Abstract
During each yeast cell cycle, ∼100,000 nicks are generated during lagging-strand DNA replication. Efficient nick processing during Okazaki fragment maturation requires the coordinated action of DNA polymerase δ (Pol δ) and the FLAP endonuclease FEN1. Misregulation of this process leads to the accumulation of double-stranded breaks and cell lethality. Our studies highlight a remarkably efficient mechanism for Okazaki fragment maturation in which Pol δ by default displaces 2–3 nt of any downstream RNA or DNA it encounters. In the presence of FEN1, efficient nick translation ensues, whereby a mixture of mono- and small oligonucleotides are released. If FEN1 is absent or not optimally functional, the ability of Pol δ to back up via its 3′–5′-exonuclease activity, a process called idling, maintains the polymerase at a position that is ideal either for ligation (in case of a DNA–DNA nick) or for subsequent engagement by FEN1 (in case of a DNA–RNA nick). Consistent with the hypothesis that DNA polymerase ε is the leading-strand enzyme, we observed no idling by this enzyme and no cooperation with FEN1 for creating a ligatable nick.
Keywords: Okazaki fragment, DNA replication, DNA polymerase, nick translation, exonuclease
The nature of semidiscontinuous DNA replication in a eukaryotic cell mandates the synthesis of short RNA-primed Okazaki fragments on the lagging strand. During each cell cycle, tens of millions of these small Okazaki fragments are synthesized in a mammalian cell. Even during replication of the much more compact yeast genome, an estimated ∼100,000 Okazaki fragments are synthesized. Each of these fragments needs to be matured into continuous double-stranded DNA with high fidelity. Unrepaired nicks lead to double-strand breaks during the next cell cycle. These breaks can be repaired by the double-strand break repair system; however, the accumulation of as few as 30 unsealed nicks can overwhelm this system in yeast and cause cell lethality (Resnick 1978). Therefore, it is essential not only that the maturation of Okazaki fragments occurs with high speed and high fidelity, but also that backup mechanisms are available for the infrequent occasions that the prevailing maturation machinery fails.
Several different models have been proposed to account for initiator RNA degradation and gap filling during the maturation of Okazaki fragments (for review, see Kao and Bambara 2003). In one proposed mechanism, the initiator RNA is degraded by the sequential action of RNase H2 and the FLAP endonuclease FEN1. A second mechanism has been proposed in which the initiator RNA is degraded by the sequential action of the nuclease/helicase Dna2 and FEN1. Based on biochemical studies, we, and others, have recently proposed a third model for Okazaki fragment maturation, in which a simple machinery of DNA polymerase δ (Pol δ) and FEN1 carries out nick translation in a processive complex with the replication clamp PCNA (proliferating cell nuclear antigen) in order to degrade the RNA primer (Maga et al. 2001; Ayyagari et al. 2003; Kao et al. 2004). In this model, Dna2 and/or RNase H2 would only be required under rare conditions of dysfunction of the maturation complex, either by mutation, scarcity of one of the factors, or by the presence of unusual DNA structures, when the simple machinery starts to fail. Whereas in our model, Dna2 serves as an essential backup factor during Okazaki fragment maturation, others have proposed a constitutive function for this enzyme at the lagging strand (Budd and Campbell 1997; Bae et al. 2001, 2002). On the other hand, analysis of the mutation spectrum of RNase H2 mutator mutants suggests that this enzyme may act in an alternate pathway of Okazaki fragment maturation, which may only be activated upon FEN1 dysfunction (Qiu et al. 1999; Chen et al. 2000).
Unexpectedly, the 3′–5′ exonuclease activity of Pol δ was also identified as an important factor in Okazaki fragment maturation in vivo and in vitro, suggesting that this nuclease activity cooperates with FEN1 in creating a ligatable nick and preventing genome instability (Jin et al. 2001, 2003). Severe phenotypic defects, indicative of a failure in the maturation of Okazaki fragments, were observed if both activities were even partially compromised. The most frequent class of mutations observed in such mutants where the removal of 5′-flaps, generated by the DNA polymerase during Okazaki fragment synthesis, is impaired are extended duplications (up to 100 bp) flanked by short direct repeats (4–8 bp) (Tishkoff et al. 1997b; Jin et al. 2001, 2004).
More than 30 years ago, it was recognized that turnover of dNTPs to dNMPs during DNA replication by bacteriophage T4 DNA polymerase could function to increase replication fidelity (Muzyczka et al. 1972). The stochastic choice that the polymerase makes between 5′–3′ extension and 3′–5′ excision, and the influence of a mismatch on this choice, has been put in a kinetic framework for several DNA polymerases (for review, see Johnson 1993). Turnover also occurs when DNA synthesis with only one dNTP present leads to repeated cycles of incorporation and degradation at the primer terminus, or when progression of the polymerase is inhibited by the immediate downstream presence of stable hairpins in the template (Topal et al. 1980; Roth et al. 1981). These latter processes have been called “idling” because dNTP → dNMP turnover is not accompanied by net DNA synthesis or degradation.
Here we report that idling by Pol δ at a nick is an important mechanism for maintaining a ligatable nick during lagging-strand DNA replication. We present experimental evidence indicating that hidden in the simple Pol δ/FEN1 machinery is a homeostatic control mechanism that maintains this machinery at the position of the nick such that extended and deleterious strand-displacement synthesis cannot readily take place. This homeostatic control depends on a fully functional 3′–5′-exonuclease activity of Pol δ. We find that idling by Pol δ through successive cycles of polymerization and 3′-degradation serves to maintain the maturation complex at the position of the nick. Importantly, idling occurs in such a fashion that 2–3 nt of the 5′-strand are opened, providing an ideal substrate for FEN1, and thereby increasing the efficiency of nick translation. Idling is an unique property of the lagging-strand DNA polymerase δ. DNA polymerase ε (Pol ε) is the proposed leading-strand enzyme (Fukui et al. 2004). In support of the model that Pol ε is not required for Okazaki fragment maturation, we find that it does not idle at a nick.
Results
Strand-displacement synthesis by Pol δ
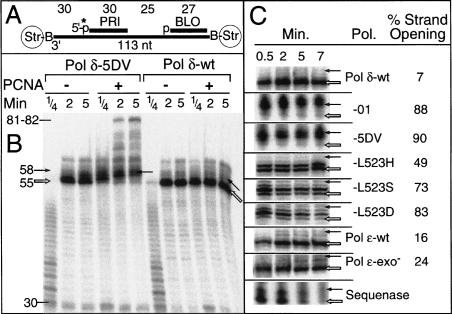
In order to accurately assess strand opening and strand-displacement synthesis by the processive PCNA–Pol δ complex, and determine the factors that contribute to it, we used a model oligonucleotide system (Table 1; Fig. 1A). The 113-nt-long oligonucleotide template is primed with a 30-mer primer (PRI) and with a downstream blocking oligo (BLO). The two oligos are separated by 25 template dT residues, and an additional 30 dT residues precede the PRI position. The single-stranded binding protein RPA is first bound to the oligo(dT) stretches, followed by loading of PCNA by replication factor C (RFC). The terminal biotin–streptavidin anchors prevent PCNA from sliding off the DNA (Kaboord and Benkovic 1993; Ayyagari et al. 2003). Upon initiating replication, 25 dATPs were incorporated by the polymerase prior to encountering the blocking oligo. Replication products longer than 55 nt are the result of strand-displacement synthesis. In general, strand-displacement synthesis by a DNA polymerase is more efficient when its exonuclease has been inactivated (e.g., see Canceill et al. 1999). This was also observed with Pol δ. Complete strand-displacement synthesis, as shown by the appearance of extension products 81–82 nt in length, was observed with the exonuclease-defective Pol δ-5DV, but only in a complex with PCNA (Fig. 1B). The wild-type enzyme was much less active in strand-displacement synthesis.
