice, the three tumors correspond to two pituitary adenocarcinomas and one hemangiosarcoma; in the (-/-;tg/·) mice, the tumors were three skin carcinomas and one pituitary adenocarcinoma
Collectively, the ability of the Ink4a/Arf transgenic allele to correct the aberrant eye phenotype of Arf-null mice, its normal expression levels in vivo, and its ability to provide significant levels of cancer protection unequivocally demonstrate that the transgenic allele is active and functional in mice.
`Super Ink4a/Arf' MEFs have increased resistance to in vitro transformation
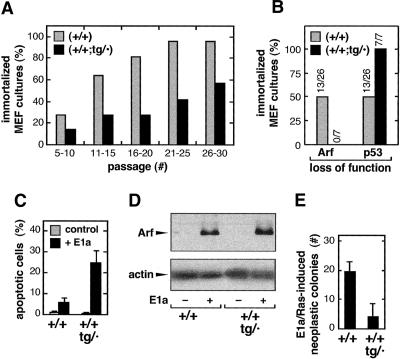
Having demonstrated the functionality of the transgenic Ink4a/Arf allele, we examined the impact of the transgene when present in combination with the normal genetic dose of two Ink4a/Arf wild-type loci. We began by comparing the immortalization and transformation properties of (+/+;tg/·) and (+/+) MEFs. “MEF senescence” (or “culture shock”) occurs after serial cultivation in vitro due to the activation of the p19Arf/p53 pathway (Zindy et al. 1997, 1998). In most cultures, the senescent population of cells is eventually overgrown by mutant cells that have stochastically lost either p19Arf or p53 function, thus giving rise to an immortal culture. Individually derived MEF preparations of (+/+;tg/·) or (+/+) genotype were serially cultivated until immortalization, and then analyzed for p19Arf or p53 function. Figure 4A represents the percentage of cultures of each genotype that had become immortal at different passage intervals. Remarkably, by passages 21–25, ∼95% of the (+/+)-derived cultures were immortal, whereas at that interval only 40% of the (+/+;tg/·)-derived cultures were immortal. Thus, spontaneous immortalization of “super Ink4a/Arf” MEFs was less efficient than in the case of wild-type MEFs. Interestingly, spontaneously immortalized wild-type MEFs lost p19Arf and p53 with similar frequencies, whereas immortalized super Ink4a/Arf MEFs always lost p53 and retained p19Arf (Fig. 4B). These observations demonstrate that the transgenic allele present in (+/+;tg/·) MEFs results in a less probable loss of p19Arf and, consequently, in a lower efficiency of immortalization.
Figure 4.
MEFs derived from super Ink4a/Arf mice have increased resistance to immortalization and oncogenic transformation. (A) Super Ink4a/Arf MEFs have increased resistance to spontaneous immortalization. Cultures derived from independent embryos, either wild-type (n = 27) or super Ink4a/Arf (n = 7) were passaged according to the 3T3 protocol. Cultures were maintained until immortalization. Bars represent the percentage of cultures that have attained immortalization at each of the indicated passage intervals. (B) Spontaneously immortalized super Ink4a/Arf MEFs preferentially lose p53 function. Spontaneously immortalized MEFs derived from wild-type (n = 26) or super Ink4a/Arf (n = 7) MEFs (see A) were analyzed for the loss of p19Arf (by immunoblot) and for the loss of p53 function (by testing the stabilization of p53 upon doxorubicin treatment). Bars represent the percentage of analyzed cultures according to their mechanism of immortalization. (C) Super Ink4a/Arf MEFs have increased sensitivity to E1a-mediated apoptosis. Early-passage MEFs of the indicated genotype were retrovirally transduced with E1a (+) or empty vector (–), and the basal level of apoptosis was measured by cytometry. Data correspond to the average and standard deviation of four assays with independent MEF cultures. (D) Super Ink4a/Arf MEFs present increased levels of p19Arf in response to E1a. MEFs from the experiment shown in C were used to prepare protein extracts, and the levels of p19Arf were measured by immunoblots. Similar results were obtained in two additional independent assays. (E) Super Ink4a/Arf MEFs have increased resistance to neoplastic transformation by cooperating oncogenes Ras and E1a. Early-passage MEFs of the indicated genotype were retrovirally transduced with a construct that simultaneously expresses oncogenic Ras and E1a. After selection, infected MEFs (2 × 105 cells) were plated and, after 3 wk, the number of neoplastic colonies was scored. Data correspond to the average and standard deviation of three assays with independent MEF cultures.
