Abstract
The possibility that single-cell organisms undergo programmed cell death has been questioned in part because they lack several key components of the mammalian cell death machinery. However, yeast encode a homolog of human Drp1, a mitochondrial fission protein that was shown previously to promote mammalian cell death and the excessive mitochondrial fragmentation characteristic of apoptotic mammalian cells. In support of a primordial origin of programmed cell death involving mitochondria, we found that the Saccharomyces cerevisiae homolog of human Drp1, Dnm1, promotes mitochondrial fragmentation/degradation and cell death following treatment with several death stimuli. Two Dnm1-interacting factors also regulate yeast cell death. The WD40 repeat protein Mdv1/Net2 promotes cell death, consistent with its role in mitochondrial fission. In contrast to its fission function in healthy cells, Fis1 unexpectedly inhibits Dnm1-mediated mitochondrial fission and cysteine protease-dependent cell death in yeast. Furthermore, the ability of yeast Fis1 to inhibit mitochondrial fission and cell death can be functionally replaced by human Bcl-2 and Bcl-xL. Together, these findings indicate that yeast and mammalian cells have a conserved programmed death pathway regulated by a common molecular component, Drp1/Dnm1, that is inhibited by a Bcl-2-like function.
Keywords: Mitochondria, Dnm1, Drp1, Fis1, Bcl-xL, apoptosis, autophagy
Programmed cell death is widely accepted as an essential process in multicellular organisms, but was generally considered to be unnecessary for single-cell organisms that do not form multicellular structures. However, it is possible that programmed death has an ancient origin to provide species of unicellular organisms with stress responses to nutrient deprivation or to guard against the spread of pathogens, and that these mechanisms were subsequently adapted to sculpt and maintain multicellular organisms. Thus, survival of a species, be it multi- or unicellular, may require programmed cell death.
Just as morphology of dying mammalian cells provided early clues of programmed death, evidence that unicellular organisms have genetic self-destruct mechanisms is based in part on morphological characteristics of dying yeast that resemble some of the characteristics of apoptotic mammalian cells (Madeo et al. 2002a). Yeast were reported to externalize phosphatidylserine on the outer leaflet of their plasma membrane and to undergo DNA degradation and chromatin condensation, all characteristics of apoptotic mammalian cells (Madeo et al. 1997, 1999). Yeast also encode orthologs of the DNA endonuclease EndoG that is involved in cell death of mammals and Caenorhabditis elegans (Ikeda and Kawasaki 2001; Li et al. 2001; Parrish et al. 2001; Wang et al. 2002), and mammalian Beclin, a Bcl-2-interacting factor that is homologous to the autophagy regulator Apg6/Vps30 in yeast (Liang et al. 1999). Despite these analogies, yeast appear to lack the Bcl-2 family proteins and caspases that constitute the core cell death machinery in mammals. However, the cell death-promoting function of plant and yeast metacaspases fuel the idea that yeast possess a protease-mediated death pathway analogous to mammals (Madeo et al. 2002b; Hoeberichts et al. 2003; Suarez et al. 2004). The pro-death Bcl-2 family member Bax also promotes yeast cell death that is inhibited by anti-apoptotic Bcl-2 and Bcl-xL when these mammalian proteins are coexpressed in yeast (Hanada et al. 1995). Furthermore, overexpression of human Bcl-2 improves the survival of yeast lacking superoxide dismutase (Longo et al. 1997), suggesting potential conservation of Bcl-2-like functions in yeast.
Mitochondria play a central role in mammalian cell death, both as an essential organelle targeted for destruction and as a perpetrator of the death pathway. Mitochondria can contribute to cell death when Bax is activated to permeabilize the mitochondrial outer membrane, resulting in release of cytochrome c and other factors that trigger caspase activation and enhance cell death (Newmeyer and Ferguson-Miller 2003). Fragmentation of tubular mitochondria into short punctate structures is a common early feature of apoptotic mammalian cells and closely correlates with Bax activation and cytochrome c release from mitochondria (Hockenbery et al. 1993; Vieira et al. 2002; Breckenridge et al. 2003). Furthermore, Bax localizes at constriction points on mitochondria in apoptotic mammalian cells (Karbowski et al. 2002). However, mitochondrial division also occurs in normal healthy cells, although at a slower rate and apparently without detrimental consequences (Karbowski et al. 2004).