Table 1.
Oligonucleotides used in this study
| Bio-V6: | Bio-3′-T10TCCCTTCCCTCTCCCTCCTCTTCTTCCCTCT24TCCAAGGTGGTTTGTTTTGGTTGGGTTGA-5′Bio |
| C12 (PRI): | 5′-AGGGAAGGGAGAGGGAGGAGAAGAAGGGAG-3′ |
| pC6 (BLO): | 5′-pGGTTCCACCAAACAAAACCAACCCAAC-3′ |
| pRC6: | 5′-pGGUUCCACCAAACAAAACCAACC-3′ |
| pC6-2G: | 5′-pTTCCACCAAACAAAACCAACCCAAC-3′ |
| pAC6: | 5′-pAGGTTCCACCAAACAAAACCAACCCAAC-3′ |
| pA2C6: | 5′-pAAGGTTCCACCAAACAAAACCAACCCAAC-3′ |
| pA4C6: | 5′-pAAAAGGTTCCACCAAACAAAACCAACCCAAC-3′ |
| pA6C6: | 5′-pAAAAAAGGTTCCACCAAACAAAACCAACCCAAC-3′ |
The underlined sequence denotes RNA. The standard primer (PRI) and blocking (BLO) oligonucleotides are indicated (see also Fig. 1A).
Figure 1.
Strand-displacement synthesis by DNA polymerases. (A) Schematic of the substrate. The 113-mer template is Strep-Bio-V6 (Table 1); PRI is the 5′-32P-labeled 30-mer primer; BLO is the 5′-phosphorylated 27-mer blocking oligo. The template strand between the PRI and BLO positions is (pT)25 (see Materials and Methods). (B) DNA synthesis by Pol δ-5DV and Pol δ-wt with or without PCNA present. The standard assay (Materials and Methods) was modified to contain only 40 mM NaCl to allow PCNA-independent synthesis to proceed more efficiently. Full-length strand-displacement products should be 83 nt, but 1–2-nt shorter products were mainly observed because the biotin–streptavidin block inhibited progression of the polymerase. The open arrows indicate extension products paused at the nick position (55 nt, position 0) and the closed arrows indicate products at the +3 position (58 nt). (C) Strand opening by various DNA polymerases in the presence of PCNA. Only a close-up of the nick region is shown for each enzyme. The exo-deficient forms of Pol δ and Pol ε also carried out complete strand-displacement synthesis, generally up to ∼30% after 7 min (data not shown). The standard assays contained 100 mM NaCl, RPA, RFC, PCNA, and a threefold molar excess of each DNA polymerase over template-primer. Assays with Sequenase were carried out on naked DNA without accessory factors. Assays were at 30°C for the indicated times and analyzed by 7 M urea/12% PAGE, followed by PhosphorImaging of the dried gel. Strand opening (as percent) is defined as 100 × [products in positions 1 + 2 + 3 + 4]/[products in positions 0 + 1 + 2 + 3 + 4] at the 5-min time point.
Interestingly, when Pol δ-5DV replicated into the double-stranded DNA region, the major replicational pause site was at positions 2–3 nt into the double-stranded DNA, whereas pausing by the wild-type was at the precise nick position. Previously, we had detected strand opening of 2–3 nt into different template-primer sequences by Pol δ-5DV, indicating that this is a general property of the polymerase (Jin et al. 2003). In this paper, we call the characteristic displacement of the nick-proximal few nucleotides strand opening, reserving strand-displacement synthesis for more extensive synthesis into the double-stranded region. Our working hypothesis is that strand opening is a reversible step for wild-type Pol δ, whereas strand displacement is not. Similar to what is observed for complete strand-displacement synthesis, the appearance of strand-opened products generated by Pol δ-5DV was also strongly stimulated by PCNA. Furthermore, the relative distribution of pause sites stayed constant once the polymerase reached the blocking oligo.
We surveyed several forms of Pol δ and two forms of Pol ε to determine the percentage of strand opening in the presence of PCNA at 100 mM NaCl. This percentage is defined as 100 × [products in positions +1 to +4]/[products in positions 0 to +4], where position 0 identifies the precise nick position (open arrows in Fig. 1B,C). During the course of the assay, all forms of Pol δ and Pol ε maintained a fairly constant ratio of pausing products between that at the nick position and those at the +1 to +4 positions (Fig. 1C). Interestingly, whereas the exonuclease-deficient forms of Pol δ, that is, Pol δ-01 and Pol δ-5DV, produced a greatly increased percentage of strand-opened products compared to wild-type, the same was not observed with the exonuclease-deficient Pol ε, which predominantly paused at the precise nick position. Pausing by Sequenase was much more transient.
Recently, we studied a series of POL3 mutator mutations at Leu 523 in the exonuclease domain (Jin et al. 2004). Of these, Pol δ-L523S and Pol δ-L523H show a close to wild-type or wild-type exonuclease activity, respectively (Table 2). Their in vivo defects, including a defect in Okazaki fragment maturation, have been proposed to result from a partial defect in switching of the template-primer between the polymerase and exonuclease domains. The L523X mutant enzymes also show increased strand opening, suggesting that the lack of observable strand opening by the wild-type enzyme may be a reflection of efficient switching from the polymerase to the 3′–5′-exonuclease domain after strand opening had occurred, followed by rapid degradation back to the nick position (Fig. 1C).
Table 2.
Overview of Pol δ mutants
| Growth with
|
||||||
|---|---|---|---|---|---|---|
| Allele | Mutation rate can1 | rad27-Δ | rad27-p | Exonuclease activity | Idling rate | Nick ligation |
| wild type | 1 | + | + | 100 | 100 | 100 |
| -DE321,323AA (01) | 19 | - | - | <0.1 | 0 | 2 |
| -D520V (5DV) | 13 | - | + | <0.1 | 0 | 2 |
| -L523H | 1.5 | - | + | 100 | 32 | 64 |
| -L523S | 20 | - | - | 68 | 10 | 6 |
| -L523D | 110 | - | n.d. | 10 | 1 | 9 |
Rapid nucleotide turnover at a nick by Pol δ
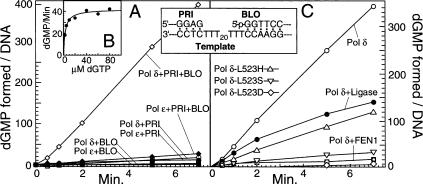
Successive cycles of 5′-strand opening followed by 3′-degradation are expected to result in overall nucleotide turnover (dNTP → dNMP) at the nick. In the oligonucleotide-based system, initial DNA replication only requires the incorporation of 25 dA residues by Pol δ. Subsequent incorporation of the next two nucleotides, two dGTPs, would require strand opening of the blocking oligo (Fig. 2). If strand opening were followed by degradation, dGMP would be liberated. Through the inclusion of radioactive dGTP in the assay, the conversion of dGTP to dGMP was measured by thin layer chromatography.