To evaluate the functional consequences of the transgene in response to oncogenes, we first measured the sensitization to apoptosis triggered by the viral oncoprotein E1a, a response that is known to depend largely on p19Arf (de Stanchina et al. 1998). Expression of E1a in (+/+;tg/·) MEFs resulted in higher levels of constitutive apoptosis and p19Arf compared to E1a-expressing (+/+) MEFs (Fig. 4C,D). Finally, we compared the susceptibility of (+/+;tg/·) and (+/+) MEFs to neoplastic transformation. For this, MEFs were retrovirally transduced with the cooperating oncogenes E1a and H-RasV12 and, after plating the same number of infected cells, we counted the resulting number of foci. As shown in Figure 4E, MEFs carrying the Ink4a/Arf transgenic allele were significantly more resistant to neoplastic transformation. In conclusion, cells endowed with the extra Ink4a/Arf function provided by the transgene are more resistant to immortalization and oncogenic transformation.
Super Ink4a/Arf mice have increased resistance to cancer
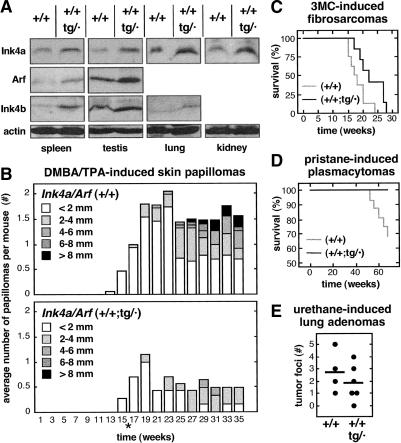
Previous to the analysis of the cancer susceptibility of super Ink4a/Arf mice, we asked whether the expression of the proteins encoded by the transgene was aberrant or abnormal in the context of a wild-type genetic background. We observed that the levels of p16Ink4a, p19Arf, and p15Ink4b were moderately, although reproducibly, increased in various tissues from super Ink4a/Arf mice compared to wild-type mice (Fig. 5A). At this point, it is pertinent to remind that the Ink4a/Arf transgene also contains the Ink4b gene encoding p15Ink4b (see Fig. 1A). Moreover, it is worth mentioning that these data were obtained with old mice (∼1.5 yr old), thus excluding the possibility that the transgene could become silent at advanced ages. Finally, it is important to emphasize that, from the available data, there is no evidence of overt deregulation of any of the proteins encoded by the transgene in a variety of tissues, thus suggesting that the transgene retains an overall normal regulation, not significantly different from the endogenous locus.
Figure 5.
Super Ink4a/Arf mice have increased resistance to cancer. (A) Moderately increased expression of p16Ink4a, p19Arf, and p15Ink4b in super Ink4a/Arf mice. Extracts from the indicated organs were obtained from mice that were ∼1.5 yr old. In some tissues, expression levels were too low and, therefore, the corresponding immunoblots are not shown. For each tissue, similar data were obtained with at least two mice of each genotype. (B) Super Ink4a/Arf mice have increased resistance to DMBA/TPA-induced skin papillomas. The figure shows the average number of papillomas at every week, and their relative size distribution. Initiation treatments (DMBA application) were applied when mice were 1 wk old and, 3 wk later, promotion treatments began (TPA applications) and continued for an additional 12 wk. At 16 wk of age (indicated by *) treatment was terminated. The size of each group of mice was as follows: (+/+), n = 15; (+/+;tg/·), n = 7. (C) Super Ink4a/Arf mice have increased resistance to 3MC-induced fibrosarcomas. Survival curves of mice treated with 3MC. The size of each group of mice was as follows: (+/+), n = 8; (+/+;tg/·), n = 7. (D) Super Ink4a/Arf mice have increased resistance to pristane-induced plasmacytomas. Survival curves of mice treated with pristane. The size of each group of mice was as follows: (+/+), n = 16; (+/+;tg/·), n = 10 (E) Super Ink4a/Arf mice have increased resistance to urethane-induced lung adenomas. Dots indicate the number of visible tumors per mouse; horizontal lines indicate the average. The size of each group of mice was as follows: (+/+), n = 4; (+/+;tg/·), n = 6.
Super Ink4a/Arf mice, carrying the two wild-type endogenous loci together with the transgenic allele, were challenged with carcinogenic treatments to evaluate their susceptibility to a variety of tumors. Groups of (+/+;tg/·) and (+/+) mice were treated with carcinogens to induce skin papillomas, fibrosarcomas, plasmacytomas, and lung adenomas (Fig. 5B–E). In all cases, (+/+;tg/·) mice were more resistant to tumors than their (+/+) littermates, although the impact of extra Ink4a/Arf activity was more prominent in the case of skin papillomas (Fig. 5B), fibrosarcomas (Fig. 5C), and plasmacytomas (Fig. 5D). Furthermore, the available data with a limited number of mice that died spontaneously also suggests a lower incidence of spontaneous tumors (Table 2). In particular, six out of 10 wild-type mice (60%) presented spontaneous tumors, whereas only two of nine super Ink4a/Arf mice (22%) had tumors upon death. Together, these results indicate that super Ink4a/Arf mice are more cancer-resistant than their wild-type littermates. In conclusion, and paralleling the super p53 mice previously generated by us (Garcia-Cao et al. 2002), additional Ink4a/Arf activity confers a generalized increased resistance to cancer.
