Abstract
The activity of adult stem cells is essential to replenish mature cells constantly lost due to normal tissue turnover. By a poorly understood mechanism, stem cells are maintained through self-renewal while concomitantly producing differentiated progeny. Here, we provide genetic evidence for an unexpected function of the c-Myc protein in the homeostasis of hematopoietic stem cells (HSCs). Conditional elimination of c-Myc activity in the bone marrow (BM) results in severe cytopenia and accumulation of HSCs in situ. Mutant HSCs self-renew and accumulate due to their failure to initiate normal stem cell differentiation. Impaired differentiation of c-Myc-deficient HSCs is linked to their localization in the differentiation preventative BM niche environment, and correlates with up-regulation of N-cadherin and a number of adhesion receptors, suggesting that release of HSCs from the stem cell niche requires c-Myc activity. Accordingly, enforced c-Myc expression in HSCs represses N-cadherin and integrins leading to loss of self-renewal activity at the expense of differentiation. Endogenous c-Myc is differentially expressed and induced upon differentiation of long-term HSCs. Collectively, our data indicate that c-Myc controls the balance between stem cell self-renewal and differentiation, presumably by regulating the interaction between HSCs and their niche.
Keywords: Hematopoietic stem cell (HSC), stem cell niche, self-renewal, c-Myc, N-cadherin
Many adult tissues, including skin epidermis, gastrointestinal epithelia, or the hematopoietic system, are regenerative and self-renewing. In such tissues, mature cells in the stratum corneum of the skin, differentiated enterocytes at the tip of the intestinal villi, or blood erythrocytes have limited lifespans, and must be continuously replaced to replenish those steadily lost by shedding or apoptosis (Watt and Hogan 2000; Fuchs et al. 2004). Life-long production of differentiated progeny relies on rare long-lived adult tissue stem cells (TSC) that have the ability to both perpetuate themselves through self-renewal and to generate all mature cell types of a particular tissue through differentiation (Weissman 2000). Probably the most well-characterized adult stem cell is the hematopoietic stem cell (HSC), which has the clonal capacity to provide life-long reconstitution of all hematopoietic lineages after transplantation into lethally irradiated mice (Till and McCulloch 1961; Cantor and Orkin 2001; Kondo et al. 2003). Using various protocols, murine HSCs can be purified close to homogeneity by fluorescent-activated cell sorting (FACS) (Smith et al. 1991; Goodell et al. 1996). All functional repopulating HSCs are contained within the c-Kit+ lin–Sca-1+ (KLS-HSC) bone marrow (BM) population (Uchida and Weissman 1992; Weissman et al. 2001). Under steady-state conditions, the total number of HSCs is kept constant, whereas differentiated progeny are continuously generated to replace mature cells. However, in response to injury (e.g., blood loss), disease (e.g., anemia, cancer), or BM stress (e.g., myelotoxic chemotherapy), total stem cell number can vary significantly, indicating that the balance between stem cell self-renewal and differentiation can adapt to physiological needs (Morrison et al. 1997; Uchida et al. 1997; Watt and Hogan 2000; Weissman 2000). This flexibility is most likely achieved by reciprocal intercellular interactions between stem cells and their specific microenvironment called the niche. The stem cell niche is defined as a subset of tissue cells and extracellular substrates that can harbor one or more stem cells controlling their self-renewal and progeny production in vivo (Trentin 1970; Schofield 1978; Watt and Hogan 2000; Spradling et al. 2001; Lin 2002; Fuchs et al. 2004). Retention of stem cells in the niche is thought to be accomplished by stem cell niche and stem-cell extracellular matrix (ECM)-ligand interactions. It has been shown in the Drosophila ovary that DE-cadherin-mediated anchoring of germ line and somatic stem cells to the niche is essential for their maintenance (Song and Xie 2002; Song et al. 2002). Putative niches have also been identified in vertebrates, including the bulge region in the skin epidermis and the stem cell-bearing base of intestinal crypts (Cotsarelis et al. 1990; Potten and Loeffler 1990; Rochat et al. 1994). In the BM, HSCs are located at the endosteal lining of the BM cavities, and recent studies show that specialized spindle-shaped N-cadherin+ osteoblasts (SNO) are a key component of the BM stem cell niche. HSCs are thought to be anchored to SNO cells via a homotypic N-cadherin interaction (Whetton and Graham 1999; Nilsson et al. 2001; Visnjic et al. 2001; Calvi et al. 2003; Zhang et al. 2003; Zhu and Emerson 2004). However, the molecules that regulate stem cell niche interactions and how these may influence the balance between self-renewal and differentiation are currently unknown.
The proto-oncogene c-myc encodes an unstable nuclear factor c-Myc, which has been implicated in regulation of a wide variety of biological processes, including control of division, apoptosis, cellular growth, angiogenesis, and differentiation (Amati et al. 1998; Schuhmacher et al. 1999; Grandori et al. 2000; Jain et al. 2002; Pelengaris et al. 2002b; Bellmeyer et al. 2003; Watnick et al. 2003). c-Myc dimerizes in the cell with the ubiquitous protein Max, and Myc/Max heterodimers control two large independent sets of target genes. One set of target genes is activated by binding of Myc/Max to E-boxes in target gene promoters, inducing recruitment of complexes containing histone acetyl-transferase activity (Eisenman 2001; Frank et al. 2001). The second set of target genes is repressed by binding of Myc/Max to the transcriptional activator Miz-1 that binds to INR elements at the transcriptional start site of target genes (Staller et al. 2001; Wanzel et al. 2003). Various approaches have led to the identification of a large number of putative Myc target genes (http://www.myc-cancer-gene.org/index.asp). We and others have previously shown that deletion of c-myc by gene targeting in mice causes midgestation lethality, and mutant embryos fail to develop a primitive hematopoietic system (Trumpp et al. 2001; Baudino et al. 2002). Here, we have utilized conditional gene targeting, as well as a gain-of-function approach, to investigate the role of c-Myc during adult hematopoiesis. Our data indicate that c-Myc, in addition to being essential for proliferation of lineage-committed hematopoietic cells in vivo, plays an unexpected role in controlling the balance between HSC self-renewal and differentiation.
Results
Loss of c-Myc during adult hematopoiesis causes severe cytopenia
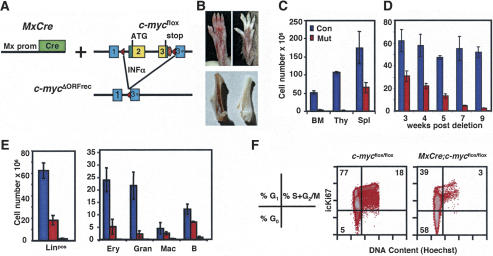
Previous studies have shown that c-Myc is required for primitive hematopoiesis (Trumpp et al. 2001). To study the function of c-Myc during maintenance of adult hematopoiesis, mice homozygous for the conditional c-mycflox allele were crossed with mice harboring the IFNα-inducible Mx–Cre transgene (Fig. 1A; Kuhn et al. 1995; Trumpp et al. 2001) to produce MxCre;c-mycflox/flox mice (mutant) and their MxCre-negative littermate controls (control). Postnatal induction of Mx–Cre after poly-I–poly-C (pI–pC) treatment results in close to 100% deletion of alleles flanked by loxP sites in liver, spleen, and BM (Kuhn et al. 1995; Radtke et al. 1999). Conversion of the c-mycflox into the c-mycΔORFrec allele, and therefore, deletion of the coding region, was monitored by real-time Taqman PCR and found to occur in 93% ± 6% of purified c-Kit+, linneg, Sca-1+ cells (KLS-HSCs). In parallel, c-myc transcription was decreased in mutant KLS-HSCs and total BM 13- and 22-fold, respectively. The Myc family member L-myc was not expressed in purified KLS-HSCs from normal or mutant mice (data not shown). N-myc expression has been reported to be expressed in embryos, embryonic stem cells, as well as in fetal and adult BM HSCs (Ivanova et al. 2002; Ramalho-Santos et al. 2002; Sperger et al. 2003). In agreement with these reports, N-myc transcripts were detectable in control and mutant-purified KLS-HSCs. The N-myc expression level was more than 100-fold below that detected in E10.5 embryos used as positive controls (data not shown). Importantly, no significant compensatory up-regulation of N-myc transcripts was detected in c-Myc-deficient KLS-HSCs by real-time PCR (data not shown) or in KLSF-HSCs by Affymetrix microarray analysis (W. Blanco, M.J. Murphy, and A. Trumpp, unpubl.).
Figure 1.
Induced deletion of c-myc in the adult bone marrow results in severe cytopenia and a decrease in proliferation of differentiated progenitors. (A) MxCre-mediated conversion of the c-mycflox allele into the c-mycΔORFrec allele by deletion of DNA between the two loxP sites in the c-mycflox locus. This includes the entire c-myc ORF (yellow). Exons 1–3 are indicated. (Red triangles) loxP sites. Cre expression is induced by INFα or pI–pC. (B) Hind-paws (top) and femurs (bottom) of control (left) and MxCre;c-mycflox/flox (right) mice 8 wk after pI–pC injection. (C) Cellularity of control (blue) and mutant (red) lymphoid organs 8 wk after deletion. (BM) Bone marrow; (Thy) thymus; (Spl) spleen. Results are mean ± SD from 12 (control) and 15 (mutant) mice. (D) Kinetics of BM cellularity from 3 to 9 wk after c-myc deletion. (E) Quantitation of linpos (left) BM cells and BM subsets (right) in controls (blue) and mutants at 3 wk (red) or 8 wk (orange) post-deletion. Between three and 12 mice were analyzed at each time point. (F) Cell cycle analysis of linpos BM cells. Total BM was surface stained to define the linpos subset, then fixed, permeabilized, and stained with Hoechst 33342 and Ki-67 (icKi67) for FACS analysis. Different cell cycle phases are indicated on the left.
By 4 wk post-deletion, MxCre;c-mycflox/flox mice develop acute anemia as apparent by their white feet and ears and a hemoglobin concentration in peripheral blood (PBL) as low as 1.6 g/dL compared with 14 g/dL in controls (Fig. 1B; data not shown). Analysis of PBL subsets shows that hematopoietic cell lineages are decreased four- to sevenfold (data not shown). All hematopoietic cell types normally present in the blood are generated from precursors located principally in the BM, spleen, and thymus. Analysis 8 wk post-c-myc-deletion shows that all three organs are greatly reduced in absolute cell number, particularly the thymus and BM, where the reduction is 50- and 20-fold, respectively (Fig. 1C). Because the BM is the primary site of adult hematopoiesis, the cellular composition of this organ was analyzed further. The long bones of mutants appear white (Fig. 1B), and kinetic analysis of BM cellularity demonstrates a continuous loss down to ≤5% of control BM numbers by 8–9 wk post-deletion of c-myc (Fig. 1D). To determine whether all BM cell lineages are affected, four-color FACS analysis was performed. Eight weeks post-deletion, differentiating and mature lineage-positive (linpos) BM cells are decreased >30-fold (Fig. 1E). This decrease is observed in all mature subsets tested (including B lymphocytes (B220), granulocytes (Gr-1), macrophages (Mac-1), and erythroblasts (TER-119) (Fig. 1E).
As c-Myc has been shown to be rate limiting for proliferation of fibroblasts and peripheral T cells (Trumpp et al. 2001), the cell cycle status of committed cell types in the BM (linpos cells) was quantitated. Surface staining to define subsets was combined with intracellular staining for DNA content (Hoechst 33342) and the proliferation marker Ki67, which is expressed in all actively dividing cells, but not in resting (G0) cells (Brown and Gatter 2002). Whereas 95% of control linpos cells are found in the active phases of the cell cycle (G1 + S + G2/M), most of the few remaining c-Myc-deficient linpos cells accumulate in G0 (Fig. 1F). These results suggest that c-Myc activity is required to maintain proliferation of lineage-committed cell types present in the BM, and provide an initial explanation for the severe cytopenia observed in response to loss of c-Myc.