Figure 2.
Nucleotide turnover at a nick. (A) Standard dGTP → dGMP turnover assays contained the Strep-Bio-V6 template either with the PRI oligo or with the BLO oligo, or with both, as indicated (see Fig. 1A). Reactions were carried out for the indicated times with Pol δ or with Pol ε. (B) The standard turnover assay, with both the PRI and BLO oligos hybridized, contained wild-type Pol δ and increasing [α-32P]dGTP, from 2.5 μM to 80 μM. Rates of turnover were calculated from time-course assays and plotted against the dGTP concentration. Half-maximal turnover rates were obtained at 3 μM dGTP. (C) The standard turnover assay, with both the PRI and BLO oligos hybridized, contained wild-type or L523X Pol δ, as indicated, or wild-type Pol δ with FEN1 or with DNA ligase I. (Inset) Sequences of the oligos at the junction positions are shown.
When only the primer oligo or the blocking oligo was hybridized to the template, very limited turnover of dGTP by Pol δ was observed. However, when both oligos were hybridized to the template, turnover was stimulated >20-fold, indicative of active idling by the polymerase at the nick (Fig. 2A). Interestingly, dGTP → dGMP turnover by Pol ε did not increase substantially when it encountered the blocking oligo. This lack of nucleotide turnover is not an artifact resulting from complete strand-displacement synthesis by Pol ε. Less than 20% of the blocking oligo was fully displaced by Pol ε during the time period of the assay (data not shown).
The turnover assays were carried out with 100 μM each of the nonradioactive dNTPs and increasing concentrations of [α-32P]dGTP. The half-maximal rate of turnover was reached at 3 μM dGTP (Fig. 2B). Therefore, all subsequent assays were carried out with 10 μM dGTP in order to be close to saturation while still retaining the sensitivity of the assay. Additional control experiments showed that the rate of idling was dependent on the concentration of the DNA polymerase. However, once saturation was reached with the polymerase in excess over DNA, addition of even a 10-fold molar excess of polymerase over DNA did not further increase the observed turnover rate, indicating that idling occurred only in the stable polymerase–DNA complex (data not shown). Therefore, all DNA polymerases were used in three- to fourfold molar excess over DNA.
Idling by Pol δ was not limited to a DNA–DNA nick. When the blocking oligo contained an initial 8 nt of RNA, efficient idling at the DNA–RNA junction was also observed (Table 3). Therefore, both gap filling and Okazaki fragment maturation can be accompanied by idling at a similar rate. Consistent with our observation that strand opening by the exonuclease-deficient Pol δ-5DV required PCNA, idling by the wild-type enzyme was abolished when PCNA was absent (Table 3). Furthermore, when DNA ligase was included in the assay, the rate of idling decreased with time (Fig. 2C), because ligation of the nick was achieved (∼60% ligation after 7 min) (data not shown). Inclusion of FEN1 in the assay completely abolished idling, indicating efficient utilization of the invaded strand by FEN1 to give nick translation.
Table 3.
Nucleotide turnover at a nick
| Turnover
|
||
|---|---|---|
| Condition | dGTP | dTTP |
| Block = pC6 (BLO) | 100 | 9 |
| Block = pRC6 (RNA8DNA15 | 97 | 14 |
| Block = pC6-2G | 16 | 57 |
| 40 mM NaCl | 132 | |
| 40 mM NaCl-PCNA | 2 | |
| Pol δ-5DV or Pol δ-01 | 0 | |
Standard turnover assays were modified as indicated. The turnover rate of dGTP with pC6 (BLO) and wild-type Pol δ at 100 mM NaCl (standard conditions) was set to 100. See Figure 3 for alignment of the different oligos. The sequence of pRC6 was identical to that of pC6, except that the eight 5′-nucleotides were RNA.
The mutator activity of pol3-L523X mutants has been proposed to result in part from a defect in switching of the template-primer between the polymerase and exonuclease domains (Jin et al. 2004). We reasoned that a similar domain switching would also be required for efficient idling, and therefore be important for high-fidelity gap filling and Okazaki fragment maturation. Of the three mutant enzymes, Pol δ-L523H is the only one to show wild-type exonuclease activity while being partially defective in switching. This enzyme idled with only 32% the rate of wild-type, indicating that idling requires efficient switching between the polymerase and exonuclease domains (Fig. 2C; Table 2). Idling by the more severe mutator Pol δ-L523S was 10% of wild type, and no significant idling by Pol δ-L523D was observed.
Strand opening is a prerequisite for nucleotide turnover
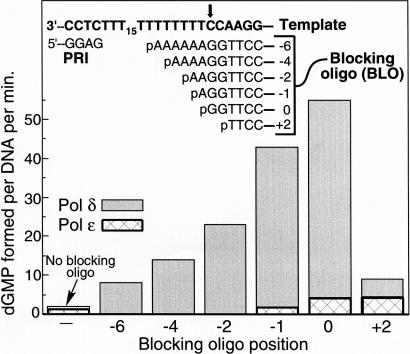
We carried out two complementary series of experiments to determine how many nucleotides on either side of the nick are subject to idling. In the first assay, turnover of dTTP, representing positions +3 and +4 past the nick, was compared to that of dGTP (positions +1 and +2). Turnover of dTTP was substantially lower than that of dGTP with both the DNA and RNA blocking oligo (Table 3). No significant turnover of dCTP, corresponding to positions +5 and +6, was observed (data not shown).
In the second set of experiments, we measured dGTP turnover using a series of blocking oligos with different 5′-end positions (Fig. 3; Table 1). When the blocking oligo was shortened by 2 nt at the 5′-end (pC6-2G in Table 1), turnover of dGTP was severely reduced, but that of dTTP was enhanced (Fig. 3; Table 3). The main conclusion from this series of experiments is that turnover by Pol δ of dNTPs corresponding to those nucleotides just prior to the double-stranded DNA block is not efficient. On the other hand, when the length of the blocking oligo was increased at the 5′-end by the successive addition of dA residues (pAC6 through pA6C6 in Table 1), a concomitant decrease in dGTP turnover was observed, indicating that the highest turnover is observed for those dNTPs corresponding to 2–3 nt into the double-stranded region. The very low nick-dependent nucleotide turnover that we observed with Pol ε was also further characterized (Fig. 3). In contrast to Pol δ, turnover catalyzed by Pol ε proceeded both via strand opening and gap formation with comparable rates.
Figure 3.
Efficient turnover by Pol δ requires strand opening. Standard dGTP → dGMP turnover assays contained the Strep-Bio-V6 template primed with the PRI oligo and one of a series of BLO oligos as shown. The C on the template indicated by the arrow is defined as position 0, and therefore blocking oligo pC6 (5′-p-GGTTCC—) has its 5′-junction at the 0 position, and the other oligos accordingly. Blocking oligo pC6-2G (Table 1) is the +2 oligo. Turnover rates with Pol δ (gray) or with Pol ε (cross-hatched) were plotted as a function of the junction position.