Table 2.
Aging-associated pathologies in Ink4a/Arf (+/+) and (+/+;tg/·) mice
| Genotype | Mousea (#, sex) | Lifespan (months) | Main pathological findings | Presence of neoplasias |
|---|---|---|---|---|
| (+/+)
|
1, F | 31 | multicentric lymphoma | yes |
| 2, M | 26 | without significant findings | no | |
| 3, M | 25 | multicentric lymphoma | yes | |
| 4, M | 24 | without significant findings | no | |
| 5, M | 24 | liver carcinoma & liver metastasis | yes | |
| 6, M | 21 | hemangiosarcoma and lymphoma | yes | |
| 7, F | 21 | ulcerative cutaneous papilloma | yes | |
| 8, F | 17 | without significant findings | no | |
| 9, F | 17 | without significant findings | no | |
| 10, M | 13 | ulcerative tricoepithelioma | yes | |
|
|
|
TOTAL MICE WITH NEOPLASIAS
|
6/10 (60%)
|
|
| (+/+;tg/·)
|
1, F | 34 | without significant findings | no |
| 2, F | 34 | without significant findings | no | |
| 3, F | 25 | cutaneous papilloma | yes | |
| 4, F | 25 | without significant findings | no | |
| 5, M | 24 | lung congestion | no | |
| 6, M | 23 | without significant findings | no | |
| 7, F | 22 | splenic lymphoma, pneumonia | yes | |
| 8, M | 13 | without significant findings | no | |
| 9, F | 12 | without significant findings | no | |
| TOTAL MICE WITH NEOPLASIAS | 2/9 (22%) |
The table includes animals that died spontaneously at an advanced age; some of them were not part of the cohorts analyzed in Figure 6B
Super Ink4a/Arf mice have normal aging
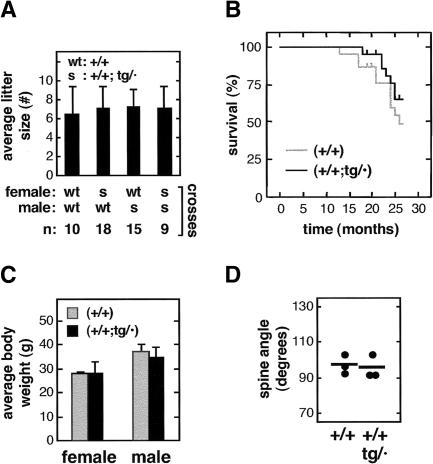
An important issue regarding tumor suppression is its interplay with aging (Campisi 2003). We have determined a number of quantitative indicators of viability, fertility, and aging. As a first indication of viability, the birth ratio of transgenic mice was normal. Specifically, (+/+) × (+/+;tg/·) crosses produced transgene-carrying mice at a ratio of 49% (total progeny analyzed, n = 235). The fertility of transgene-carrying mice was also normal for both sexes as deduced from their average litter size (Fig. 6A). Observation of cohorts of mice for more than 2 yr revealed similar survival rates for wild type and super Ink4a/Arf (Fig. 6B). The slight increase in survival observed in super Ink4a/Arf mice was not significant (log-rank test, p = 0.3), although it may reflect to some extent the lower incidence of spontaneous neoplasias in aged super Ink4a/Arf mice (see Table 2). In this regard, it is important to emphasize that the presence of neoplasias in old moribund mice does not necessarily imply that the neoplasia is the cause of death. We also recorded the weight of (+/+) and (+/+;tg/·) mice, and we did not observe significant differences between them, neither when the mice were young (data not shown) nor when they reached 2 yr of age (Fig. 6C). Finally, we evaluated biomarkers of aging. Specifically, the angle of the spine (lordokyphosis) was similar between wild-type and super Ink4a/Arf mice 2 yr of age (Fig. 6D). Moreover, analysis of bone density and skin structure did not show any significant difference between wild-type and super Ink4a/Arf mice at 2 yr of age (data not shown). Altogether, our findings indicate that the fertility, viability, aging, and lifespan of super Ink4a/Arf (+/+;tg/·) mice are normal.
Figure 6.