Phenotypic LT-HSCs accumulate in c-Myc-deficient bone marrow
In contrast to the decrease in linpos cells, lineage-negative (linneg) precursors defined by their lack of surface expression of lineage markers (CD3, CD4, CD8, B220, Mac-1, Gr-1, Ter119, and NK1.1) are increased around threefold in the mutants 3 wk after c-myc deletion (Fig. 2A). In addition to early committed precursors of all hematopoietic lineages, the linneg population also contains the c-Kit+ (CD117) Sca-1+ subpopulation highly enriched for hematopoietic stem cells (KLS-HSC) (Uchida and Weissman 1992; Weissman et al. 2001). Unexpectedly, c-Myc-deficient BM is enriched >20-fold in KLS-HSCs compared with control BM (Fig. 2B). Due to the net decrease in BM cells, this enrichment corresponds to a two- to threefold increase in the absolute number of KLS-HSCs (Fig. 2E). As it has recently been shown that functional LT-HSC activity is exclusively contained within the Flk2/Flt3Rneg (CD135neg) subset of the KLS population (c-Kit+linnegSca1+Flk2neg (KLSF) (Adolfsson et al. 2001; Christensen and Weissman 2001), the proportion of KLS cells expressing this receptor was determined. As shown in Figure 2C, the relative ratio of Flk2neg/Flk2+ KLS cells increases from 1:1 (50%/50%) to >7:1 (88%/12%) in the c-Myc-deficient KLS population compared with controls. In addition, a larger percentage of c-Myc-deficient KLS-HSCs express low levels of CD90 (Thy1), further confirming their primitive undifferentiated character (Fig. 2D; Christensen and Weissman 2001). Quantitation of KLSF cells 3 wk post-deletion reveals a four-fold net increase compared with controls (Fig. 2F). These results show that cells with a LT-HSC phenotype accumulate, whereas committed progenitors and differentiated cell types are lost after elimination of c-Myc activity in the BM.
Figure 2.
Accumulation of hematopoietic stem cells (HSC) in c-Myc-deficient BM. (A) Quantitation of the linneg population in control (blue) and mutant BM either 3 wk (red) or 8 wk (orange) post-deletion. Results are mean ± SD of 12 (control) or six (mutant) mice. (B) BM was stained for lineage markers, gated on the linneg subset, and further stained for c-Kit (CD117) and Sca-1. Numbers on the plot are the frequency of cells in the indicated regions. Hematopoietic stem cells are contained within the c-Kit+ linneg Sca-1+ (KLS cells) population. Control (left) and mutant (right). An unusual linneg, c-Kitlow, Sca-1low population (which is also CD45+, IL-7Rneg, CD135neg and expressing high levels of the integrins α2, α4, β1, and β2) appears in the mutants (data not shown). Despite extensive analysis, this population does not correspond to any characterized cell type. (C) c-Kit (CD117) vs. CD135 (Flk2/Flt3R) expression on linneg Sca-1+ BM. Numbers on the plot are the frequency of cells in the indicated (c-Kit+ linneg Sca-1+) regions. Control (left) and mutant (right). (D) CD117 vs. CD90 (Thy1) expression linneg Sca-1+ BM. (E) Total number of KLS cells per two femurs 3, 4, and 5 wk post-c-myc-deletion in control (blue) and mutant (red) BM. Results are mean ± SD of five mice per time point. (F) Number of KLSF (KLS Flk2– cells) 3 wk post-deletion. Results are mean ± SD of three mice each. (G) Cell cycle analysis of KLS cells. Cells were stained with Hoechst 33342 and Ki67 (icKi67) and analyzed by FACS. The different cell cycle phases are indicated in the scheme at the left. Numbers are the proportion of cells in each phase. (H) In vivo BrdU-labeling kinetics of KLSF-HSCs isolated from control (blue) and mutant (red) mice 3–4 wk post-deletion of c-myc. Results are mean ± SD from three mice of each genotype per time point.
Proliferation and survival of HSCs in vivo is c-Myc independent
To determine the mechanism by which c-Myc-deficient KLSF-HSCs accumulate in the BM, the effect of c-Myc loss on proliferation and survival of KLS-HSCs was determined. c-Myc is known to play an important role in the control of proliferation and survival of various cell types (Grandori et al. 2000; Trumpp et al. 2001; Pelengaris et al. 2002a). Either process, if altered, could provide a possible explanation for the accumulation of HSCs in c-Myc-deficient BM. However, the apoptotic rate of c-Myc-deficient KLS-HSCs (as determined by Annexin V/7AAD staining) is unchanged in mutant compared with control cells, further supporting the suggestion that the observed accumulation is not due to increased survival of c-Myc-deficient KLS-HSCs (data not shown). In contrast to linpos BM cells that require c-Myc for proliferation (Fig. 1F), no change in the cell cycle profile as analyzed by DNA content is observed in mutant compared with control KLS-HSCs. In addition, the size of the quiescent (G0) pool in the KLS-HSC population as determined by the absence of expression of the proliferation marker Ki67 was unchanged (Ki67neg 2n DNA) (Fig. 2G). To determine the turnover rate of KLSF-HSCs in vivo, the kinetics (2 h, 15 h, 3 d, 5 d) of BrdU uptake was determined by five-color flow cytometry. No significant difference could be detected at any time point, suggesting that normal and mutant KLSF-HSCs have a very similar turnover rate in vivo (Fig. 2H). Taken together, these data show that the accumulation of c-Myc-deficient HSCs cannot be explained by increased proliferation or survival. It is interesting to note that HSCs proliferate in a c-Myc-independent manner in vivo. It is possible that HSCs, like ES-cells, may lack a functional G1–S checkpoint, and thus, would not require Myc activity (Burdon et al. 2002). Alternatively, this could simply be due to the fact that N-Myc is still expressed in mutant HSCs. This issue needs to be addressed by a conditional c-Myc/N-Myc double knockout.
BM devoid of c-Myc activity reconstitutes HSCs, but not more mature hematopoietic lineages
Mice lacking c-Myc become cytopenic and accumulate cells with an HSC phenotype in the BM. Because proliferation and survival are unchanged in mutant HSCs, an alternative explanation for their observed accumulation may be a defect in HSC differentiation. To directly test this possibility, c-Myc-deficient HSCs were analyzed for their reconstitution capacity in vivo. Groups of four lethally irradiated mice were injected with a range of doses of total BM isolated from c-Myc-deficient or control animals. While as few as 0.63 × 106 control BM cells were able to fully reconstitute 100% of irradiated hosts, no survivors were observed with doses of up to 2.5 × 106 c-Myc-deficient BM (data not shown). This inability to reconstitute suggests that c-Myc is required for either HSC function or progenitor expansion, or both. To distinguish between these possibilities and to eliminate any contribution from a mutant BM microenvironment, competitive mixed BM chimeras, in which the fate of mutant HSCs could be followed in mice harboring a functional hematopoietic system with a wild-type BM microenvironment were prepared. Lethally irradiated CD45.1+ mice were transplanted with a mixture of CD45.1+ wild-type BM, together with either mutant or control CD45.2+ BM. Analysis of chimeric mice 1–4 mo after transplantation revealed very poor contribution of c-Myc-deficient donor cells in peripheral organs (Fig. 3A; data not shown). However, while control donor HSCs generated normal proportions of stem and progenitor cell types, phenotypic LT-HSCs (KLS CD4neg CD11bneg), ST-HSCs (KLS CD4neg CD11blow); and MPPs (KLS, CD4low CD11blow) of c-Myc-deficient origin accumulate (Fig. 3B,C). Quantitation of these precursor populations in mixed chimeras shows an ∼20-fold increase in LT-HSCs in c-Myc-deficient BM compared with control. After normalization of input KLS cells (twofold increase in mutant compared with control at 3 wk post-deletion, Fig. 2E), the net increase in donor derived c-Myc-deficient LT-HSCs after reconstitution remains elevated by a factor of 10 (Fig. 3C). Both ST-HSCs and MPPs are also increased, but to a lesser extent (Fig. 3C). In contrast, CMPs (c-Kithi Sca-1neg CD34+ and CD16/32+) and CLPs (c-Kitlow Sca-1+ CD127+), as well as differentiated cell types such as granulocytes and B cells of mutant origin, are virtually absent (Fig. 3A,C; Kondo et al. 1997; Akashi et al. 2000). These results directly show that transferred c-Myc-deficient BM cells are unable to reconstitute early committed progenitors or mature hematopoietic lineages, whereas at the same time, HSCs with a predominant LT-HSC phenotype accumulate in situ.
Figure 3.
Differentiation potential of c-Myc-deficient HSCs in vivo or in vitro. (A) The development of HSCs in vivo was determined by injecting control or mutant CD45.2+ BM cells into groups of lethally irradiated wild-type CD45.1+ host mice together with competing wild-type CD45.1+ BM cells. (Left) After 2 mo, splenocytes were analyzed by flow cytometry for the contribution of each donor population by expression of CD45.1 and CD45.2. The presence of differentiated cell types (granulocytes, Gr1+; and B lymphocytes, B220+) derived from CD45.2+ donor cells (control, blue; and mutant, red) was quantitated. (Right) Results are mean ± SD from five mice per group. (B) The Sca-1/CD117 phenotype of Linneg CD45.2+ BM in control and mutant chimeras 2 mo post-reconstitution. While the majority of mutant cells show a KLS-HSC phenotype, they often express slightly reduced levels of c-Kit (see also Fig. 2B). (C) Number of different HSC and progenitor subsets derived from CD45.2+ Linneg donor cells in the chimeras shown in A and B, and defined as CD117+Sca1neg (CMPs), CD117loSca1loCD127+ (CLPs), CD117+Sca1+CD4loCD11blo (MPPs), CD117+Sca1+CD11blo (ST-HSCs), and CD117+Sca1+CD11b– (LT-HSCs) (Weissman 2000). Mutant donor BM 3 wk post-deletion contains twofold more KLS cells compared with control BM (Fig. 2E); therefore, the net increase in HSCs is about half that indicated. (D) Differentiation potential of KLS cells in vitro. FACS sorted KLS cells from control (left) and mutant (right) BM were isolated and grown in stem cell medium containing a cytokine cocktail including mSCF, mTPO, mFlt3L, IL-6, IL-7, Il-11, GM-CSF, and EPO. After 7 d, cultures were photographed using phase contrast. FACS analysis of expression of lineage markers on bulk cultured KLS cells. (Top) Gr-1 (granulocytes) versus CD11b (macrophages). (Bottom) B220 (B lymphocytes) (control, blue; mutant, red). Numbers on plots are the proportion of cells in indicated regions. (E) Genotyping by PCR of cultured control (C) and mutant (M) cells from D after 7 d. Control cells were positive for the c-mycflox allele (flox) and the DNA control (18s), whereas the mutant (M) cells were negative for the c-mycflox allele (flox), but positive for the c-mycΔORF/rec (Δ) allele as expected. (F, left) In vitro differentiation of purified KLS cells. LinnegCD117+Sca1+ BM cells were FACS sorted from control and c-Myc-deficient mice and cultured for 9 d after transfection with either MYC–IRES–huCD2 or huCD2 control viruses. Expression of mature hematopoietic cell markers (Gr1, CD11b, Ter119, and CD19) is shown on huCD2+ cells in control (huCD2 alone virus in control cells, dark-blue line), c-Myc-deficient cells transfected with the huCD2 control virus (solid red histogram), or c-Myc-deficient cells transfected with the MYC–IRES–huCD2 virus (light-blue line). (Right) Expansion of c-Myc-deficient (KO) or control (C) KLS-HSC cultures 9 d after infection with the huCD2 control virus or with the MYC–IRES–huCD2 virus (c-MYC). Data is expressed as fold increase over input cell number. This is a representative example of one of two experiments giving similar results.