Idling maintains the Pol δ machinery at a ligatable nick position
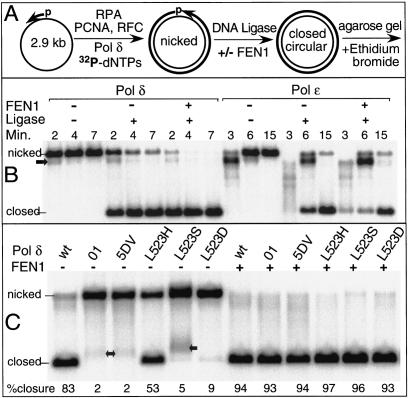
Idling by Pol δ appears to shuttle the DNA substrate primarily between a strand-displacement product and a precise nick. Therefore, in the presence of DNA ligase I, idling should allow efficient ligation of DNA replication products. We used a circular SS DNA substrate primed with a 5′-phosphorylated primer to assay the accuracy of gap-filling synthesis and nick closure (Ayyagari et al. 2003). DNA ligase-catalyzed closure of the nick resulting from precise gap filling produces covalently closed circular DNA that has a unique migration position when electrophoresed through an agarose gel in the presence of ethidium bromide (Fig. 4A). Replication by Pol δ in the presence of DNA ligase I produced predominantly covalently closed DNA after 4 min of reaction (Fig. 4B). Addition of FEN1 increased the efficiency of closure to virtually 100%. Replication by Pol ε in the presence of DNA ligase also produced covalently closed double-stranded circles albeit at a lower rate. However, addition of FEN1 did not significantly alter the efficiency of closure, suggesting that Pol ε and FEN1 do not act synergistically at a nick.
Figure 4.
Gap filling and ligation by Pol δ and Pol ε. Assays were as described in Materials and Methods. (A) The scheme shows the proposed sequential action of the factors. However, in the assay PCNA was first loaded by RFC onto RPA-coated primed DNA. Subsequently, Pol δ or Pol ε was added together with DNA ligase and FEN1, where indicated. (B) Comparison of Pol δ and Pol ε. Replication was for the indicated times at 30°C. PCNA was increased to a 10-fold excess over DNA to increase processivity of Pol ε (Burgers 1991). The arrow indicates a replicational pause that is particularly strong with Pol ε. (C) Comparison of Pol δ mutants. All assays were for 7 min at 30°C. The replication products indicated by the arrows are due to aberrant strand displacement. The generation of these products is suppressed in the presence of FEN1.
Studies of the Pol δ mutants revealed that only the L523H mutant produced covalently closed DNA with some efficiency (53%), compared to 83% for wild type. The other mutants yielded little or no covalently closed DNA, suggesting that those enzymes did not generate or maintain ligatable nicks (Fig. 4C). The severity of the observed defect in the production of covalently closed DNA by these DNA polymerases is a function of their increased strand-opening capacity (Fig. 1B). However, when FEN1 was also added to the assay, proper processing of the displaced strand produced ligatable nicks with near full efficiency for all forms of Pol δ.
Coupled activity of Pol δ and FEN1 during nick translation
Given the propensity of Pol δ and its exonuclease-deficient mutants to rapidly displace 2–3 nt of DNA, we expected that nick translation in the presence of FEN1 would primarily result in the release of small oligonucleotides. In this assay the blocking oligo was 5′-labeled to allow detection of FEN1-dependent degradation products made specifically during nick translation (Fig. 5A). Incubation of the substrate with Pol δ and dNTPs did not liberate small labeled products, whereas incubation with just FEN1 liberated the expected mononucleotide dGMP and very low amounts of the trinucleotide (Harrington and Lieber 1994). Surprisingly, however, when both Pol δ and FEN1 were included so that nick translation could take place, dGMP was the main reaction product and larger products, di- to tetranucleotides, made up a total of only 5% of the products specifically released by the combined action of Pol δ and FEN1 (Fig. 5A).
Figure 5.
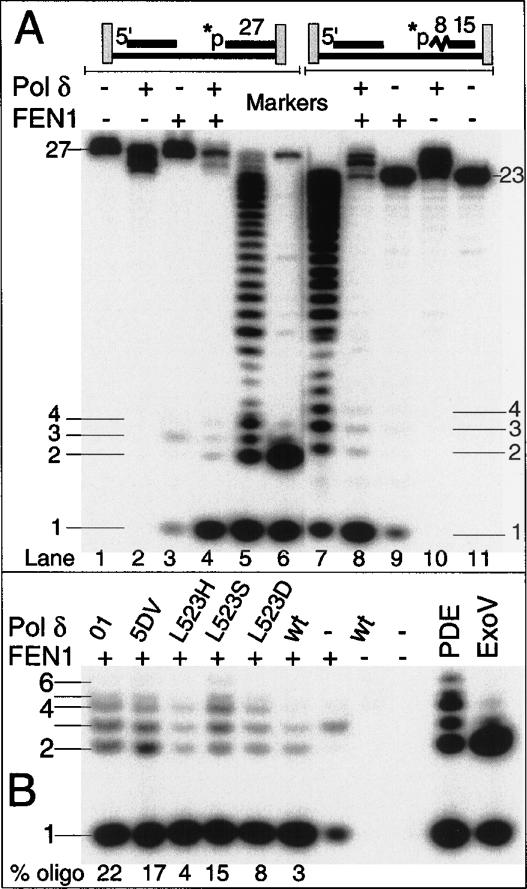
Coupled action of Pol δ and FEN1 at a nick. Nick translation assays were as described in Material and Methods. (A) Analysis of nick translation products at a 5′-DNA or a 5′-RNA nick. Nick translation was carried out for 1 min at 30°C. Prolonged incubation, up to 5 min, did not significantly change the distribution of the small oligonucleotide and mononucleotide products (data not shown). Control reactions carried out under otherwise identical conditions were as indicated. In the marker lanes, the DNA oligo was partially digested with snake phosphodiesterase (PDE, lane 5), or with exonuclease V (ExoV, lane 6), which produces a 5′-dinucleotide (Burgers et al. 1988), and the RNA–DNA oligo with PDE (lane 7). (B) Comparison of Pol δ mutants in nick translation. The indicated forms of Pol δ were used on the DNA oligo substrate. Only the mononucleotide to hexanucleotide region is shown. None of the DNA polymerases showed products in this region in the absence of FEN1.
FEN1 also efficiently cuts RNA tails, and therefore nick translation into downstream RNA should proceed similarly (Murante et al. 1996). Previous studies had shown that Pol δ and FEN1 carry out a remarkable efficient RNA degradation coupled to DNA replication of model substrates such as shown in Figure 4 in which the primer was RNA (Jin et al. 2003). However, how RNA degradation was accomplished during maturation has not yet been determined. A comparable distribution of products was observed when the nick translation machinery encountered an RNA rather than a DNA primer; ∼8% of the specific product were oligoribonucleotides while 92% was GMP. These results indicate that the functional coupling between Pol δ and FEN1 is extremely efficient.