Normal viability and aging of super Ink4a/Arf mice. (A) Super Ink4a/Arf mice have normal fertility. Average litter size and standard deviation of the indicated crosses. The number of litters scored for each type of cross is indicated (n). (B) Super Ink4a/Arf mice have normal lifespan. Cohorts of wild-type (n = 22) and super Ink4a/Arf (n = 22) littermate mice were followed for a period of 27 mo. The figure shows a Kaplan-Meier representation of the survival of the two groups of mice. (C) Old super Ink4a/Arf mice have normal weight. Average weight and standard deviation of the indicated mice. All mice were 2 yr of age. The size of each group of mice was as follows: (+/+) females, n = 5; (+/+) males, n = 8; (+/+;tg/·) females, n = 7; (+/+;tg/·) males, n = 9. (D) Similar lordokyphosis in old super Ink4a/Arf and wild-type mice. Dots indicate the angle of the dorsal spine measured from X-radiographs of individual mice at 2 yr of age, as described (Garcia-Cao et al. 2002). Short lines indicate the average. The size of each group of mice was as follows: (+/+), n = 3; (+/+;tg/·), n = 3.
Discussion
In this work, we generated a mouse model with increased functionality of the Ink4a/Arf tumor suppressor locus. The transgene introduced into these mice contains three tumor suppressor genes (Ink4a, Arf, and Ink4b) which produce moderately increased levels of the corresponding proteins, and retains its main function as a sensor of oncogenic stress. However, quantitative analyses, both in vivo and in cultured cells, indicate that the functionality of the transgene is partially attenuated compared to the endogenous allele. This attenuation of the transgene is likely due to the absence of positive modulatory elements or, alternatively, of insulator elements that prevent negative effects from the surrounding chromatin. In any case, the relevant feature of our transgenic allele is that it confers significant levels of tumor suppression. In fact, super Ink4a/Arf mice, carrying the transgene in addition to the endogenous alleles, are more resistant to the development of a variety of chemically induced tumor types and have a lower incidence of spontaneous tumors.
The analysis of in vitro cultured MEFs suggests two mechanisms that explain the additional tumor suppression provided by the transgene. First, oncogenic stress triggers more potent anti-oncogenic responses in super Ink4a/Arf MEFs, as indicated by the increased apoptotic levels elicited by E1a, or by the decreased number of neoplastic foci produced by the cooperating oncogenes Ras and E1a. Additionally, the transgene constitutes an extra gene copy that makes less probable the stochastic loss of Ink4a/Arf. This latter concept is illustrated by the observation that spontaneously immortalized super Ink4a/Arf MEFs retained p19Arf expression but lost p53 in all of the cases analyzed, a situation that contrasts with wild-type MEFs that lose p19Arf or p53 with similar frequencies. Therefore, the cellular effect of the Ink4a/Arf transgene has two aspects: stronger anti-oncogenic responses, and lower frequency of stochastic loss of function.
The super Ink4a/Arf mice described here not only contribute to the understanding of cancer susceptibility; they are also relevant regarding the interplay between tumor suppression and aging. Three mouse models were previously generated based on modifications of the p53 gene. Two of those models are based on truncations of p53 that produce abnormally regulated p53 activity and result in shortened lifespan (Tyner et al. 2002; Maier et al. 2004). In contrast, the super p53 mice we previously generated carry an additional copy of an intact, not manipulated, p53 gene and, importantly, these mice are cancer-resistant while retaining normal lifespan (Garcia-Cao et al. 2002). The present super Ink4a/Arf mice provide an independent demonstration of the concept that it is possible to increase cancer resistance without a negative impact on viability. A unifying explanation for the presence or absence of an aging phenotype in these mouse models with increased tumor suppression is that the proposed trade between tumor suppression and aging (Campisi 2003) only operates when tumor suppressors are activated beyond a certain threshold or when they are deregulated, but not when they are moderately increased and retain their basic regulatory features.
The concept of increased tumor suppression in the absence of detrimental effects is relevant in the context of the recently appreciated variability in the expression levels among the different alleles of a same gene (Yan and Zhou 2004). Small variations in the DNA, even at distant, noncoding locations may underlie this variability in expression. In addition, it is worth noting that expression levels can vary due to the presence of duplications in the genome. These duplications appear to vary between individuals and imply a diversity of loci across the genome, probably in a random manner, and resulting in copy number variations of the genes involved (Carter 2004; Iafrate et al. 2004; Sebat et al. 2004). A well characterized example of natural copy number variation is the gene for the CCL3-L1 chemokine that may vary in Caucasians from zero to six copies per diploid genome and which translates in a corresponding increase in the expression of this chemokine in vivo (Townson et al. 2002). Based on the phenotype of the super Ink4a/Arf and super p53 mice, we anticipate the existence of human allelic variants or genomic duplications that result in moderately increased levels of expression of tumor suppressor genes and that are associated with a genetically inherited resistance to cancer without accompanying detrimental effects, such as premature aging.