This failure to generate expanding progenitors is similar to what is observed in MxCre;c-mycflox/flox mutants; however, the accumulation of HSCs is significantly more pronounced in the mixed chimeras. Most importantly, in the latter situation, mutant HSCs specifically accumulate in a wild-type mouse, which develops a normal hematopoietic system and carries a normal stromal microenvironment. Together with the finding that KLS-HSC proliferation and survival are c-Myc independent (Fig. 2G,H), this provides compelling evidence that the failure to generate early progenitors is due to the inability of mutant HSCs to differentiate. Instead, they accumulate, suggesting that the balance in these cells is shifted toward self-renewal at the expense of differentiation (Fig. 7A, left, see below). Thus, c-Myc activity is essential to maintain functional HSCs in vivo.
Figure 7.
(A) Model for the regulation of HSC fate by c-Myc-controlled adhesion to the stem cell niche. (Top) A quiescent HSC expressing low c-Myc levels is retained in the stem cell niche consisting of spindle-shaped N-cadherin+ osteoblasts (SNO) (Zhang et al. 2003) embedded in stromal fibroblasts. HSCs are anchored to SNO cells via homotypic N-cadherin and LFA-1/ICAM interaction. In addition, expression of α2β1-integrin and α5β1-integrin connects HSCs to the specialized extracellular matrix (ECM) in the niche. In response to mitogenic signals, HSCs enter the cell cycle and generate two daughter cells. (a) In the absence of c-Myc-induction integrins and other putative cell-adhesion molecules remain highly expressed in both daughter cells retaining them in the niche, thereby promoting expansion of HSCs at the expense of differentiation. (b) Induction of c-Myc in only one of the daughter cells causes down-regulation of cell-adhesion molecules and generates asymmetry with one HSC retained in the niche and one leaving the niche, promoting differentiation into committed progenitors (CP). In this homeostatic situation, the stem cell pool is maintained while differentiated progeny are produced. (c) High c-Myc expression in both daughter cells (e.g., ectopic c-MYC expression in HSCs) results in repression of cell-adhesion molecules and departure of both cells from the niche. This leads to the production of two CPs and hence progressive exhaustion of the stem cell pool due to differentiation. (B) Model for the regulation of hematopoiesis by c-Myc. (a) The wild-type (WT) hematopoietic system. Under homeostatic conditions, long-term hematopoietic stem cells (LT-HSCs) express low levels of c-Myc, ensuring self-renewal. In a subset of LT-HSCs, c-Myc expression increases, inducing differentiation toward a short-term hematopoietic stem cell (ST-HSC) fate. Continued differentiation leads to the loss of self-renewal activity, and early progenitor cells of different hematopoietic cell types become transient-amplifying cells (TA-cells), which rapidly expand through proliferation while continuing to differentiate into more and more lineage-restricted progenitors. High c-Myc levels are maintained in TA-cells, ensuring continued cell cycle progression. At the onset of terminal differentiation, c-Myc is down-regulated to allow permanent cell cycle exit and progression toward terminal differentiation. (b) Induced elimination of c-Myc in the fully developed adult hematopoietic system results in two distinct effects. First, TA-cells (progenitors) stop expanding, as c-Myc is required to maintain cells in an active cell cycle. Second, c-Myc-deficient LT-HCSs self-renew, but fail to initiate normal differentiation, leading to their accumulation as long as niche space is available. Together, this leads to loss of all hematopoietic cell types with the exception of HSCs. (c) Enforced expression of c-Myc in LT-HSCs promotes differentiation at the expense of self-renewal, resulting in stem cell exhaustion. In the absence of stem cell activity, all hematopoietic cell types are lost over time due to normal cellular turnover.
c-Myc-deficient HSCs can differentiate ex vivo
The balance between self-renewal and differentiation of stem cells is thought to be controlled by interaction of stem cells with their niches (Watt and Hogan 2000; Spradling et al. 2001; Fuchs et al. 2004). To test whether the effect of c-Myc on HSC differentiation in vivo is direct or is dependent on their location in the BM microenvironment/niche, KLS-HSCs were isolated and bulk cultured in vitro in the presence of a differentiation permitting cytokine cocktail containing IL-3, IL-11, SCF, TPO, Flt3L, GM-CSF, EPO (Akashi et al. 2000). After 1 wk, cells were harvested and analyzed for the appearance of linpos cells (Fig. 3D). In control cultures, differentiation into several different lineages is accompanied by a >100-fold increase in cell number (50,000 → 5,000,000). In contrast, only minimal or no expansion occurred in c-Myc-deficient cultures, consistent with the fact that c-Myc is required for proliferation of differentiating transient-amplifying (TA) cells. However, despite this observed lack of expansion, morphologically differentiated cells expressing myeloid and lymphoid markers such as Gr-1 and CD11b or B220 and CD19, were present in c-Myc-deficient cultures (as in control cultures), indicating that c-Myc-deficient HSCs can differentiate in vitro in contrast to what is observed in vivo (Fig. 3D). To rule out that c-Myc-deficient cells were overgrown with rare cells that escaped Cre-mediated deletion, cultured cells were genotyped by PCR and found to be c-mycΔORFrec/ΔORFrec (Fig. 3E). Interestingly, single-cell sorted c-Myc-deficient KLS-HSCs fail to divide, suggesting that c-Myc is essential for division in vitro, whereas HSCs in the BM proliferate in a c-Myc-independent manner (M.J. Murphy and A. Trumpp, unpubl.).
To show that mutant KLS-HSCs can be rescued by ectopic expression of c-MYC, mutant, and control, KLS-HSCs were sorted and infected with a mouse stem cell virus (MSCV) expressing human c-MYC in conjunction with a huCD2 reporter gene (MYC–IRES–huCD2) or with a control virus expressing only huCD2 (Deftos et al. 1998). After culturing the transduced KLS-cells for 9 d as described above, cells were counted and differentiation was determined by expression of the lineage markers Ter119, Gr1/CD11b, and CD19. Whereas the differentiation potential of c-Myc-deficient HSCs was similar in the presence or absence of ectopic c-MYC (Fig. 3F), mutant cultures expressing c-MYC expanded about 20 times compared with those infected with the control virus (Fig. 3F). This result shows that the failure of mutant KLS-cells to proliferate in vitro can be rescued by ectopic c-MYC. In addition, these experiments show that mutant KLS-HSCs are not an abnormal population of cells expressing stem cell markers by chance. Most importantly, the ability of c-Myc-deficient HSCs to undergo multilineage differentiation (but not expansion) in vitro suggests that the defect in vivo (no normal differentiation, but proliferation/self-renewal) is indirect and potentially due to a misregulated interaction with the local niche microenvironment.
c-Myc-deficient HSCs home to the stem cell niche at the BM endosteum
BM stem cell niches have recently been shown to be part of the endosteal lining of the internal bone cavities (Nilsson et al. 2001; Askenasy et al. 2002). In addition, it has recently been demonstrated that osteoblastic cells are a key component of the BM niche (Calvi et al. 2003; Zhang et al. 2003). One explanation for the dichotomy in differentiation capacity of c-Myc-deficient HSCs observed in vitro and in vivo, and for their increased self-renewal and consequent accumulation in vivo, could be their retention in the differentiation preventive microenvironment of the niche. This hypothesis would require that c-Myc-deficient stem cells be localized in the stem cell niche in vivo. Thus, immature purified progenitors (linneg cells) were labeled with the fluorescent dye CFSE and injected i.v. into sublethally irradiated wild-type recipients. HSCs present in this precursor population are predicted to home to stem cell niches located at the BM endosteum (Nilsson et al. 2001). Therefore, long bones were isolated 15 h later, and CFSE+ cells were localized on decalcified trabecular bone sections using fluorescence microscopy. Bone sections were stained for osteopontin, an extracellular matrix phosphoglycoprotein secreted by osteoblasts (Fig. 4A, panels 1,2) and BMPR1α (Fig. 4A, panels 3,4), which is expressed on the surface of osteoblasts (Denhardt and Guo 1993; ten Dijke et al. 1994; Zhang et al. 2003). As shown in Figure 4A, control and mutant progenitor/stem cells can home to the endosteum of the trabecular bone and are in close proximity or in direct contact with osteoblasts. Zhang et al. (2003) has shown that a subset of spindle-shaped N-cadherin-expressing osteoblasts (SNO) are an essential component of the stem cell niche, and has further suggested that N-cadherin-expressing HSCs are anchored to SNO via N-cadherin-mediated homotypic interactions (Zhang et al. 2003). To visualize SNO-cells and N-cadherin-expressing early progenitors present at the endosteum (presumably HSCs), trabecular bone sections were stained for N-cadherin expression. As shown in Figure 4B, panels 7–15, c-Myc-deficient N-cadherin-expressing early progenitors can home to the endosteum similar to control progenitors (Fig. 4B, panels 1–6). Most mutant progenitors at the endosteum are strongly stained for N-cadherin, while control cells are only weakly stained or not at all, suggesting that N-cadherin expression is up-regulated on stem cells in the absence of c-Myc. Importantly, the majority of mutant N-cad+ cells are in direct contact with SNO cells (Fig. 4B, panels 9,12,15). While not quantitative, these data show that c-Myc-deficient early progenitors are able to home to the BM endosteal stem cell niche and are in direct contact with SNO niche cells putatively via increased levels of N-cadherin.
Figure 4.
Localization of precursors in the N-cadherin-expressing bone marrow stem cell niche. Linneg precursors from control (Co) and c-Myc-deficient (Mu) BM were stained with CFSE (green, *) and i.v. injected into sub-lethally irradiated wild-type mice. (A) Trabecular bone sections are stained with DAPI (blue) and anti-osteopontin (panels 1,2, red) or anti-BMPRIα (panels 3,4, red) to visualize osteoblasts. White lines trace the edge of the endosteum. Yellow arrowheads denote osteopontin deposits. (BM) bone marrow (B, middle row) Trabecular bone sections are stained with anti-N-cadherin (red) to visualize SNO-cells (spindle-shaped N-cadherin+ osteoblasts) in the niche. (Bo) Bone; (En) endosteum. White lines trace the edge of the endosteum. Arrowheads denote CFSE+ cells (green). (Bottom row) Images were merged using Adobe Photoshop software. (Panels 1–3) Control precursors localized at the N-cadherin+ endosteum. (Panels 4–6) Control cell with low expression of N-cadherin localized in the center of the bone marrow. (Panels 7–12) Two frames showing c-Myc-deficient precursor cells expressing high levels of N-cadherin localized to the N-cadherin+ endosteum. (Panels 12–15) Two CFSE+ mutant precursor cells attached to the N-cadherin+ endosteum. Whereas the cell at the top expresses N-cadherin only at low levels, the cell at the bottom (panels 14,15,and inset in panel 15) expresses high levels of N-cadherin and is in direct contact with a SNO-cell.
Up-regulation of adhesion molecules on c-Myc-deficient HSCs
Since our data indicate that c-Myc-deficient HSCs may have a deregulated interaction with their BM stem cell niche, surface expression of 31 adhesion molecules was compared on control and mutant KLS-HSCs by four-color FACS analysis (Table 1). Interestingly, both the percentage of N-cad+ cells, as well as the expression level of N-cadherin increased significantly in mutant KLS-HSCs compared with controls (Fig. 5A), supporting the bone immunohistochemistry shown in Figure 4. In contrast, E-cadherin is not expressed on KLS cells, and only a minor population (5%) express VE-cadherin at similar levels in normal and mutant cells (Table 1). In addition to N-cadherin, members of the integrin family of adhesion receptors have also been implicated in homing and mobilization of HSCs into and out of the BM stem cell niche (Whetton and Graham 1999). Leukocyte function antigen 1 (LFA-1), a heterodimer consisting of the αL-integrin light chain (CD11a) and β2-integrin heavy chain (CD18) (Hogg et al. 2002) is consistently up-regulated around threefold at both mRNA (data not shown) and surface protein levels in the absence of c-Myc (Fig. 5B; Table 1). In addition, c-Myc-deficient HSCs express slightly increased levels of α2-integrin (CD49b), α5-integrin (CD49e), and β1-integrin (CD29) (Fig. 5B). As listed in Table 1, many other adhesion molecules tested were either not detected on the surface of HSCs or were unchanged in the mutants. The greatly increased levels of some adhesion molecules on HSCs lacking c-Myc activity is consistent with the hypothesis of deregulated interaction of mutant HSCs with SNO niche cells, and suggests that the mutants fail to differentiate due to their inability to detach from the differentiation preventive niche.