The distribution of nick translation products was also analyzed with the Pol δ mutants. With the exception of Pol δ-L523H, each one of the mutants released a much higher percentage of oligonucleotides than wild type (Fig. 5B). The exonuclease-defective Pol δ-01 and the switching-defective Pol δ-L523S released even hexanucleotide products at a low frequency and a total of 22% oligonucleotides was released during nick translation by Pol δ-01 and FEN1.
Discussion
The hypothesis that the 5′-nuclease activity of FEN1 and the 3′-exonuclease activity of Pol δ act cooperatively during Okazaki fragment maturation and simple gap filling stemmed from the observation that double mutants in which mutations in RAD27(FEN1) are combined with exonuclease mutations in POL3 (Pol δ) are either lethal, or show a strong genetic instability phenotype indicative of Okazaki fragment maturation defects (Jin et al. 2001, 2004). Preliminary DNA replication studies revealed a cooperation between the Pol δ exonuclease and FEN1 in forming a ligatable nick (Jin et al. 2003). However, the biochemical principle underlying this hypothesis remained unclear. Here, we provide compelling evidence that the main function of the 3′–5′-exonuclease in maturation is to prevent uncontrolled strand-displacement synthesis by the polymerase, through idling. Unlike other DNA polymerases, Pol δ uniquely carries out limited strand-opening synthesis when downstream double-stranded RNA–DNA is encountered. By doing so, the substrate for FEN1 cutting is generated on the fly, allowing rapid and efficient degradation of the RNA portion of an Okazaki fragment to take place. DNA ligase I does not catalyze ligation of an RNA–DNA nick (Ayyagari et al. 2003). This specificity ensures that the entire RNA portion of an Okazaki fragment is degraded before ligation can occur. However, once the RNA has been degraded, closure can occur as early as a few nucleotides past the RNA–DNA junction (Ayyagari et al. 2003).
Role of strand opening in Okazaki fragment maturation
Given the degree of strand opening effected by Pol δ, we expected that the products formed during a coupled nick translation reaction would mostly consist of di- and trinucleotides. Surprisingly, the degradation products observed were mainly mononucleotides, regardless of whether nick translation proceeded through RNA or DNA (Fig. 5A). Therefore, during lagging-strand DNA replication the RNA primer is likely degraded through repeated cycles of strand opening and FEN1 action. Interestingly, inactivation of the exonuclease activity of Pol δ generated both longer oligonucleotides and a higher proportion of oligonucleotides to mononucleotides, consistent with the increased strand opening observed with these enzymes (Fig. 1B). However, even then, mononucleotides still made up the majority of the degradation products (Fig. 5B). An alternative explanation is that the presence of FEN1 in the maturation complex may modify the strand-opening capability of Pol δ to that of only a single nucleotide, which is then rapidly cut by FEN1. Currently, our data do not allow us to distinguish between these two mechanisms.
Why is strand opening by Pol δ not much more extensive so that it can exceed the RNA portion of an Okazaki fragment, ∼10 nt in length (Badaracco et al. 1985)? In that case, the entire RNA portion of an Okazaki fragment could be removed in one single-coupled reaction by Pol δ and FEN1. Our studies indicate, for the first time, that strand opening is actually not even allowed to proceed for more than 1 nt when FEN1 is present (Fig. 5A). The strong coupling between FEN1 and Pol δ indicates that lagging-strand replication proceeds in such an efficient manner that nucleotides are cleaved by FEN1 as soon as they are displaced by Pol δ. Even when increased strand opening is catalyzed by an exonuclease-deficient form of Pol δ, the tight coupling to FEN1 cutting is still maintained to a large extent (Fig. 5B). Owing to the high coupling efficiency, it is perhaps not surprising that the exonuclease-deficient forms of Pol δ do not show a large defect in Okazaki fragment maturation in vivo, unless FEN1 is also compromised (Jin et al. 2001, 2004). For instance, neither the single pol3-L523H mutant, which shows a minor defect in switching between the polymerase and exonuclease domains, nor the single rad27-p mutant, which specifies FEN1 compromised for binding PCNA, shows an increase in duplication mutations. However, the double mutant shows a large increase of extended duplications flanked by short direct repeats, indicative of a failure in the maturation of Okazaki fragments (Jin et al. 2004).
Another important consideration is that if more extensive strand displacement were allowed to take place, it would increase the chance of protein binding to the displaced SS RNA or DNA or the formation of secondary structures in the SS tail, both of which inhibit access by FEN1 (Murante et al. 1995; Gordenin et al. 1997). The critical tail length in vivo that would allow protein binding and consequent FEN1 inhibition is unknown, but in the reconstituted in vitro systems with purified proteins, it is 25–30 nt, the binding site size of RPA (Bae et al. 2001; Ayyagari et al. 2003; Kao et al. 2004). In such cases, the Dna2 nuclease/helicase could act to trim down this 5′-tail, but this must be considered a fail-safe rather than a standard response. Therefore, it appears that the cell has opted for very tight control and safety by allowing only a few nucleotides to be displaced in a single action cycle.
Idling by Pol δ prevents extensive strand displacement
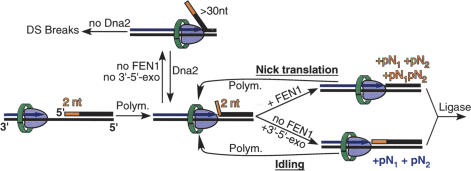
Uncontrolled strand-displacement synthesis forms a potential problem associated with the strand-opening approach to maturation, and can lead to the formation of double-strand breaks and duplication products (Jin et al. 2004). To curb extended strand displacement, Pol δ uses a control mechanism that consists of successive rounds of limited strand opening and 3′-degradation, called idling. This process cycles the DNA substrate between a nick, which can be ligated, and a strand-opening product, which can be cut by FEN1. Notably, the extent of strand opening of 2–3 nt, as determined with the exonuclease-deficient forms of Pol δ (Fig. 1C), matches the number of nucleotides in the double-stranded DNA that undergo most efficient idling (2–3 nt) (Fig. 3). Figure 6 shows our model in which a ligatable nick can be maintained either by idling or by nick translation.
Figure 6.
A binary choice model for Pol δ to maintain a ligatable nick. A DNA gap and displacement of 2 nt by Pol δ is shown. During Okazaki fragment maturation, Pol δ and FEN1 go through multiple cycles of displacement synthesis and FLAP cutting (upper cycle, nick translation) until all RNA has been degraded. pN1pN2 is the oligonucleotide released during nick translation, whereas pN1 and pN2 indicate mononucleotides released either during nick translation (orange) or during idling (purple).
Under our experimental conditions, nucleotide turnover occurred at a rate of ∼50 dGMPs/min produced per DNA substrate. Assuming that two dGTP residues are incorporated and two dGMPs produced during each idling cycle with this DNA substrate (Fig. 3), idling occurs at a rate of 0.4 cycles/sec. Under comparable conditions, nick translation by the concerted action of Pol δ and FEN1 occurs at a rate of 1.7 nt/sec (Ayyagari et al. 2003). Consequently, nucleotide turnover is expected to be suppressed in the presence of FEN1 because nick translation dominates. Unfortunately, in the experimental system we used to measure idling, the end of the template is rapidly reached during nick translation. Therefore, the observed suppression of nucleotide turnover in the presence of FEN 1 is more a reflection of the limitation of the system than a true measurement of nucleotide turnover during nick translation (Fig. 2B).