Materials and methods
Transgenesis
A large genomic DNA segment (77 kb) from C57BL6 mice, containing Ink4b, Arf, and Ink4a and cloned into the PAC (P1 artificial chromosome) vector pAd10SacBII was kindly provided by Gordon Peters (Cancer Research UK, London). The terminal sequences (∼500 bp) were determined with primers complementary to the insert-flanking elements of the vector, namely, SP6-PAC (5′-ATTTAGGTGACACTATAG-3′) and T7-PAC (5′-TAATACGACTCACTATAGGG-3). For transgenesis, the Ink4b/Arf/Ink4a-containing PAC was digested with SfiI, thus linearizing the PAC and resulting in the following structure: SfiI-SP6-[Ink4b/Arf/Ink4a]-T7-vector-SfiI. Pronuclei of fertilized oocytes, derived from intercrosses between (C57BL6 × CBA)F1 mice, were injected with ∼2 pL of a DNA solution containing the linearized PAC (Garcia-Cao et al. 2002). The resulting offspring were analyzed for the presence of the transgene using PCR reactions that detect the terminal transgenic sequences adjacent to the SP6 or T7 flanking elements: for the SP6 terminus, primers SP6-PAC (see above) and SP6-tg (5′-AG ATGGGTTTAGTGCAGA-3′); and for the T7 terminus, T7-PAC (see above) and either one of the following two primers T7-tg1 (5′-AGTCAGAGGGTGGATGGG-3′) or T7-tg2 (5′-AAGCACTGAACCACCAAA-3′). From a total of 28 mice derived from microinjected oocytes, three were identified as carriers by PCR, and two of them transmitted the transgene through the germline. After the appropriate crosses to put the transgene into a genetic background that was null for the endogenous Ink4a and Arf alleles, it was found by PCR that one of the transgenic lines had an internal deletion in the transgene affecting exon 2 of Ink4a/Arf and, therefore, did not express any of the two proteins. Thus, only one transgenic line was successfully generated, and this allele was named tg.
Mouse breeding and monitoring
The founder Ink4a/Arf (+/+;tg/·) mouse had a mixed genetic background C57BL6:CBA (50%:50%), and its progeny were backcrossed for at least three generations with pure C57BL6 mice of either Ink4a/Arf (+/+) or Ink4a/Arf (–/–) genotype (Serrano et al. 1996). In this manner, all of the mice used in this study shared a genetic background that is at least 93.75% C57BL6. Mice were housed at the pathogen-free barrier areas of the Spanish National Center of Biotechnology and the Spanish National Cancer Center, where they were observed on a daily basis. Upon signs of morbidity or overt tumors, mice were euthanized in accordance with the Guidelines for Humane Endpoints for Animals Used in Biomedical Research. Mouse tissues were fixed in 10% buffered formalin and embedded in paraffin, and 4-mm sections were stained with Harris hematoxylin and eosin according to standard procedures. Histological analysis of the major organs and tissues was performed in all mice.
Mice were genotyped by PCR. The presence of the transgene was detected using the following pairs of primers (see their sequences above): SP6-PAC and SP6-tg, T7-PAC and T7-tg1, and T7-PAC and T7-tg2. The wild-type and transgenic Ink4a/Arf loci were detected using primers against exon 2, specifically, mp16F (5′-ATGATGATGGGCAACGTTC-3′) and mp16R (5′-CAAATATCGCACGATGTC-3′). The Ink4a/Arf-null allele (Serrano et al. 1996) was detected using primers Neo038 (complementary to the neo-cassette, 5′-CTATCAGGACATAGCG TTGG-3′) and R1 (complementary to flanking genomic DNA, 5′-AGTGAGAGTTTGGGGACAGAG-3′). Finally, we also genotyped the gene encoding tyrosinase-related protein 1, Tyrp1, for the presence of the brown mutation, because the Ink4a/Arf-null allele is genetically linked to the Tyrp1(brown) mutant allele (Sviderskaya et al. 2003). In this manner, it was possible to discriminate (+/–;tg/·)and(–/–;tg/·) genotypes. A segment of the coding region of Tyrp1 was amplified using primers Tyrp1F (5′-CTGCGATGTCTGCACTGATGACTT-3′) and Tyrp1R (5′-AGGGTATCTCGTACTCTTCCAAGGAT-3′), which amplify a product of 126 bp. The presence of a diagnostic TaqI site present in the wild-type allele but absent in the Tyrp1(brown) allele (Zdarsky et al. 1990) was detected by subsequent TaqI digestion of the amplified product.
Assays with MEFs
Isolation, culture, and assays with mouse embryo fibroblasts (MEFs) were done as described (Pantoja and Serrano 1999; Palmero and Serrano 2001). Oncogenic Ras (H-rasV12) was ectopically expressed from retroviral vectors pWZL–hygro and pLPC–puro, oncoprotein E1a was ectopically expressed from vector pLPC–puro, and simultaneous expression of E1a and oncogenic Ras was achieved with the construct pLPC–E1a–IRES–Ras (Serrano et al. 1997). For detailed methods see Supplemental Material.