Table 1.
Expression of cell-adhesion molecules on KLS-HSCs
| CD designation | Molecule | Control | Change |
|---|---|---|---|
| CD2 | LFA-2 | NE | U |
| CD11a | αL integrin | + | ↑↑% |
| CD11b* | αM integrin | +/- | ↑*% |
| CD11c | αX integrin | NE | U |
| CD18b | β2 integrin | + | ↑↑% |
| CD24 | HSA | +/- | U |
| CD29 | β1 integrin | + | ↑ |
| CD34 | CD34 | +/- | ↑*% |
| CD43 | leukosialin | + | U |
| CD44 | PgP1, hyaluronan | + | U |
| CD49a | α1 integrin | NE | U |
| CD49b | α2 integrin | + | ↑ |
| CD49d | α4 integrin | NE | U |
| CD49e | α5 integrin | + | ↑ |
| CD49f | α6 integrin | + | U |
| CD51 | αV integrin | NE | U |
| CD54 | ICAM-1 | + | U |
| CD61 | β3 integrin | NE | U |
| CD62L | L-selectin | + | U |
| CD90* | Thy-1 | +/- | ↑*% |
| CD93 | O-sialoglycoprotein | NE | U |
| CD102 | ICAM-2 | NE | U |
| CD103 | αE integrin | NE | U |
| CD104 | β4 integrin | NE | U |
| CD105 | endoglin | NE | U |
| CD106 | VCAM-1 | + | U |
| CD144 | VE-cadherin | +/- | U |
| CD184 | CXCR4 | + | U |
| — | β7 integrin | NE | U |
| — | E-cadherin | NE | U |
| — | N-cadherin | +/- | ↑↑ |
Four-color FACS analysis of control and c-Myc deficient KLS-cell (gated as linneg cKit+ Scal+). (NE) not expressed; (U) unchanged; (+/-) low expression on a % of cells; (+) intermediate to high expression on most cells; (↑%) change in the proportion of positive cells; (↑*%) expected increase due to enrichment of LT-HSC in mutants; (↑ or ↑↑) up-regulated in both % and mean fluorescence intensity (MFI).
Figure 5.
Up-regulation of N-cadherin and several integrins on c-Myc-deficient HSCs. Expression of N-cadherin on c-Myc-deficient KLS-HSCs as determined by FACS. (A, filled histogram) c-Myc-deficient KLS-HSCs; (solid line overlay) control KLS-HSCs; (dotted line overlay) negative control (omission of N-cadherin antibody). Horizontal arrow indicates positive N-cadherin staining. (B) Up-regulation of LFA-1 (αLβ2 integrin), α5and β1 integrins on c-Myc-deficient KLS-HSCs. Histogram analysis of CD11a (αL integrin), CD18 (β2 integrin), CD29 (β1 integrin), CD49d (α4 integrin), CD49b (α2 integrin), CD49e (α5 integrin), CD62L (L-selectin), and CXCR4. (Filled histogram) Mutants; (overlaid line) control.
Deregulated overexpression of c-Myc in HSCs causes repression of cell-adhesion molecules and loss of self-renewal activity
Loss of c-Myc function results in failure of differentiation, presumably by retention of HSCs in the BM niche due to up-regulated expression of adhesion molecules. Therefore, HSCs unable to down-regulate c-Myc should increase differentiation at the expense of self-renewal accompanied by down-regulation of adhesion molecules. To test this hypothesis, wild-type linneg precursors were infected with MYC–IRES–huCD2 or with a control virus expressing only huCD2. After 4 d, cells were harvested and surface expression of N-cadherin in the linneg Sca-1+huCD2+ population was assessed. As predicted, N-cadherin levels were reduced almost to background levels in c-MYC-transduced cells (Fig. 6A), showing that c-MYC alone is sufficient to repress N-cadherin expression on HSCs. Similarly, c-MYC overexpression also caused down-regulation of both chains of the LFA-1 receptor, αL- and β2-integrins, whereas β1- and α4-integrin expression were essentially unchanged (Fig. 6B). These data show that c-Myc represses cell-adhesion molecules involved in the interactions between HSCs and SNO niche cells, and furthermore, suggest that c-Myc expression levels might be crucial for the control of these interactions.
Figure 6.
Down-regulation of N-cadherin expression in response to ectopic expression of c-MYC. (A) Wild-type linneg BM infected with MYC–IRES–huCD2 virus (c-MYC overexpression, filled histogram) or huCD2 control virus (wild-type, solid line). Dotted overlay shows a negative control-omitting N-cadherin antibody. Histograms are gated on huCD2+ linneg Sca-1+ cells after 4 d in culture. (B) Integrin expression in response to ectopic expression of c-MYC. Wild-type linneg cells were isolated by flow cytometry and either infected with MYC–IRES–huCD2 (filled histogram), or huCD2 control virus (overlaid line). After 7 d, cells were harvested and the huCD2+ linneg subset analyzed for expression of indicated integrins by FACS. (C) Ectopic expression of c-MYC in vivo. Wild-type Linneg BM, highly enriched for KLS-HSCs, was isolated and infected as in A and B. c-MYC overexpression (MYC–IRES–huCD2) (dotted lines), or control expressing huCD2 alone (solid lines). Infected BM was transferred together with wild-type BM into lethally irradiated recipients to generate mixed BM chimeras. At times indicated, the percent huCD2+ cells in donor phenotype PBLs or BM was assessed by FACS analysis. Each line represents data from an individual mouse. (D) Percent huCD2+ cells in KLSF cells after BM reconstitution in vivo as in C, except that starting linneg BM cells were obtained from H2K–BCL-2 transgenic mice in which HSCs express BCL-2. Control chimeras (BCL-2) and c-MYC overexpression chimeras (c-MYC/BCL-2). Results shown are from three (BCL-2) and four (c-Myc/BCL-2) mice.
To address whether c-Myc overexpression results in defects in stem cell function, wild-type CD45.2+ linneg BM was infected with the MYC–IRES–huCD2 or huCD2 control viruses and used to generate mixed BM chimeras together with wild-type CD45.1+ BM cells. Initially, the contribution of c-MYC-expressing cells was similar to that of control grafts, as determined by the percentage of huCD2+ cells in peripheral blood cells (PBLs) 2 wk after transplantation (Fig. 6C). By 4 wk post-transplantation, huCD2+ donor-derived c-MYC overexpressing PBLs comprised 45.8% ± 10.2% Ter119+ (erythroid lineage), 22.8% ± 11.4% Gr1+ (granulocytes), and 38.2% ± 14.9% B220+ (B cells), similar to huCD2+ control donor-derived BM (data not shown). However, compared with 2 wk post-transfer, the proportion of c-MYC-expressing huCD2+ cells was significantly decreased (Fig. 6C), and became virtually undetectable (<1%) in BM, spleen, and PBLs of the CD45.1+ recipients after 12 wk (Fig. 6C; data not shown). Moreover, huCD2+ linneg Sca-1+ donor cells overexpressing c-MYC were absent in BM after 12 wk (data not shown). As expected, engraftment of the cells infected with the huCD2 control virus remained stable even after 12 wk post-transplantation (Fig. 6C). These results directly demonstrate that although enforced expression of c-MYC allows homing and multilineage differentiation, long-term self-renewal activity of HSCs is progressively lost.
Whereas the kinetics of HSC loss supports a premature differentiation mechanism, it is still formally possible that c-Myc-overexpressing HSCs are lost due to apoptosis. To address the effects of c-Myc overexpression on HSC survival in vitro, wild-type linneg BM cells were infected with the MYC–IRES–huCD2 or huCD2 control viruses. After 72 h in culture, the percentage of apoptotic cells within the huCD2+ KLS-HSC populations was determined by 7-AAD/Annexin V staining. No increase in apoptotic (Annexin V+) cells was observed in c-MYC-overexpressing KLS-HSCs in comparison to controls. In fact, these cells showed slightly better survival, suggesting that c-MYC overexpression does not induce apoptosis in cultured KLS-HSC cells (data not shown).
c-Myc-induced apoptosis has been previously shown to be blocked by expression of BCL-2 (Pelengaris et al. 2002a; Nilsson and Cleveland 2003). To further exclude the possibility that c-MYC-overexpressing HSCs are lost in vivo due to apoptosis, we generated mixed BM chimeras in which H2K–BCL-2 transgenic linneg BM cells infected with the MYC–IRES–huCD2 or huCD2 control viruses were transferred together with wild-type total BM. In these chimeras, repopulating virus-transduced (but not wild-type) HSCs express a BCL-2 transgene (Domen et al. 2000). Two weeks post-transfer, huCD2+ cells were detected in PBLs of both experimental and control mice (data not shown). At 6 wk post-transfer, the BM was analyzed for the proportion of huCD2+ KLSF-HSCs. As shown in Figure 6D, 14.6% ± 2% of control KLSF cells were huCD2+. In contrast, only around 1.5% ± 0.5% of c-MYC/BCL-2-expressing KLSF cells were huCD2+, showing that even in the presence of BCL-2, c-MYC-overexpressing HSCs fail to long-term self-renew and are eventually lost (Fig. 6D). In addition, 5/8 experimental mice developed pre-B-cell lymphomas originating from cells coexpressing c-MYC and BCL-2, similar to what has been previously reported (Strasser et al. 1990; Cory et al. 1999). However, it is important to note that c-Myc-expressing KLSF cells are virtually absent whether tumors develop or not. Thus, despite initial engraftment (differentiation) and inhibition of apoptosis, c-Myc overexpressing HSCs apparently fail to self-renew, strongly suggesting that this is due to premature differentiation.
Expression of endogenous c-myc is induced upon onset of LT-HSC differentiation
Our genetic studies show that total loss of c-Myc or deregulated gain of c-MYC activity shift the balance toward HSC self-renewal or differentiation, respectively (Fig. 7A). In a homeostatic situation, division of a wild-type HSC generates two daughter cells, with one remaining an HSC, whereas the other initiates differentiation. If endogenous c-Myc is involved in this decision process in normal HSCs, its expression should be negative/low in the HSC remaining in the niche, but would be higher in the daughter cell that starts to differentiate. To test this hypothesis, c-myc transcripts were quantified in FACS-sorted HSC subsets by real-time RT–PCR. The results show that although c-myc is expressed at low levels in KLSF-HSCs (LT-HSCs), it is expressed in FLK2+ KLS-HSCs (ST-HSCs/MPPs) at 2.3-fold ± 1.3 (P = 0.008) higher levels. Collectively, our results indicate that the balance between HSC self-renewal and differentiation is controlled by c-Myc expression levels (Fig. 7A). The effects of c-Myc throughout hematopoiesis are summarized in Figure 7B.