Relationship between strand opening, idling, and ligatability of a nick
How does the DNA polymerase initiate strand opening, and how is the extent of strand opening regulated by the polymerase? Based on a structural comparison between phage T7 RNA polymerase and the Klenow fragment of Escherichia coli DNA polymerase I, a helix in the finger domain has been implicated in strand separation of downstream DS DNA (Steitz and Yin 2004). Release of pyrophosphate after nucleotide incorporation is correlated with a conformational change in the finger domain that may be coupled to strand separation. The closest structural model for Pol δ is that of the bacteriophage RB69 DNA polymerase, which unfortunately does not present a clue regarding strand separation. However, in the structure of the replicating complex, the downstream template strand exits the active site through a groove made by the N-terminal domain and the exonuclease domain (Franklin et al. 2001). Therefore, it is not unreasonable to assume that the exonuclease domain of the structurally analogous Pol δ may participate in regulating the degree of strand opening at a nick. Our study of two exonuclease-deficient forms of Pol δ with different strand-opening signatures supports this hypothesis. Strand opening of 2–3 nt predominates for Pol δ-5DV, whereas 3–4-nt strand-opening products predominate for Pol δ-01 (Fig. 1C). Pause sites at the +1 position predominate for the three L523X mutants. Since the L523D mutant enzyme does not idle, it appears that in this mutant strand opening by only 1 nt is most common. It is therefore likely that the different structural perturbations associated with these different mutations affect putative interactions with the downstream double-stranded DNA. Whatever the mechanism may be that dictates pausing of Pol δ after limited strand opening, it is likely that this is not determined by the length of the displaced 5′-tail. In previous studies we have shown that the extent of opening into the double-stranded region by Pol δ-5DV was the same, regardless of whether the downstream 5′-strand was fully hybridized or if it contained a 10-nt noncomplementary 5′-tail (Jin et al. 2003).
The study of exonuclease-proficient but switching-defective Pol δ-L523X mutants has also allowed us to establish a relationship between strand opening, idling, and maintenance of a ligatable nick. Both the L523H and L523S mutants of Pol δ have wild-type or close to wild-type exonuclease activity, respectively (Table 2). Switching of the DNA from the polymerase to the exonuclease domain after misincorporation is more defective in the L523S than in the L523H mutant, making the former a stronger mutator (Jin et al. 2004). Strand opening increased from 7% with wild-type Pol δ to 49% with L523H and 73% with L523S. Associated idling rates were 50 nt/sec, 16 nt/sec, and 5 nt/sec, and nick ligatability decreased from 83% to 53% and 5%, respectively, for these enzymes (Figs. 1C, 4; Table 2). Therefore, both proofreading and idling require the same action by the DNA polymerase, that is, rapid switching of the DNA substrate, either a mismatch or a strand-opened product, from the polymerase to the exonuclease domain. These in vitro phenotypes are reflected in the cell, where we found that pol3-L523S is not only synthetic lethal with rad27-Δ, but also with a mild mutation in FEN1 (rad27-p) that decreases its affinity for PCNA. On the other hand, pol3-L523H is only synthetic lethal with rad27-Δ, although the viable pol3-L523H rad27-p double mutant accumulates duplication mutations indicative of defects in Okazaki fragment maturation (Jin et al. 2004). In view of the fact that RAD27 deletion mutants are viable but double mutants that also delete a related nuclease EXO1 are not, we assume that Exo1 can substitute, albeit less efficiently, for FEN1 during nick translation (Tishkoff et al. 1997a; Tran et al. 2002). However, this assumption still requires direct experimental verification. Apparently, the functional interactions between Pol δ and Exo1, while sufficient for viability, are so stressed that even a very mild perturbation of the exonuclease such as in pol3-L523H is sufficient to cause lethality.
Idling is specific for the lagging-strand DNA polymerase
These studies allow us to compare the functions of Pol δ and Pol ε in Okazaki fragment maturation and gap filling. Elegant genetic experiments with mutator DNA polymerases have shown that Pol δ and Pol ε are opposite-strand DNA polymerases (Shcherbakova and Pavlov 1996; Karthikeyan et al. 2000). The assignment of Pol ε as the leading-strand enzyme follows primarily strong experimental support for Pol δ as the lagging-strand enzyme, although in vitro replication studies with Xenopus extracts provide suggestive evidence for Pol ε as the leading-strand enzyme (Fukui et al. 2004; for review, see Kao and Bambara 2003). In this study we find that the weak idling carried out by Pol ε is as likely to proceed by degradation from the nick, leading to a small gap, than by strand opening (Figs. 2, 3). This would make Pol ε inefficient at nick translation. Indeed, in a gap-filling assay, we find no evidence for a functional interaction between FEN1 and Pol ε (Fig. 4B).
Both Pol δ and Pol ε are B class DNA polymerases, all of which have an invariant aspartate residue in the ExoIII motif (D520 in Saccharomyces cerevisiae) that is essential for exonuclease catalysis (Soengas et al. 1992; Jin et al. 2001). The Pol δ-5DV enzyme used in this study is mutated at D520. Several amino acids near this catalytic aspartate are invariant in all 26 sequenced Pol δ catalytic subunits, deposited in GenBank, but not in other B class DNA polymerases (Jin et al. 2004). L523 is one of these invariant Pol δ-specific amino acids. It is unlikely to be directly involved in exonuclease catalysis, but may well be important for a specific Pol δ-related function, such as the ability of strand opening and active site switching. Certainly, it appears that the acquisition of strand opening and idling are unique evolutionary properties acquired by Pol δ to deal with the recurring challenge of Okazaki fragment maturation during lagging-strand DNA synthesis. Given these properties, it may also have been developed as the preferred enzyme for regular gap filling, for example, during base excision repair (Blank et al. 1994; Jin et al. 2003).
Materials and methods
Materials
DNA Ligase I, Pol δ, and its mutants were purified from yeast overproduction strains as previously described (Burgers and Gerik 1998; Ayyagari et al. 2003; Jin et al. 2003, 2004). Pol ε and the exonuclease-deficient form of Pol ε (containing Pol2–4) were also overproduced and purified from yeast (Chilkova et al. 2003; Shcherbakova et al. 2003). Replication protein A (RPA), PCNA, Replication factor C (RFC), and FEN1 were purified from E. coli overproduction strains (Henricksen et al. 1994; Ayyagari et al. 2003). All other enzymes were obtained commercially.
All oligonucleotides were obtained from Integrated DNA Technologies and purified by polyacrylamide gel electrophoresis or HPLC before use. The 113-nt 5′- and 3′-biotinylated template Bio-V6 (Table 1) was prepared by hybridizing two half oligos to a bridging primer followed by ligation with T4 DNA ligase and purification by preparative urea-PAGE (see Fig. 1A). The oligos were hybridized to the template at 70°C. After hybridization, streptavidin was added in a twofold molar excess.