RNA and protein analysis
For real-time quantitative RT–PCR, total RNA from tissues was extracted with Trizol (Life Technologies). Reverse transcription was performed using random priming and Superscript Reverse Transcriptase (Life Technologies) according to the manufacturer's guidelines. Real-time PCR was performed using an ABI PRISM 7700 (Applied Biosystems) by using DNA Master SYBR Green I mix (Applied Biosystems). PCR reaction mixtures were incubated at 95°C for 5 min followed by 40 PCR cycles (5 sec at 95°C, 90 sec at 68°C). For each sample, Ink4a-specific and Arf-specific transcripts were quantified as the cycle threshold difference (ΔCt) to β-actin mRNA. All reactions were carried out in triplicate per assay, and at least two independent assays were performed per value obtained. The primer sequences used were as follows: for Ink4-specific mRNA, p16-F (complementary to exon 1α, 5′-GCCCAACGCCCCGAACTCTTTC-3′) and either p16-R1 (5′-GCGACGTTCCCAGCGGTACACA-3′) or p16-R2 (5′-TGAAGCTATGCCCGTCGGTCTG-3′), both complementary to exon 2; for Arf-specific mRNA, ARF-F (complementary to exon 1β, 5′-TGGGTCGCAGGTTCTTGGTCAC-3′) and ARF-R (complementary to exon 2, 5′-CGACGTTCCCAGCG GTACACAA-3′); and for β-actin, ACT-F (5′-GGCACCACA CCTTCTACAATG-3′) and ACT-R (5′-GTGGTGGTGAAGCT GTAGCC-3′).
Immunoblots were performed following standard procedures. Typically, equal amounts of protein (30 μg) were separated on 15% SDS polyacrylamide gels and blotted onto nitrocellulose membranes (Bio-Rad). For detection of murine p16Ink4a, we used rabbit polyclonal antibody M156 (Santa Cruz), working dilution 1:1000; for detection of murine p19Arf, we used rabbit polyclonal antibody R562 (AbCam), 1:100; and, for β-actin, we used mouse monoclonal antibody AC-15 (Sigma), 1:10,000. For p15Ink4b, we used a rabbit polyclonal antibody produced by the laboratory of Mariano Barbacid (CNIO, Madrid) and generously provided to us. As secondary antibodies, we used HRP-linked anti-rabbit or HRP-linked anti-mouse (DAKO), both at a 1:2000 dilution. Detection was performed by chemiluminescence using ECL (Amersham).
Chemical carcinogenesis
For DMBA-induced carcinogenesis, cohorts of Ink4a/Arf (+/–), (–/–), or (–/–;tg/·) mice at post-natal day 3–7 received a single application of 50 μL of an acetone solution containing 0.5% DMBA (7,12-dimethylbenz[a]anthracene, Sigma) applied to the dorsal surface (total amount of DMBA applied: 250 μg) as described (Serrano et al. 1996). For pristane-induced carcinogenesis, 2-mo-old mice were injected three times, at 60-d intervals, with 0.5 mL of pristane (2,6,10,14-tetra-methyl-pentadecane, Sigma) as described (Zhang et al. 2001). For intramuscular 3MC-induced carcinogenesis, 3–6-mo-old mice received a single intramuscular injection at one of the rear legs of a 40 μL solution containing 3MC, at a concentration of 25 μg/μL and dissolved in sesame oil (Sigma) as described (Wexler and Rosenberg 1979). Mice were observed on a daily basis until tumors reached a diameter >1.5 cm. For DMBA/TPA-induced carcinogenesis, mice at post-natal day 3–7 received a single application of 250 μg of DMBA (see above) and, 3 wk later, mice were treated during a 12-wk time course with periodic applications, three times per week, of 5 μg of TPA (12-O-tetradecanoyl-phorbol-13-acetate, Sigma) dissolved in acetone, as described (Sotillo et al. 2001). For urethane-induced carcinogenesis, 6-wk-old female mice were injected intraperitoneally with a single dose of 1 mg/g of body weight of urethane (Sigma) (Sharpless et al. 2001). With the exception of these last assays, in all other assays the different groups of mice were composed of similar numbers of males and females.
Acknowledgments
We thank Gordon Peters (CRUK, London) and Mariano Barbacid (CNIO, Madrid) for providing us with reagents. We are indebted to Maribel Muñoz and Santiago Rodriguez for excellent mouse colony management and animal care, and to Elisa Santos and Jessica Freire for mouse genotyping. A.M. and A.E. were recipients of a predoctoral fellowship from the Spanish Ministry of Education. This work was partly funded by the Spanish Ministry of Science and Technology (SAF2002-03402), and by the European Union (QLRT-2000-02084, QLRT-2000-00616, INTACT).
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.310304.