Discussion
In this study, we provide genetic evidence confirming the expected involvement of c-Myc during the expansion of committed progenitors in the adult hematopoietic system. In addition, and in contrast to progenitors, we have also uncovered a novel role for c-Myc during the first steps of HSC differentiation. This function of c-Myc is only evident in HSCs located within BM niches, but not if grown in vitro, where stem cell niches are absent. c-Myc controls (represses) the expression of specific integrins and the HSC anchor N-cadherin, suggesting a model in which c-Myc controls the balance between self-renewal and differentiation by modulating migration and/or adhesion of HSCs to the niche (Fig. 7A).
c-Myc function in stem-cell differentiation
Although the model shown here proposes a function for c-Myc in the first differentiation steps of stem cells, the data may also be explained (at least at first glance) by a role of c-Myc in proliferation alone. In this study, we demonstrate that, whereas HSCs proliferate in a c-Myc-independent manner, late and potentially also early progenitors require c-Myc activity for cell cycle progression. Thus, early progenitors would not expand in the absence of c-Myc, making them virtually invisible, and generating the false impression of HSCs not undergoing differentiation. In such a situation, there is no a priori reason for HSC numbers to increase. However, deletion of c-Myc leads to a specific increase of LT-HSCs. Because the mutants develop severe anemia, one could predict feedback mechanisms that may lead to HSC accumulation. However, in mixed BM chimeras, where c-Myc-deficient HSCs develop in the context of a normal hematopoietic system that includes wild-type stem cell niches, an increase of mutant HSCs is still observed. In fact, the increase in HSCs is even more pronounced in BM chimeras compared with mutant mice. Thus, we strongly favor a model in which c-Myc has a dual role. Initially, it is essential to induce the first differentiation steps in HSCs, whereas in committed progenitors, c-Myc function is required for cell cycle progression and expansion (Fig. 7B).
Recently, data obtained in other systems also provided evidence of c-Myc influencing progenitor differentiation independent of its function in division and survival. For example, during Xenopus, neural crest induction knock down of c-Myc results in the failure to induce expression of early neural crest markers, causing a subsequent block in the formation of neural crest-derived structures (Bellmeyer et al. 2003). Development and maintenance of intestinal and colonic mucosa are controlled by the canonical Wnt/β-catenin/TCF4 pathway. Recent studies in colorectal carcinoma cell lines suggest that c-Myc and its downstream target gene p21CIP1 are key effectors of this pathway, consequently controlling self-renewal, expansion, and differentiation of mucosal progenitors (He et al. 1998; van de Wetering et al. 2002). A role for c-Myc in intestinal stem cell driven maintenance of mucosa is further suggested by expression profiling of microdissected early progenitor populations from intestinal crypts in which a number of genes either controlling or controlled by c-Myc have been identified (Stappenbeck et al. 2003). Complementary to the BM results presented here, where forced expression of c-MYC in HSCs leads to their differentiation and subsequent loss, over-expression of c-MYC in the stem cell-containing basal layer of the murine epidermis results in severe epidermal defects (including epidermal loss) thought to be caused by the loss of epidermal stem cells (Arnold and Watt 2001; Waikel et al. 2001). The latter result is in direct contrast to a different c-Myc-overexpressing transgenic mouse line, in which c-Myc was targeted to a more differentiated progenitor population. In this second model, precancerous epidermal lesions develop (Pelengaris et al. 1999). These contrasting results in the skin are consistent with our model, suggesting that c-Myc has distinct roles in stem and progenitor cell types (Fig. 7B). Collectively, these data suggest that c-Myc is not only involved in the first differentiation steps of HSCs, but may have similar roles in other self-renewing tissues, including the intestinal mucosa and the skin epidermis.
Role of c-Myc in HSC–niche interactions
The differentiation defect of c-Myc-deficient HSCs in the BM does not appear to be intrinsic, as in vitro-cultured FACS-purified HSCs were able to differentiate along myeloid and lymphoid lineages. Interestingly, this occurs in the absence of significant proliferation, a phenomenon previously reported by Fairbairn et al. (1993), who showed that multipotent progenitors can differentiate without proliferation if cultured in the absence of cytokines, but in the presence of BCL-2. Re-expression of constitutive MYC activity in mutant HSCs rescued the in vitro proliferation defect without affecting multilineage differentiation. The contrasting behavior of HSCs under- or overexpressing c-Myc in vitro and in vivo reported in this study, together with the identification of c-Myc-regulated specific cell adhesion molecules strongly suggests that c-Myc controls interaction between HSCs and their stem cell niche environment.
While the concept of the stem cell niche was first suggested several decades ago (Trentin 1970; Schofield 1978), experimental evidence for the location and cellular composition of stem cell niches in higher organisms is still in its infancy. Genetic studies in the Drosophila ovary have demonstrated that germ-line stem cells are required to remain attached via an adhesion anchor (DE-cadherin) to the niche cells (Cap cells). Detachment from the niche induces stem cell differentiation and loss of self-renewal (Spradling et al. 2001; Song and Xie 2002; Song et al. 2002). BM stem cell niches are thought to be located in the endosteal lining of trabecular BM cavities, with ostoblastic cells being a crucial component of the stem cell-maintaining niche (Nilsson et al. 2001; Calvi et al. 2003; Visnjic et al. 2004). Zhang et al. (2003) have further demonstrated that spindle-shaped N-cadherin+ osteoblasts (SNO) fulfill the function of BM niche cells. Because a fraction of HSCs also express N-cadherin, it is likely (in analogy to the situation in the Drosophila ovarian niche) that HSCs are anchored to SNO cells via homotypic N-cadherin junctions (Song and Xie 2002; Song et al. 2002; Zhang et al. 2003). Our findings that c-Myc-deficient HSCs are in direct contact with SNO cells, and furthermore express increased levels of surface N-cadherin, strongly support the notion that mutant stem cells are retained in the niche, and c-Myc-mediated down-regulation of adhesion molecules is necessary for HSCs to exit the stem cell niche. Our model suggests that upon division of a c-Myc-deficient HSC, both daughter cells maintain an HSC fate, thereby contributing to expansion of the stem cell pool at the expense of differentiated cell types (Fig. 7A). This model is further supported by the fact that HSCs that cannot down-regulate c-MYC display no surface N-cadherin and c-MYC+N-cadherinneg HSCs progressively lose their self-renewal capacity, presumably due to failure of retention in the BM niche. It is interesting in this respect that the Angiopoietin-1 (Ang-1)/Tie2-signaling pathway maintains repopulating quiescent HSCs in the niche, presumably by activating cell adhesion molecules such as N-cadherin (Arai et al. 2004). This raises the possibility that c-Myc (and N-Myc) may be negative mediators downstream of the Tie2 pathway repressing N-cadherin and activating the cell cycle. In ES-cells, it has been shown that lack of c-Myc causes a marked up-regulation of Ang-1 (Baudino et al. 2002). If this were also the case in HSCs, one could hypothesize that the Myc and the Ang-1/Tie2-signaling pathways are intricately linked in the control of HSC–niche interactions, ultimately regulating HSC function.
In addition to the more recent implication of cadherins in stem-cell niche interactions, there is ample evidence to support an important role for integrins in adult stem cell function (Jones and Watt 1993; Pruijt et al. 1998; Whetton and Graham 1999; Hynes 2002; Lapidot and Petit 2002; Velders et al. 2002; Watt 2002). In the hematopoietic system, integrins have been particularly implicated in controlling HSC migration from niches into the circulation (mobilization) and from the circulation into niches (homing). For example, HSCs deficient in β1-integrin fail to engraft lethally irradiated mice due to impaired homing and retention in the BM (Potocnik et al. 2000). Experiments using blocking antibodies have also implicated αL, α4, α5, and β2 integrins in the complex mobilization process (Pruijt et al. 1998; Asaumi et al. 2001; Velders et al. 2002). It is intriguing, in this respect, that in addition to controlling N-cadherin expression on HSCs, c-Myc also represses expression of several members of the integrin family of adhesion receptors, namely VLA-5 (α5β1 integrin, fibronectin receptor), α2β1 integrin (collagen receptor), and LFA-1 (αLβ2 integrin). Integrins and cadherins have been previously shown to synergistically regulate migration and mobility of cells (Huttenlocher et al. 1998). Whether these adhesion molecules also cooperate in controlling the function of HSCs, and whether they only function as adhesion receptors or also as signaling molecules, is still an open question. In addition, the exact function of N-cadherin in HSC–niche interactions remains to be elucidated by tissue-specific knockouts, as embryos lacking N-cadherin fail to develop past midgestation (Radice et al. 1997). However, the connections between c-Myc, Ang-1/Tie2, and N-cadherin, as well as that of c-Myc and integrin signaling, raises the intriguing possibility of a complex network linking adhesion to specific signaling cascades in stem cells attached to their niche (Jamora and Fuchs 2002; Schwartz and Ginsberg 2002; Perez et al. 2003; Arai et al. 2004).
The total number of existing stem cell niches in the BM, as well as their individual size, is currently unknown. In this context, it is interesting to note that KLS-HSC numbers in mutants increase only two- to threefold, whereas an accumulation of up to 10-fold is observed in mixed BM chimeras. The difference may be explained by the fact that whereas stem-cell niches are wild type in chimeras, they are partially c-Myc-deficient in mutants, due to MxCre activity in BM stromal cells (M.J. Murphy and A. Trumpp, unpubl.). The concept that c-Myc may also be required for (long-term) BM niche maintenance is currently under investigation. It is also possible that c-Myc-deficient HSCs positively impact on (wild-type) niche size. In any event, our data suggest that stem cell niches in the BM are limiting and crucial for maintaining HSCs in the undifferentiated state, a conclusion also recently reached by others (Akashi et al. 1999; Calvi et al. 2003; Zhang et al. 2003).
Concluding remarks
Although c-myc is the first proto-oncogene described to control stem cell homeostasis, some of its target genes and proteins that collaborate with Myc during tumorigenesis have recently been implicated in stem cell function. For example, the polycomb protein Bmi-1 collaborates with c-Myc during lymphomagenesis, and has been shown to be essential for maintenance of adult HSCs (Jacobs et al. 1999; Park et al. 2003). The CDK inhibitor p21CIP, which is repressed by c-Myc, controls HSC proliferation, and is furthermore required for maintenance of long-term self-renewal (Cheng et al. 2000; van de Wetering et al. 2002). This suggests that part of c-Myc`s effects in HSCs could be mediated by p21 repression. Because c-Myc has been postulated to be an effector of canonical Wnt signaling and also appears to be connected to the Ang-1/Tie2-signaling pathway, this protein is now evolving as a ringmaster in regulating adult stem cell function in vivo (Baudino et al. 2002; van de Wetering 2002; Arai et al. 2004). It is thus crucial to elucidate which signaling pathways are responsible for the tight control of c-Myc expression in stem/progenitor cells. Irrespective of what niche signals fine tune c-Myc expression during the constantly changing conditions of BM homeostasis in vivo, it appears that this oncoprotein is a key element that fulfils the function of a homeostat, determining the balance between stem cell self-renewal and differentiation.
Materials and methods
Generation of c-Myc-deficient mice
c-mycflox/flox (control) mice as previously described (Trumpp et al. 2001) were crossed with the Mx–cre transgenic mice (Kuhn et al. 1995) to obtain MxCre;c-mycflox/flox (mutant) mice. IFNα-induced deletion was effected by five i.p. injections of polyI–polyC (pI–pC), each 2 d apart as previously described (Radtke et al. 1999). Unless otherwise stated, all control mice were littermates of mutant mice and were all treated with pI–pC. Deletion was assessed by Southern Blot analysis and Taqman PCR on total BM and FACS-sorted KLS BM as previously described (Trumpp et al. 2001). Taqman RT–PCR was also performed to analyze c-myc, L-myc, and N-myc expression in the BM and KLS subsets. Primers and conditions were as described (Trumpp et al. 2001), except for L-myc, where the following primers were used: LmycF (ACGGCACTCGTCTGGAA) and LmycR (GT GACTGGCTTTCGGATGTC). LFA-1 was detected using the following primers: LFA-1F (ATTTTCCTGGCGCTCTACAA) and LFA-1R (TCCATTTGGAACACCTCCAT). All cDNAs were normalized using β2microglobulin (β2mF [GTGTATGC TATCCAGAAAACCC] and β2mR [TCACATGTCTCGATC CCAGTAG]) expression. All DNA samples were normalized using the 18s rRNA gene (probe: 5′6-FAM/3′TAMRA, GTG TATGCTATCCAGAAAACC; m18sF, ATTAAGTCCCTGCC CTTTGTACAC; m18sR, CCGAGGGCCTCACTAAACC).