Idling assays
Standard 80 μL assays contained 20 mM Tris-HCl at pH 7.8, 1 mM dithiothreitol, 100 μg/mL bovine serum albumin, 8 mM MgAc2, 0.5 mM ATP, 100 mM NaCl, 10 μM (unless otherwise indicated) the [α-32P]dNTP for which turnover was being measured, 100 μM the other dNTPs, 580 fmol of DNA, 3 pmol of RPA, 1 pmol of RFC, and 2 pmol of PCNA. The DNA was preincubated with RPA, PCNA, and RFC for 30 sec at 30°C, and the reaction was started by adding the relevant DNA polymerase (1.5–3 pmol). Incubations were at 30°C. Aliquots of 10 μL were quenched by the addition of 5 μL of stop buffer (25 mM EDTA, 1% SDS, and 5 mM each of the relevant nonradioactive dNTP and dNMP). Reaction products were analyzed by thin layer chromatography on PEI/cellulose in 0.5 M LiCl/HCOOH, followed by PhosphorImager analysis.
Strand-displacement and nick translation assays
The idling assays were modified as follows. Either primer C12 (strand-displacement assays in Fig. 1), or oligo pC6 or pRC6 (nick translation assays in Fig. 5) was labeled at the 5′-end using T4 polynucleotide kinase (NEB) and [γ-32P]ATP. The labeled oligo, together with a fivefold excess of the other nonlabeled oligo, was hybridized to a twofold molar excess, over the labeled oligo, of template Bio-V6. All other reaction conditions were the same, except that 100 μM each nonradioactive dNTPs were included. Reactions were analyzed on 12% (strand displacement) or 20% (nick translation) denaturing polyacrylamide gels. The gels were dried and exposed in a PhosphorImager. Quantitation was carried out using ImageQuant software (Amersham).
Maturation assays
Standard 25-μL assays contained 20 mM Tris-HCl at pH 7.8; 1 mM dithiothreitol; 100 μg/mL bovine serum albumin; 8 mM MgAc2; 1 mM ATP; 100 μM each dATP, dCTP, and dGTP; 10 μM [α-32P]dTTP; 100 mM NaCl; 100 fmol of single-stranded bluescript SKII+ DNA primed with SKdc10 (5′-p-ACGACGTTG TAAAACGACGGCCAGTGAGCG); 10 pmol of RPA; 250 fmol of PCNA; 200 fmol of RFC; 200 fmol of Pol δ wild-type or mutant or 300 fmol of wild-type Pol ε; and when indicated, 250 fmol of FEN1 and/or 500 fmol of DNA ligase. The DNA was preincubated with RFC, PCNA, and RPA for 30 sec at 30°C, and the reaction was started by the addition of the rest of the proteins in a mixture. Incubations were for the indicated times at 30°C. The products were analyzed by electrophoresis on a 1% agarose gel containing 0.5 μg/mL ethidium bromide. The gels were dried and analyzed on a PhosphorImager. ImageQuant software was used to quantitate the data. The images in the figures were contrast-enhanced for visualization purposes.
Acknowledgments
We thank John Majors and Dmitry Gordenin for critical discussions during the course of this work. This work was supported in part by grant GM32431 from the National Institutes of Health (to P.M.B.); by grants from the Swedish Research Council, the Swedish Cancer Society, the Magnus Bergwalls Stiftelse, and the Medical Faculty of Umeå University (to E.J.); the Sigma Foundation (to P.G.); and the Kempe Foundation (to N.S.).
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1252304.
References
- Ayyagari R., Gomes, X.V., Gordenin, D.A., and Burgers, P.M. 2003. Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 and DNA2. J. Biol. Chem. 278: 1618-1625. [DOI] [PubMed] [Google Scholar]
- Badaracco G., Bianchi, M., Valsasnini, P., Magni, G., and Plevani, P. 1985. Initiation, elongation and pausing of in vitro DNA synthesis catalyzed by immunopurified yeast DNA primase: DNA polymerase complex. EMBO J. 4: 1313-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae S.H., Bae, K.H., Kim, J.A., and Seo, Y.S. 2001. RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature 412: 456-461. [DOI] [PubMed] [Google Scholar]
- Bae S.H., Kim, D.W., Kim, J., Kim, J.H., Kim, D.H., Kim, H.D., Kang, H.Y., and Seo, Y.S. 2002. Coupling of DNA helicase and endonuclease activities of yeast Dna2 facilitates Okazaki fragment processing. J. Biol. Chem. 277: 26632-26641. [DOI] [PubMed] [Google Scholar]
- Blank A., Kim, B., and Loeb, L.A. 1994. DNA polymerase δ is required for base excision repair of DNA methylation damage in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 91: 9047-9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd M.E. and Campbell, J.L. 1997. A yeast replicative helicase, Dna2 helicase, interacts with yeast FEN-1 nuclease in carrying out its essential function. Mol. Cell. Biol. 17: 2136-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgers P.M.J. 1991. Saccharomyces cerevisiae Replication factor C. II. Formation and activity of complexes with the proliferating cell nuclear antigen and with DNA polymerases delta and epsilon. J. Biol. Chem. 266: 22698-22706. [PubMed] [Google Scholar]
- Burgers P.M. and Gerik, K.J. 1998. Structure and processivity of two forms of Saccharomyces cerevisiae DNA polymerase delta. J. Biol. Chem. 273: 19756-19762. [DOI] [PubMed] [Google Scholar]
- Burgers P.M.J., Bauer, G.A., and Tam, L. 1988. Exonuclease V from Saccharomyces cerevisiae. A 5′ → 3′-deoxyribonuclease that produces dinucleotides in a sequential fashion. J. Biol. Chem. 263: 8099-8105. [PubMed] [Google Scholar]
- Canceill D., Viguera, E., and Ehrlich, S.D. 1999. Replication slippage of different DNA polymerases is inversely related to their strand displacement efficiency. J. Biol. Chem. 274: 27481-27490. [DOI] [PubMed] [Google Scholar]
- Chen J.Z., Qiu, J., Shen, B., and Holmquist, G.P. 2000. Mutational spectrum analysis of RNase H(35) deficient Saccharomyces cerevisiae using fluorescence-based directed termination PCR. Nucleic Acids Res. 28: 3649-3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilkova O., Jonsson, B.H., and Johansson, E. 2003. The quaternary structure of DNA polymerase epsilon from Saccharomyces cerevisiae. J. Biol. Chem. 278: 14082-14086. [DOI] [PubMed] [Google Scholar]
- Franklin M.C., Wang, J., and Steitz, T.A. 2001. Structure of the replicating complex of a Pol α family DNA polymerase. Cell. 105: 657-667. [DOI] [PubMed] [Google Scholar]
- Fukui T., Yamauchi, K., Muroya, T., Akiyama, M., Maki, H., Sugino, A., and Waga, S. 2004. Distinct roles of DNA polymerases δ and ε at the replication fork in Xenopus egg extracts. Genes Cell 9: 179-191. [DOI] [PubMed] [Google Scholar]
- Gordenin D.A., Kunkel, T.A., and Resnick, M.A. 1997. Repeat expansion—All in a flap? Nat. Genet. 16: 116-118. [DOI] [PubMed] [Google Scholar]