References
- Balmain A. 2002. Cancer as a complex genetic trait: Tumor susceptibility in humans and mouse models. Cell 108: 145-152. [DOI] [PubMed] [Google Scholar]
- Campisi J. 2003. Cancer and ageing: Rival demons? Nat. Rev. Cancer 3: 339-349. [DOI] [PubMed] [Google Scholar]
- Carter N.P. 2004. As normal as normal can be? Nat. Genet. 36: 931-932. [DOI] [PubMed] [Google Scholar]
- de Stanchina E., McCurrach, M.E., Zindy, F., Shieh, S.Y., Ferbeyre, G., Samuelson, A.V., Prives, C., Roussel, M.F., Sherr, C.J., and Lowe, S.W. 1998. E1A signaling to p53 involves the p19(ARF) tumor suppressor. Genes & Dev. 12: 2434-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Cao I., Garcia-Cao, M., Martin-Caballero, J., Criado, L.M., Klatt, P., Flores, J.M., Weill, J.C., Blasco, M.A., and Serrano, M. 2002. `Super p53' mice exhibit enhanced DNA damage response, are tumor resistant and age normally. EMBO J. 21: 6225-6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate A.J., Feuk, L., Rivera, M.N., Listewnik, M.L., Donahoe, P.K., Qi, Y., Scherer, S.W., and Lee, C. 2004. Detection of large-scale variation in the human genome. Nat. Genet. 36: 949-951. [DOI] [PubMed] [Google Scholar]
- Latres E., Malumbres, M., Sotillo, R., Martin, J., Ortega, S., Martin-Caballero, J., Flores, J.M., Cordon-Cardo, C., and Barbacid, M. 2000. Limited overlapping roles of P15(INK4b) and P18(INK4c) cell cycle inhibitors in proliferation and tumorigenesis. EMBO J. 19: 3496-3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo H.S., Wang, Z., Hu, Y., Yang, H.H., Gere, S., Buetow, K.H., and Lee, M.P. 2003. Allelic variation in gene expression is common in the human genome. Genome Res. 13: 1855-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe S.W. and Sherr, C.J. 2003. Tumor suppression by Ink4a-Arf: Progress and puzzles. Curr. Opin. Genet. Dev. 13: 77-83. [DOI] [PubMed] [Google Scholar]
- Maier B., Gluba, W., Bernier, B., Turner, T., Mohammad, K., Guise, T., Sutherland, A., Thorner, M., and Scrable, H. 2004. Modulation of mammalian life span by the short isoform of p53. Genes & Dev. 18: 306-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M. and Barbacid, M. 2001. To cycle or not to cycle: A critical decision in cancer. Nat. Rev. Cancer 1: 222-231. [DOI] [PubMed] [Google Scholar]
- McKeller R.N., Fowler, J.L., Cunningham, J.J., Warner, N., Smeyne, R.J., Zindy, F., and Skapek, S.X. 2002. The Arf tumor suppressor gene promotes hyaloid vascular regression during mouse eye development. Proc. Natl. Acad. Sci. 99: 3848-3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmero I. and Serrano, M. 2001. Induction of senescence by oncogenic Ras. Methods Enzymol. 333: 247-256. [DOI] [PubMed] [Google Scholar]
- Palmero I., McConnell, B., Parry, D., Brookes, S., Hara, E., Bates, S., Jat, P., and Peters, G. 1997. Accumulation of p16INK4a in mouse fibroblasts as a function of replicative senescence and not of retinoblastoma gene status. Oncogene 15: 495-503. [DOI] [PubMed] [Google Scholar]
- Palmero I., Pantoja, C., and Serrano, M. 1998. p19ARF links the tumour suppressor p53 to Ras. Nature 395: 125-126. [DOI] [PubMed] [Google Scholar]
- Pantoja C. and Serrano, M. 1999. Murine fibroblasts lacking p21 undergo senescence and are resistant to transformation by oncogenic Ras. Oncogene 18: 4974-4982. [DOI] [PubMed] [Google Scholar]
- Ramirez R.D., Morales, C.P., Herbert, B.S., Rohde, J.M., Passons, C., Shay, J.W., and Wright, W.E. 2001. Putative telomere-independent mechanisms of replicative aging reflect inadequate growth conditions. Genes & Dev. 15: 398-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruas M. and Peters, G. 1998. The p16INK4a/CDKN2A tumor suppressor and its relatives. Biochim. Biophys. Acta 1378: F115-F177. [DOI] [PubMed] [Google Scholar]
- Sebat J., Lakshmi, B., Troge, J., Alexander, J., Young, J., Lundin, P., Maner, S., Massa, H., Walker, M., Chi, M., et al. 2004. Large-scale copy number polymorphism in the human genome. Science 305: 525-528. [DOI] [PubMed] [Google Scholar]