Analysis of endogenous c-myc expression in HSCs by real-time RT–PCR
RNA from wild-type LT-HSCs and ST-HSCs (sorted as described below from pools of six mice) was extracted using the RNeasy mini kit (Qiagen). cDNA was generated using Stratascript Reverse Transcriptase (Stratagene). Subsequently, samples were normalized using β-2 microglobulin, and c-myc levels were determined using the primers/probes described above on both a Light Cycler (Roche) and a Taqman GeneAmp5700 Sequence Decector (Applied Biosystems). This experiment was performed five times on different days and the mean ± SD determined (both Light Cycler and the GeneAmp5700).
PBL analysis
For FACS analysis of PBLs, five drops of blood were collected into a tube containing Heparin, diluted in PBS, and centrifuged over a Lympholyte M gradient (Cedarlane Laboratories). PBLs were harvested from the interface, washed, and FACS stained as described below. Hemaglobin concentration was determined on PBLs by the Veterinaermedizinisches Labor of the University of Zurich, using a CELL-DYN Haematology Instrument.
BM preparation, analysis, and culture
BM was taken from the long bones of hind- and forelegs and prepared by standard procedures. All cell suspensions were filtered through a nylon mesh filter (70 μm) prior to FACS analysis to prevent clumps. Linneg BM was prepared by first staining with a cocktail of FITC or biotin-conjugated mAbs against lineage markers (CD3, CD4, CD8, CD11b, CD161, B220, Gr1, and Ter119). After washing, labeled cells were removed by incubation with Sheep anti-Rat IgG-coated M450 Dynabeads (Dynal Biotech) at a bead-to-target cell ratio of either 10 or 20:1. Linneg cells were harvested and purity checked by FACS. A second round of bead depletion was performed when required, otherwise remaining linpos cells were gated out on the FACS during analysis or sorting. To isolate KLS cells (linneg, cKit+Sca1+), lin-depleted BM was subsequently stained with mAbs to Sca1 and cKit (see below), and FACS sorted using a FACS Star+ Flow Cytometer (Becton Dickinson), at 1000 per well directly into 96-well flat-bottomed culture plates containing Stem Pro-34SFM medium (GIBCO Invitrogen Corp.) supplemented with L-glutamine (GIBCO), mSCF (50 ng/mlL, mTPO (25 ng/mL), mFlt3L (30 ng/mL), and IL-6 (105 μ/mL) all purchased from Research Diagnostics Inc. Individual wells were observed microscopically daily for 14 d, and cell number and morphology was scored. Medium was partially exchanged every 3 d. For in vitro bulk cultures, KLS-HSC cells were sorted (as above) and seeded at between 10 and 40 K per well in 96 flat-well plates in stem-cell medium (as above) supplemented with additional cytokines as follows: IL-11 (20 ng/mL), GM-CSF (50 ng/mL), and EPO (3 ng/mL) (Research Diagnostics). Cells were cultured for 7–9 d and subsequently harvested, surface stained, and analyzed by four- or five-color FACS. For infection, KLS-HSCs or Linneg BM were prepared and infected as described below. Medium was changed 12 h after infection, and cells were cultured for 5–9 d prior to FACS analysis. To isolate LT-HSCs and ST-HSCs, wild-type Linneg BM was prepared from 7-wk-old C57BL/6 mice (Harlan Olac), and stained as described above with mAbs to CD117, Sca-1, and CD135. cKit+Sca1+ CD135+ (ST-HSC) and cKit+Sca1+ CD135– (LT-HSC) were sorted on a five-color FACS DIVA Flow Cytometer (Becton Dickinson). For each individual experiment, the BM of six mice was pooled.
Monoclonal antibodies and FACS analysis
The following monoclonal antibody conjugates were purchased from eBiosciences: CD117 (c-kit R, ACK2)-PE, -PE-Cy5, -PE-Cy7, and -APC-Cy7; CD127 (IL7Rα chain, A7R34)-biotin, -PE and -PE-Cy5; CD135 (Flt3R/Flk2, A2F10)-PE; CD11b (M1/70)-PE-Cy5; Sca-1 (Ly-6A/E, D7)-FITC, -PE, and -APC; Ter 119-PE; B220 (RA3-6B2)-PE-Cy5; CD49b (α2 integrin, HMa2)-biotin; CD49d (α4 integrin, R1-2)-biotin; CD51 (αV integrin, RMV-7) protein; CD34 (RAM34)-biotin; CD43 (αIIb integrin, S7)-biotin; CD54 (ICAM-1, YN1/1.7.4)-biotin; and CD106 (VCAM-1, 429)-biotin. Anti-E-cadherin-FITC (36), CD9 (KMC8)-biotin; CD29 (Ha2/5)-biotin and purified; CD49a (α1 integrin, HA3/18)-biotin; CD49e (α5 integrin, 5H10-27)-biotin; CD61 (β3 integrin, 2C9.G2)-biotin; CD104 (β4 integrin, 346.11A)-biotin; Sca-1 (2B8)-biotin; β7 integrin (M293)-biotin; CD49f (α6 integrin, GoH3)-PE; and CD184 (CXCR4, 2B11)-PE were purchased from BD Biosciences. B220-PE was purchased from Caltag. Gr-1 (Ly-6G, RB6-8C5)-FITC, biotin and -Alexa 647, Ter 119-FITC, B220 (RA3-6B2)-FITC and biotin, CD11b-FITC and biotin, CD4 (clone GK1.5)-FITC and biotin, CD8α (53.6.7)-FITC and biotin, CD161 (NK1.1, PK136)-FITC and biotin, CD3ε (17A2)-FITC and biotin, CD11a (αL integrin, FD44.8)-biotin, CD18 (β2-integrin, FD18.5)-biotin, CD45.1 (A20.1)-FITC, -biotin, -PE or -Alexa 647, CD45.2 (ALI-4A2)-FITC, -biotin, -PE or -Alexa 647, CD24 (HSA, M1/69)-biotin, and CD44 (Ly24/PgP1, IM.781)-biotin were purified and conjugated in this laboratory following standard protocols. Alexa 647 conjugates were prepared using the appropriate Alexa protein labeling kits (Molecular Probes). CD144-purified protein (Rat anti-mouse VE-cadherin, 11D4.1) was purchased from BD Biosciences and revealed with Goat anti-Rat Ig-PE (Caltag), followed by blocking with Rat Ig (Jackson ImmunoResearch) prior to staining with lineage cocktail-FITC, CD117-PECy5, and Sca 1-APC. Affinity-purified rabbit anti-human N-cadherin (YS) antibody was purchased from ImmunoBiological Laboratories and revealed with Goat anti-Rabbit Ig-biotin (purified and conjugated in this laboratory). Blocking and further staining was performed as described for VE-Cadherin. In the experiments in which c-MYC was overexpressed using retroviral constructs, N-cadherin was detected using the Rabbit Zenon-biotin labeling kit (Molecular Probes). Streptavidin-APC (Molecular Probes), Streptavidin PE-Cy5 and PE-Cy5.5 (eBiosciences), or Streptavidin PE-Cy7 and Streptavidin-PE (Caltag) were used to reveal biotin conjugates. Four- and five-color FACS analysis (FITC, PE, PE-Cy5, or PE-Cy5.5, PE-Cy7, and APC or Alexa 647) were performed using a FACSCalibur Flow Cytometer (Becton Dickinson) or a FACS Canto Flow Cytometer (Becton Dickinison) respectively, and data was analyzed using CellQuest software. Five-color FACS sorting (FITC, PE, PE-Cy5, APC, and APC-Cy7) was performed on a FACS DIVA Flow Cytometer (Becton Dickinson).
Cell cycle analysis and BrdU uptake
Simultaneous cell cycle analysis and four-color surface staining was performed. Briefly, BM cells were labeled with mAb conjugates (FITC, PE, PE-Cy5, and PE-Cy7) to surface markers, fixed in 2% paraformaldehyde in PBS, washed, permeabilized with 0.5% Saponin in PBS/3% FCS, and stained with Hoechst 33342 (Molecular Probes) at 20 μg/mL for 5 min. After washing, cells were analyzed on a LSR Flow Cytometer (Becton Dickinson). Doublets were eliminated using the DDM unit. To distinguish between G1 and G0 phases of the cell cycle, surface labeling using either anti-Ki67-FITC or an isotype control FITC conjugate was added with the Hoechst, and the incubation prolonged to 30 min. BrdU uptake studies were performed as previously described (Wilson et al. 2001) using the BrdU staining kit (BD Biosciences).
Bone marrow chimeras
CD45.2+ C57BL/6 female mice (Harlan Olac) and CD45.1+ C57BL/6 female mice (The Jackson Laboratory) were purchased and maintained in the ISREC animal facility. H2K–BCL-2 transgenic mice were obtained from Dr. J. Domen (Duke University Medical Center, Durham, NC). Competitive BM reconstitution was performed as previously described (Radtke et al. 1999). Briefly, a mixture of 10 × 106 MxCre;c-mycflox/flox BM (CD45.2+) and 1 × 106 wild-type BM (CD45.1+) was i.v. transferred into CD45.1+ lethally irradiated (1000 rads) recipient mice that had been treated with anti-NK1.1 mAb 48 h previously. Mice were maintained on antibiotic (Bactrim, Roche, Basel) containing water and long-term reconstitution of peripheral blood, BM, and lymphoid organs analyzed 2–3 mo later. For retroviral infections, CD45.2+ C57BL/6 or H2K–BCL-2 transgenic mice received 0.15 mg/g body weight 5-fluorouracil (Sigma-Aldrich) i.p. 3 d prior to sacrificing. Linneg BM highly enriched for KLS-HSCs was prepared as described above, and cells were infected with either pMI–IRES–huCD2 or pMI–MYC–IRES–huCD2 viruses. Around 500 × 103 cells were infected with virus using polybrene (4 μg/mL, Sigma) in Stem Pro-34SFM medium (described above). Cells were spun for 15 min at 1800rpm and incubated at 37°C for a further 2 h. Cells were subsequently i.v. transferred into lethally irradiated (1000 rads) CD45.1+ recipients. A total of 15 h later, a second dose of 500 × 103-infected BM cells, which had undergone a second round of viral infection with the same virus and been maintained in stem-cell medium was injected i.v. A further 24 h later, 0.5 × 106 wild-type CD45.1+ BM was transferred. PBLs from these chimeric mice were analyzed 2 and 4 wk post-injection, and BM and lymphoid organs analyzed 12 wk later for the presence of donor (CD45.2+) cells successfully infected with virus (huCD2+).