- Harrington J.J. and Lieber, M.R. 1994. The characterization of a mammalian DNA structure-specific endonuclease. EMBO J. 13: 1235-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henricksen L.A., Umbricht, C.B., and Wold, M.S. 1994. Recombinant replication protein A: Expression, complex formation, and functional characterization. J. Biol. Chem. 269: 11121-11132. [PubMed] [Google Scholar]
- Jin Y.H., Obert, R., Burgers, P.M., Kunkel, T.A., Resnick, M.A., and Gordenin, D.A. 2001. The 3′ → 5′ exonuclease of DNA polymerase δ can substitute for the 5′ flap endonuclease Rad27/Fen1 in processing Okazaki fragments and preventing genome instability. Proc. Natl. Acad. Sci. 98: 5122-5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y.H., Ayyagari, R., Resnick, M.A., Gordenin, D.A., and Burgers, P.M. 2003. Okazaki fragment maturation in yeast. II. Cooperation between the polymerase and 3′–5′-exonuclease activities of Pol δ in the creation of a ligatable nick. J. Biol. Chem. 278: 1626-1633. [DOI] [PubMed] [Google Scholar]
- Jin Y.H., Garg, P., Stith, C.M., Al Refai, H., Sterling, J., Weston, L., Kunkel, T., Resnick, M.A., Burgers, P.M., and Gordenin, D.A. 2004. The multiple biological roles for the 3′–5′-exonuclease of DNA polymerase δ require switching between the polymerase and exonuclease domains. Mol. Cell. Biol. (in press). [DOI] [PMC free article] [PubMed]
- Johnson K.A. 1993. Conformational coupling in DNA polymerase fidelity. Annu. Rev. Biochem. 62: 685-713. [DOI] [PubMed] [Google Scholar]
- Kaboord B.F. and Benkovic, S.J. 1993. Rapid assembly of the bacteriophage T4 core replication complex on a linear primer/template construct. Proc. Natl. Acad. Sci. 90: 10881-10885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao H.I. and Bambara, R.A. 2003. The protein components and mechanism of eukaryotic Okazaki fragment maturation. Crit. Rev. Biochem. Mol. Biol. 38: 433-452. [DOI] [PubMed] [Google Scholar]
- Kao H.I., Veeraraghavan, J., Polaczek, P., Campbell, J.L., and Bambara, R.A. 2004. On the roles of Saccharomyces cerevisiae Dna2p and Flap endonuclease 1 in Okazaki fragment processing. J. Biol. Chem. 279: 15014-15024. [DOI] [PubMed] [Google Scholar]
- Karthikeyan R., Vonarx, E.J., Straffon, A.F., Simon, M., Faye, G., and Kunz, B.A. 2000. Evidence from mutational specificity studies that yeast DNA polymerases δ and ε replicate different DNA strands at an intracellular replication fork. J. Mol. Biol. 299: 405-419. [DOI] [PubMed] [Google Scholar]
- Maga G., Villani, G., Tillement, V., Stucki, M., Locatelli, G.A., Frouin, I., Spadari, S., and Hubscher, U. 2001. Okazaki fragment processing: modulation of the strand displacement activity of DNA polymerase δ by the concerted action of replication protein A, proliferating cell nuclear antigen, and flap endonuclease-1. Proc. Natl. Acad. Sci. 98: 14298-14303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murante R.S., Rust, L., and Bambara, R.A. 1995. Calf 5′ to 3′ exo/endonuclease must slide from a 5′ end of the substrate to perform structure-specific cleavage. J. Biol. Chem. 270: 30377-30383. [DOI] [PubMed] [Google Scholar]
- Murante R.S., Rumbaugh, J.A., Barnes, C.J., Norton, J.R., and Bambara, R.A. 1996. Calf RTH-1 nuclease can remove the initiator RNAs of Okazaki fragments by endonuclease activity. J. Biol. Chem. 271: 25888-25897. [DOI] [PubMed] [Google Scholar]
- Muzyczka N., Poland, R.L., and Bessman, M.J. 1972. Studies on the biochemical basis of spontaneous mutation. A comparison of the deoxyribonucleic acid polymerases of mutator, antimutator, and wild type strains of bacteriphage T4. J. Biol. Chem. 247: 7116-7122. [PubMed] [Google Scholar]
- Qiu J., Qian, Y., Frank, P., Wintersberger, U., and Shen, B. 1999. Saccharomyces cerevisiae RNase H(35) functions in RNA primer removal during lagging-strand DNA synthesis, most efficiently in cooperation with Rad27 nuclease. Mol. Cell. Biol. 19: 8361-8371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick M.A. 1978. Similar responses to ionizing radiation of fungal and vertebrate cells and the importance of DNA doublestrand breaks. J. Theor. Biol. 71: 339-346. [DOI] [PubMed] [Google Scholar]
- Roth A.C., Nossal, N.G., and Englund, P.T. 1981. Rapid hydrolysis of deoxynucleoside triphosphates accompanies DNA synthesis by T4 DNA polymerase and T4 accessory proteins 44/62 and 45. J. Biol. Chem. 257: 1267-1273. [PubMed] [Google Scholar]
- Shcherbakova P.V. and Pavlov, Y.I. 1996. 3′ → 5′ exonucleases of DNA polymerases ε and δ correct base analog induced DNA replication errors on opposite DNA strands in Saccharomyces cerevisiae. Genetics 142: 717-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakova P.V., Pavlov, Y.I., Chilkova, O., Rogozin, I.B., Johansson, E., and Kunkel, T.A. 2003. Unique error signature of the four-subunit yeast DNA polymerase ε. J. Biol. Chem. 278: 43770-43780. [DOI] [PubMed] [Google Scholar]
- Soengas M.S., Esteban, J.A., Lazaro, J.M., Bernad, A., Blasco, M.A., Salas, M., and Blanco, L. 1992. Site-directed mutagenesis at the Exo III motif of ϕ 29 DNA polymerase; overlapping structural domains for the 3′–5′ exonuclease and strand-displacement activities. EMBO J. 11: 4227-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz T.A. and Yin, Y.W. 2004. Accuracy, lesion bypass, strand displacement and translocation by DNA polymerases. Philos. Trans. R Soc. Lond. B Biol. Sci. 359: 17-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff D.X., Boerger, A.L., Bertrand, P., Filosi, N., Gaida, G.M., Kane, M.F., and Kolodner, R.D. 1997a. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc. Natl. Acad. Sci. 94: 7487-7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff D.X., Filosi, N., Gaida, G.M., and Kolodner, R.D. 1997b. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair [see comments]. Cell 88: 253-263. [DOI] [PubMed] [Google Scholar]
- Topal M.D., DiGuiseppi, S.R., and Sinha, N.K. 1980. Molecular basis for substitution mutations. Effect of primer terminal and template residues on nucleotide selection by phage T4 DNA polymerase in vitro. J. Biol. Chem. 255: 11717-11724. [PubMed] [Google Scholar]
- Tran P.T., Erdeniz, N., Dudley, S., and Liskay, R.M. 2002. Characterization of nuclease-dependent functions of Exo1p in Saccharomyces cerevisiae. DNA Repair 1: 895-912. [DOI] [PubMed] [Google Scholar]