- Serrano M. and Blasco, M.A. 2001. Putting the stress on senescence. Curr. Opin. Cell Biol. 13: 748-753. [DOI] [PubMed] [Google Scholar]
- Serrano M., Lee, H., Chin, L., Cordon-Cardo, C., Beach, D., and DePinho, R.A. 1996. Role of the INK4a locus in tumor suppression and cell mortality. Cell 85: 27-37. [DOI] [PubMed] [Google Scholar]
- Serrano M., Lin, A.W., McCurrach, M.E., Beach, D., and Lowe, S.W. 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88: 593-602. [DOI] [PubMed] [Google Scholar]
- Sharpless N.E., Bardeesy, N., Lee, K.H., Carrasco, D., Castrillon, D.H., Aguirre, A.J., Wu, E.A., Horner, J.W., and DePinho, R.A. 2001. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature 413: 86-91. [DOI] [PubMed] [Google Scholar]
- Sharpless N.E., Kannan, K., Xu, J., Bosenberg, M.W., and Chin, L. 2003. Both products of the mouse Ink4a/Arf locus suppress melanoma formation in vivo. Oncogene 22: 5055-5059. [DOI] [PubMed] [Google Scholar]
- Sharpless N.E., Ramsey, M.R., Balasubramanian, P., Castrillon, D.H., and DePinho, R.A. 2004. The differential impact of p16(INK4a) or p19(ARF) deficiency on cell growth and tumorigenesis. Oncogene 23: 379-385. [DOI] [PubMed] [Google Scholar]
- Smogorzewska A. and de Lange, T. 2002. Different telomere damage signaling pathways in human and mouse cells. EMBO J. 21: 4338-4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillo R., Garcia, J.F., Ortega, S., Martin, J., Dubus, P., Barbacid, M., and Malumbres, M. 2001. Invasive melanoma in Cdk4-targeted mice. Proc. Natl. Acad. Sci. 98: 13312-13317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sviderskaya E.V., Gray-Schopfer, V.C., Hill, S.P., Smit, N.P., Evans-Whipp, T.J., Bond, J., Hill, L., Bataille, V., Peters, G., Kipling, D., et al. 2003. p16/cyclin-dependent kinase inhibitor 2A deficiency in human melanocyte senescence, apoptosis, and immortalization: Possible implications for melanoma progression. J. Natl. Cancer Inst. 95: 723-732. [DOI] [PubMed] [Google Scholar]
- Townson J.R., Barcellos, L.F., and Nibbs, R.J. 2002. Gene copy number regulates the production of the human chemokine CCL3-L1. Eur. J. Immunol. 32: 3016-3026. [DOI] [PubMed] [Google Scholar]
- Tyner S.D., Venkatachalam, S., Choi, J., Jones, S., Ghebranious, N., Igelmann, H., Lu, X., Soron, G., Cooper, B., Brayton, C., et al. 2002. p53 mutant mice that display early ageing-associated phenotypes. Nature 415: 45-53. [DOI] [PubMed] [Google Scholar]
- Wexler H. and Rosenberg, S.A. 1979. Pulmonary metastases from autochthonous 3-methylcholanthrene-induced murine tumors. J. Natl. Cancer Inst. 63: 1393-1395. [PubMed] [Google Scholar]
- Yan H. and Zhou, W. 2004. Allelic variations in gene expression. Curr. Opin. Oncol. 16: 39-43. [DOI] [PubMed] [Google Scholar]
- Yan H., Dobbie, Z., Gruber, S.B., Markowitz, S., Romans, K., Giardiello, F.M., Kinzler, K.W., and Vogelstein, B. 2002a. Small changes in expression affect predisposition to tumorigenesis. Nat. Genet. 30: 25-26. [DOI] [PubMed] [Google Scholar]
- Yan H., Yuan, W., Velculescu, V.E., Vogelstein, B., and Kinzler, K.W. 2002b. Allelic variation in human gene expression. Science 297: 1143. [DOI] [PubMed] [Google Scholar]
- Zdarsky E., Favor, J., and Jackson, I.J. 1990. The molecular basis of brown, an old mouse mutation, and of an induced revertant to wild type. Genetics 126: 443-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.L., DuBois, W., Ramsay, E.S., Bliskovski, V., Morse III, H.C., Taddesse-Heath, L., Vass, W.C., DePinho, R.A., and Mock, B.A. 2001. Efficiency alleles of the Pctr1 modifier locus for plasmacytoma susceptibility. Mol. Cell Biol. 21: 310-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zindy F., Quelle, D.E., Roussel, M.F., and Sherr, C.J. 1997. Expression of the p16INK4a tumor suppressor versus other INK4 family members during mouse development and aging. Oncogene 15: 203-211. [DOI] [PubMed] [Google Scholar]
- Zindy F., Eischen, C.M., Randle, D.H., Kamijo, T., Cleveland, J.L., Sherr, C.J., and Roussel, M.F. 1998. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes & Dev. 12: 2424-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]