Localization of progenitor/stem cells in situ
The localization assay was performed as described (Nilsson et al. 2001). Linneg BM was prepared and resuspended to 106 cells/mL in DMEM/0.5% FCS containing 10 μM 5-(and-6)-carboxy-fluorescein diacetate succinimidyl ester (CFSE) (Molecular probes) and incubated at 37°C for 10 min (Trumpp et al. 2001). Ice-cold PBS with 20% FCS was added to stop staining. After washing in PBS, cells from one donor mouse were resuspended in PBS/0.5% FCS and injected i.v. into one recipient that had received 300 rads 24 h prior. Fifteen hours later, the mice were sacrificed and muscle and tissue cleaned from the fore- and hindlimbs before the bones were placed in PBS/4% PFA for 16–18 h at 4°C. After decalcification (Al.Cl2, Acetic acid, HCl 7%:5%.8.5% w/v) for 24–48 h at 4°C, the bones were embedded in paraffin according to standard protocols. A total of 0–20 μm longitudinal (for observation) or 5 μm cross-sections (for immunostaining) were cut and mounted using Vectashield (Vector Laboratories). Immunohistochemistry was performed as described (Trumpp et al. 1992). Briefly, sections were deparaffinized and antigen retrieved at 70°C in 10 mM citrate buffer (pH 6.0) for 3–4 h. Slides were cooled for 20 min, blocked in 2% mouse serum, and 10% normal goat serum in PBS (blocking buffer) for 30 min, incubated in polyclonal Rabbit anti-human N-cadherin (as above) or monoclonal mouse anti-osteopontin (clone Akm2A1) (Santa Cruz Biotechnology) or BMPRIa (ALK3) (gift from Dr. C.H. Heldin, Ludwig Institute for Cancer Research, Uppsala, Sweden) in a 1:100 dilution of blocking buffer at 4°C overnight in a humidified chamber. Slides were rinsed three times in PBS/0.3% Triton X-100, incubated in goat anti-rabbit Ig-Alexa-546 (Molecular Probes) for N-cadherin and ALK3 staining, or with Goat anti-mouse Ig-Cy3 (Jackson Laboratories) for Osteopontin for 1 h at room temperature in 1:100 blocking buffer. Slides were rinsed three to five times in water, mounted in fluorescent mounting medium. Analysis was performed under a DC200 Leitz LEICA DMIRB fluorescent microscope equipped with filters for FITC (578 nm), Texas Red (610 nm). Cell nuclei were localized by staining with DAPI (Molecular Probes).
Acknowledgments
We thank Céline Agosti for excellent technical assistance; Christelle Dubey for animal husbandry and genetic screening; Drs. Bettina Ernst and Anita Wolfer for initial help with setting up bone marrow chimeras; Pierre Zaech and Frédéric Grosjean for FACS sorting; Dr. Jos Domen for the H2K–BCL2 transgenic mice; Dr. Michael Bevan for the huCD2 retroviral construct; Dr. Carl Henrik Heldin for the ALK3 antibody; and Catherine Pythoud, Estelle Sauberli, and the ISREC MIM core facility for histology. We thank Drs. Michel Aguet and Freddy Radtke for critical reading of the manuscript. This work was supported in part by grants to A.T. from the Swiss National Science Foundation, the Swiss Cancer League, and the Leenards Foundation. A.T. is member of the EMBO Young Investigator Programme. M.B. was supported by a fellowship from Roche and K.K. from the UBS Optimus Foundation.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.313104.
References
- Adolfsson J., Borge, O.J., Bryder, D., Theilgaard-Monch, K., Astrand-Grundstrom, I., Sitnicka, E., Sasaki, Y., and Jacobsen, S.E. 2001. Upregulation of Flt3 expression within the bone marrow Lin(–)Sca1(+)c-kit(+) stem cell compartment is accompanied by loss of self-renewal capacity. Immunity 15: 659-669. [DOI] [PubMed] [Google Scholar]
- Akashi K., Kondo, M., Cheshier, S., Shizuru, J., Gandy, K., Domen, J., Mebius, R., Traver, D., and Weissman, I.L. 1999. Lymphoid development from stem cells and the common lymphocyte progenitors. Cold Spring Harb. Symp. Quant. Biol. 64: 1-12. [DOI] [PubMed] [Google Scholar]
- Akashi K., Traver, D., Miyamoto, T., and Weissman, I.L. 2000. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 404: 193-197. [DOI] [PubMed] [Google Scholar]
- Amati B., Alevizopoulos, K., and Vlach, J. 1998. Myc and the cell cycle. Front. Biosci. 3: D250-D268. [DOI] [PubMed] [Google Scholar]
- Arai F., Hirao, A., Ohmura, M., Sato, H., Matsuoka, S., Takubo, K., Ito, K., Koh, G.Y., and Suda, T. 2004. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell 118: 149-161. [DOI] [PubMed] [Google Scholar]
- Arnold I. and Watt, F.M. 2001. c-Myc activation in transgenic mouse epidermis results in mobilization of stem cells and differentiation of their progeny. Curr. Biol. 11: 558-568. [DOI] [PubMed] [Google Scholar]
- Asaumi N., Omoto, E., Mahmut, N., Katayama, Y., Takeda, K., Shinagawa, K., and Harada, M. 2001. Very late antigen-5 and leukocyte function-associated antigen-1 are critical for early stage hematopoietic progenitor cell homing. Ann. Hematol. 80: 387-392. [DOI] [PubMed] [Google Scholar]
- Askenasy N., Zorina, T., Farkas, D.L., and Shalit, I. 2002. Transplanted hematopoietic cells seed in clusters in recipient bone marrow in vivo. Stem Cells 20: 301-310. [DOI] [PubMed] [Google Scholar]
- Baudino T.A., McKay, C., Pendeville-Samain, H., Nilsson, J.A., Maclean, K.H., White, E.L., Davis, A.C., Ihle, J.N., and Cleveland, J.L. 2002. c-Myc is essential for vasculogenesis and angiogenesis during development and tumor progression. Genes & Dev. 16: 2530-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellmeyer A., Krase, J., Lindgren, J., and LaBonne, C. 2003. The protooncogene c-Myc is an essential regulator of neural crest formation in Xenopus. Dev. Cell 4: 827-839. [DOI] [PubMed] [Google Scholar]
- Brown D.C. and Gatter, K.C. 2002. Ki67 protein: The immaculate deception? Histopathology 40: 2-11. [DOI] [PubMed] [Google Scholar]
- Burdon T., Smith, A., and Savatier, P. 2002. Signalling, cell cycle and pluripotency in embryonic stem cells. Trends Cell. Biol. 12: 432-438. [DOI] [PubMed] [Google Scholar]
- Calvi L.M., Adams, G.B., Weibrecht, K.W., Weber, J.M., Olson, D.P., Knight, M.C., Martin, R.P., Schipani, E., Divieti, P., Bringhurst, F.R., et al. 2003. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature 425: 841-846. [DOI] [PubMed] [Google Scholar]
- Cantor A.B. and Orkin, S.H. 2001. Hematopoietic development: A balancing act. Curr. Opin. Genet. Dev. 11: 513-519. [DOI] [PubMed] [Google Scholar]
- Cheng T., Rodrigues, N., Shen, H., Yang, Y., Dombkowski, D., Sykes, M., and Scadden, D.T. 2000. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science 287: 1804-1808. [DOI] [PubMed] [Google Scholar]
- Christensen J.L. and Weissman, I.L. 2001. Flk-2 is a marker in hematopoietic stem cell differentiation: A simple method to isolate long-term stem cells. Proc. Natl. Acad. Sci. 98: 14541-14546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cory S., Vaux, D.L., Strasser, A., Harris, A.W., and Adams, J.M. 1999. Insights from Bcl-2 and Myc: Malignancy involves abrogation of apoptosis as well as sustained proliferation. Cancer Res. 59: 1685s-1692s. [PubMed] [Google Scholar]
- Cotsarelis G., Sun, T.T., and Lavker, R.M. 1990. Label-retaining cells reside in the bulge area of pilosebaceous unit: Implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell 61: 1329-1337. [DOI] [PubMed] [Google Scholar]
- Deftos M.L., He, Y.W., Ojala, E.W., and Bevan, M.J. 1998. Correlating notch signaling with thymocyte maturation. Immunity 9: 777-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denhardt D.T. and Guo, X. 1993. Osteopontin: A protein with diverse functions. FASEB J. 7: 1475-1482. [PubMed] [Google Scholar]
- Domen J., Cheshier, S.H., and Weissman, I.L. 2000. The role of apoptosis in the regulation of hematopoietic stem cells: Overexpression of Bcl-2 increases both their number and repopulation potential. J. Exp. Med. 191: 253-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenman R.N. 2001. Deconstructing myc. Genes & Dev. 15: 2023-2030. [DOI] [PubMed] [Google Scholar]
- Fairbairn L.J., Cowling, G.J., Reipert, B.M., and Dexter, T.M. 1993. Suppression of apoptosis allows differentiation and development of a multipotent hemopoietic cell line in the absence of added growth factors. Cell 74: 823-832. [DOI] [PubMed] [Google Scholar]
- Frank S.R., Schroeder, M., Fernandez, P., Taubert, S., and Amati, B. 2001. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes & Dev. 15: 2069-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E., Tumbar, T., and Guasch, G. 2004. Socializing with the neighbors: Stem cells and their niche. Cell 116: 769-778. [DOI] [PubMed] [Google Scholar]
- Goodell M.A., Brose, K., Paradis, G., Conner, A.S., and Mulligan, R.C. 1996. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J. Exp. Med. 183: 1797-1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandori C., Cowley, S.M., James, L.P., and Eisenman, R.N. 2000. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell. Dev. Biol. 16: 653-699. [DOI] [PubMed] [Google Scholar]
- He T.C., Sparks, A.B., Rago, C., Hermeking, H., Zawel, L., da Costa, L.T., Morin, P.J., Vogelstein, B., and Kinzler, K.W. 1998. Identification of c-MYC as a target of the APC pathway. Science 281: 1509-1512. [DOI] [PubMed] [Google Scholar]
- Hogg N., Henderson, R., Leitinger, B., McDowall, A., Porter, J., and Stanley, P. 2002. Mechanisms contributing to the activity of integrins on leukocytes. Immunol. Rev. 186: 164-171. [DOI] [PubMed] [Google Scholar]
- Huttenlocher A., Lakonishok, M., Kinder, M., Wu, S., Truong, T., Knudsen, K.A., and Horwitz, A.F. 1998. Integrin and cadherin synergy regulates contact inhibition of migration and motile activity. J. Cell. Biol. 141: 515-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes R.O. 2002. Integrins: Bidirectional, allosteric signaling machines. Cell 110: 673-687. [DOI] [PubMed] [Google Scholar]
- Ivanova N.B., Dimos, J.T., Schaniel, C., Hackney, J.A., Moore, K.A., and Lemischka, I.R. 2002. A stem cell molecular signature. Science 298: 601-604. [DOI] [PubMed] [Google Scholar]
- Jacobs J.J., Scheijen, B., Voncken, J.W., Kieboom, K., Berns, A., and van Lohuizen, M. 1999. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes & Dev. 13: 2678-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M., Arvanitis, C., Chu, K., Dewey, W., Leonhardt, E., Trinh, M., Sundberg, C.D., Bishop, J.M., and Felsher, D.W. 2002. Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science 297: 102-104. [DOI] [PubMed] [Google Scholar]
- Jamora C. and Fuchs, E. 2002. Intercellular adhesion, signalling and the cytoskeleton. Nat. Cell. Biol. 4: E101-E108. [DOI] [PubMed] [Google Scholar]
- Jones P.H. and Watt, F.M. 1993. Separation of human epidermal stem cells from transit amplifying cells on the basis of differences in integrin function and expression. Cell 73: 713-724. [DOI] [PubMed] [Google Scholar]
- Kondo M., Weissman, I.L., and Akashi, K. 1997. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell 91: 661-672. [DOI] [PubMed] [Google Scholar]
- Kondo M., Wagers, A.J., Manz, M.G., Prohaska, S.S., Scherer, D.C., Beilhack, G.F., Shizuru, J.A., and Weissman, I.L. 2003. Biology of hematopoietic stem cells and progenitors: Implications for clinical application. Annu. Rev. Immunol. 21: 759-806. [DOI] [PubMed] [Google Scholar]
- Kuhn R., Schwenk, F., Aguet, M., and Rajewsky, K. 1995. Inducible gene targeting in mice. Science 269: 1427-1429. [DOI] [PubMed] [Google Scholar]
- Lapidot T. and Petit, I. 2002. Current understanding of stem cell mobilization: The roles of chemokines, proteolytic enzymes, adhesion molecules, cytokines, and stromal cells. Exp. Hematol. 30: 973-981. [DOI] [PubMed] [Google Scholar]
- Lin H. 2002. The stem-cell niche theory: Lessons from flies. Nat. Rev. Genet. 3: 931-940. [DOI] [PubMed] [Google Scholar]
- Morrison S.J., Wright, D.E., and Weissman, I.L. 1997. Cyclophosphamide/granulocyte colony-stimulating factor induces hematopoietic stem cells to proliferate prior to mobilization. Proc. Natl. Acad. Sci. 94: 1908-1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson J.A. and Cleveland, J.L. 2003. Myc pathways provoking cell suicide and cancer. Oncogene 22: 9007-9021. [DOI] [PubMed] [Google Scholar]
- Nilsson S.K., Johnston, H.M., and Coverdale, J.A. 2001. Spatial localization of transplanted hemopoietic stem cells: Inferences for the localization of stem cell niches. Blood 97: 2293-2299. [DOI] [PubMed] [Google Scholar]
- Park I.K., Qian, D., Kiel, M., Becker, M.W., Pihalja, M., Weissman, I.L., Morrison, S.J., and Clarke, M.F. 2003. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 423: 302-305. [DOI] [PubMed] [Google Scholar]
- Pelengaris S., Littlewood, T., Khan, M., Elia, G., and Evan, G. 1999. Reversible activation of c-Myc in skin: Induction of a complex neoplastic phenotype by a single oncogenic lesion. Mol. Cell 3: 565-577. [DOI] [PubMed] [Google Scholar]
- Pelengaris S., Khan, M., and Evan, G. 2002a. c-MYC: More than just a matter of life and death. Nat. Rev. Cancer 2: 764-776. [DOI] [PubMed] [Google Scholar]
- ____. 2002b. Suppression of Myc-induced apoptosis in β cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell 109: 321-334. [DOI] [PubMed] [Google Scholar]
- Perez O.D., Mitchell, D., Jager, G.C., South, S., Murriel, C., McBride, J., Herzenberg, L.A., Kinoshita, S., and Nolan, G.P. 2003. Leukocyte functional antigen 1 lowers T cell activation thresholds and signaling through cytohesin-1 and Jun-activating binding protein 1. Nat. Immunol. 4: 1083-1092. [DOI] [PubMed] [Google Scholar]
- Potocnik A.J., Brakebusch, C., and Fassler, R. 2000. Fetal and adult hematopoietic stem cells require β1 integrin function for colonizing fetal liver, spleen, and bone marrow. Immunity 12: 653-663. [DOI] [PubMed] [Google Scholar]
- Potten C.S. and Loeffler, M. 1990. Stem cells: Attributes, cycles, spirals, pitfalls and uncertainties. Lessons for and from the crypt. Development 110: 1001-1020. [DOI] [PubMed] [Google Scholar]
- Pruijt J.F., van Kooyk, Y., Figdor, C.G., Lindley, I.J., Willemze, R., and Fibbe, W.E. 1998. Anti-LFA-1 blocking antibodies prevent mobilization of hematopoietic progenitor cells induced by interleukin-8. Blood 91: 4099-4105. [PubMed] [Google Scholar]
- Radice G.L., Rayburn, H., Matsunami, H., Knudsen, K.A., Takeichi, M., and Hynes, R.O. 1997. Developmental defects in mouse embryos lacking N-cadherin. Dev. Biol. 181: 64-78. [DOI] [PubMed] [Google Scholar]
- Radtke F., Wilson, A., Stark, G., Bauer, M., van Meerwijk, J., MacDonald, H.R., and Aguet, M. 1999. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity 10: 547-558. [DOI] [PubMed] [Google Scholar]
- Ramalho-Santos M., Yoon, S., Matsuzaki, Y., Mulligan, R.C., and Melton, D.A. 2002. `Stemness': Transcriptional profiling of embryonic and adult stem cells. Science 298: 597-600. [DOI] [PubMed] [Google Scholar]
- Rochat A., Kobayashi, K., and Barrandon, Y. 1994. Location of stem cells of human hair follicles by clonal analysis. Cell 76: 1063-1073. [DOI] [PubMed] [Google Scholar]
- Schofield R. 1978. The relationship between the spleen colony-forming cell and the haemopoietic stem cell. Blood Cells 4: 7-25. [PubMed] [Google Scholar]
- Schuhmacher M., Staege, M.S., Pajic, A., Polack, A., Weidle, U.H., Bornkamm, G.W., Eick, D., and Kohlhuber, F. 1999. Control of cell growth by c-Myc in the absence of cell division. Curr. Biol. 9: 1255-1258. [DOI] [PubMed] [Google Scholar]
- Schwartz M.A. and Ginsberg, M.H. 2002. Networks and crosstalk: Integrin signalling spreads. Nat. Cell. Biol. 4: E65-E68. [DOI] [PubMed] [Google Scholar]
- Smith L.G., Weissman, I.L., and Heimfeld, S. 1991. Clonal analysis of hematopoietic stem-cell differentiation in vivo. Proc. Natl. Acad. Sci. 88: 2788-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X. and Xie, T. 2002. DE-cadherin-mediated cell adhesion is essential for maintaining somatic stem cells in the Drosophila ovary. Proc. Natl. Acad. Sci. 99: 14813-14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Zhu, C.H., Doan, C., and Xie, T. 2002. Germline stem cells anchored by adherens junctions in the Drosophila ovary niches. Science 296: 1855-1857. [DOI] [PubMed] [Google Scholar]
- Sperger J.M., Chen, X., Draper, J.S., Antosiewicz, J.E., Chon, C.H., Jones, S.B., Brooks, J.D., Andrews, P.W., Brown, P.O., and Thomson, J.A. 2003. Gene expression patterns in human embryonic stem cells and human pluripotent germ cell tumors. Proc. Natl. Acad. Sci. 100: 13350-13355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A., Drummond-Barbosa, D., and Kai, T. 2001. Stem cells find their niche. Nature 414: 98-104. [DOI] [PubMed] [Google Scholar]
- Staller P., Peukert, K., Kiermaier, A., Seoane, J., Lukas, J., Karsunky, H., Moroy, T., Bartek, J., Massague, J., Hanel, F., et al. 2001. Repression of p15INK4b expression by Myc through association with Miz-1. Nat. Cell. Biol. 3: 392-399. [DOI] [PubMed] [Google Scholar]
- Stappenbeck T.S., Mills, J.C., and Gordon, J.I. 2003. Molecular features of adult mouse small intestinal epithelial progenitors. Proc. Natl. Acad. Sci. 100: 1004-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser A., Harris, A.W., Bath, M.L., and Cory, S. 1990. Novel primitive lymphoid tumours induced in transgenic mice by cooperation between myc and bcl-2. Nature 348: 331-333. [DOI] [PubMed] [Google Scholar]
- ten Dijke P., Yamashita, H., Ichijo, H., Franzen, P., Laiho, M., Miyazono, K., and Heldin, C.H. 1994. Characterization of type I receptors for transforming growth factor-β and activin. Science 264: 101-104. [DOI] [PubMed] [Google Scholar]
- Till J.E. and McCulloch, E.A. 1961. A direct measurement of the radiation sensitivity of normal mouse bone marrow cells. Radiat. Res. 14: 213-222. [PubMed] [Google Scholar]
- Trentin J. 1970. Regulation of hematopoietic stem cells. Appleton-Century-Crofts, New York.
- Trumpp A., Blundell, P.A., de la Pompa, J.L., and Zeller, R. 1992. The chicken limb deformity gene encodes nuclear proteins expressed in specific cell types during morphogenesis. Genes & Dev. 6: 14-28. [DOI] [PubMed] [Google Scholar]
- Trumpp A., Refaeli, Y., Oskarsson, T., Gasser, S., Murphy, M., Martin, G.R., and Bishop, J.M. 2001. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature 414: 768-773. [DOI] [PubMed] [Google Scholar]
- Uchida N. and Weissman, I.L. 1992. Searching for hematopoietic stem cells: Evidence that Thy-1.1lo Lin– Sca-1+ cells are the only stem cells in C57BL/Ka-Thy-1.1 bone marrow. J. Exp. Med. 175: 175-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N., Friera, A.M., He, D., Reitsma, M.J., Tsukamoto, A.S., and Weissman, I.L. 1997. Hydroxyurea can be used to increase mouse c-kit+Thy-1. 1(lo)Lin-/loSca-1(+) hematopoietic cell number and frequency in cell cycle in vivo. Blood 90: 4354-4362. [PubMed] [Google Scholar]
- van de Wetering M., Sancho, E., Verweij, C., de Lau, W., Oving, I., Hurlstone, A., van der Horn, K., Batlle, E., Coudreuse, D., Haramis, A.P., et al. 2002. The β-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell 111: 241-250. [DOI] [PubMed] [Google Scholar]
- Velders G.A., Pruijt, J.F., Verzaal, P., van Os, R., van Kooyk, Y., Figdor, C.G., de Kruijf, E.J., Willemze, R., and Fibbe, W.E. 2002. Enhancement of G-CSF-induced stem cell mobilization by antibodies against the β 2 integrins LFA-1 and Mac-1. Blood 100: 327-333. [DOI] [PubMed] [Google Scholar]
- Visnjic D., Kalajzic, I., Gronowicz, G., Aguila, H.L., Clark, S.H., Lichtler, A.C., and Rowe, D.W. 2001. Conditional ablation of the osteoblast lineage in Col2.3δtk transgenic mice. J. Bone Miner. Res. 16: 2222-2231. [DOI] [PubMed] [Google Scholar]
- Visnjic D., Kalajzic, Z., Rowe, D.W., Katavic, V., Lorenzo, J., and Aguila, H.L. 2004. Hematopoiesis is severely altered in mice with an induced osteoblast deficiency. Blood 103: 3258-3264. [DOI] [PubMed] [Google Scholar]
- Waikel R.L., Kawachi, Y., Waikel, P.A., Wang, X.J., and Roop, D.R. 2001. Deregulated expression of c-Myc depletes epidermal stem cells. Nat. Genet. 28: 165-168. [DOI] [PubMed] [Google Scholar]
- Wanzel M., Herold, S., and Eilers, M. 2003. Transcriptional repression by Myc. Trends Cell. Biol. 13: 146-150. [DOI] [PubMed] [Google Scholar]
- Watnick R.S., Cheng, Y.N., Rangarajan, A., Ince, T.A., and Weinberg, R.A. 2003. Ras modulates Myc activity to repress thrombospondin-1 expression and increase tumor angiogenesis. Cancer Cell 3: 219-231. [DOI] [PubMed] [Google Scholar]
- Watt F.M. 2002. Role of integrins in regulating epidermal adhesion, growth and differentiation. EMBO J. 21: 3919-3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt F.M. and Hogan, B.L. 2000. Out of Eden: Stem cells and their niches. Science 287: 1427-1430. [DOI] [PubMed] [Google Scholar]
- Weissman I.L. 2000. Stem cells: Units of development, units of regeneration, and units in evolution. Cell 100: 157-168. [DOI] [PubMed] [Google Scholar]
- Weissman I.L., Anderson, D.J., and Gage, F. 2001. Stem and progenitor cells: Origins, phenotypes, lineage commitments, and transdifferentiations. Annu. Rev. Cell. Dev. Biol. 17: 387-403. [DOI] [PubMed] [Google Scholar]
- Whetton A.D. and Graham, G.J. 1999. Homing and mobilization in the stem cell niche. Trends Cell. Biol. 9: 233-238. [DOI] [PubMed] [Google Scholar]
- Wilson A., MacDonald, H.R., and Radtke, F. 2001. Notch 1-deficient common lymphoid precursors adopt a B cell fate in the thymus. J. Exp. Med. 194: 1003-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Niu, C., Ye, L., Huang, H., He, X., Tong, W.G., Ross, J., Haug, J., Johnson, T., Feng, J.Q., et al. 2003. Identification of the haematopoietic stem cell niche and control of the niche size. Nature 425: 836-841. [DOI] [PubMed] [Google Scholar]
- Zhu J. and Emerson, S.G. 2004. A new bone to pick: Osteoblasts and the haematopoietic stem-cell niche. Bioessays 26: 595-599. [DOI] [PubMed] [Google Scholar]