The machinery responsible for mitochondrial fission in healthy cells was identified in yeast as a complex of three proteins, Dnm1, Mdv1/Net2, and Fis1 (Jensen et al. 2000; Shaw and Nunnari 2002; Cerveny and Jensen 2003). Deletion of any one of these three causes yeast mitochondria to become fused into a large network. Drp1, the human homolog of yeast Dnm1, is also required for normal mitochondrial fission in mammalian cells (Sesaki and Jensen 1999; Frank et al. 2001; Yoon et al. 2003). Furthermore, a dominant negative mutant of human Drp1 (K38A/E) inhibits mammalian cell death and the excessive mitochondrial fragmentation that occurs in apoptotic cells (Frank et al. 2001; Smirnova et al. 2001; Breckenridge et al. 2003). Dominant negative Drp1 also inhibits other events associated with cell death including cytochrome c release from mitochondria and mitochondrial membrane depolarization (Frank et al. 2001; Breckenridge et al. 2003). Together, these studies suggest that division of mitochondria or other function of Drp1 can play a causal role in mammalian programmed cell death.
Dnm1 was reported to interact with the WD40 repeat domain of Mdv1/Net2 and with the Fis1 protein (Tieu and Nunnari 2000; Cerveny et al. 2001; Tieu et al. 2002; Cerveny and Jensen 2003; Yoon et al. 2003). There are no obvious homologs of Mdv1/Net2 in mammals (outside the WD40 repeat domain), but Fis1 is highly conserved from plants to humans (Mozdy et al. 2000; Tieu and Nunnari 2000). Fis1 of Saccharomyces cerevisiae is an 18-kDa protein that is anchored to the outer mitochondrial membrane by a C-terminal hydrophobic-basic domain characteristic of Bcl-2, Tom 5, and other proteins targeted to the outer mitochondrial membrane (Mozdy et al. 2000; Kaufmann et al. 2003). Consistent with its role in yeast, depletion of endogenous human Fis1 decreases mitochondrial fission (James et al. 2003; Yoon et al. 2003; Stojanovski et al. 2004).
Given the link between mitochondrial fission and programmed cell death, we investigated the possibility that the yeast mitochondrial fission machinery could regulate programmed cell death in yeast. Our results provide evidence for a conserved cell death mechanism mediated by yeast Dnm1 and human Drp1 homologs. Contrary to prediction, yeast Fis1 inhibits rather than induces cell death in yeast. However, this seemingly incongruent outcome is resolved by the finding that Fis1 also limits mitochondrial fission induced by Dnm1, providing new insight into the connection between mitochondrial fission and programmed cell death.
Results
Fis1 inhibits programmed cell death in yeast
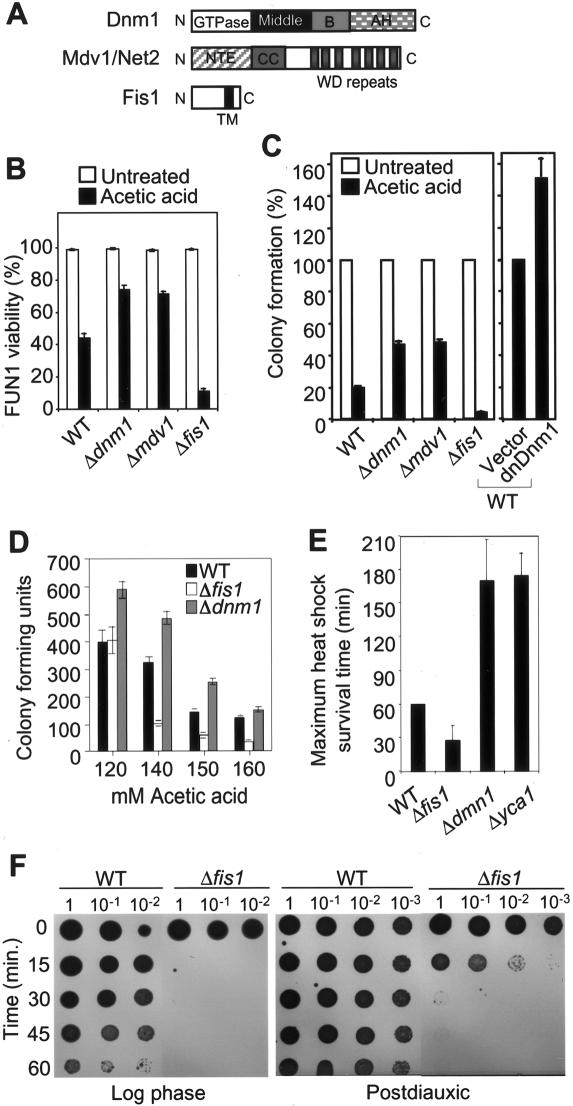
To explore the possibility that yeast undergo programmed cell death through mechanisms involving mitochondrial fission, yeast lacking the mitochondrial fission factor Fis1 were treated with hydrogen peroxide, a source of reactive oxygen species and a potent death stimulus in yeast (Madeo et al. 1999). Fis1-deficient yeast had significantly reduced viability compared with wild-type cells when treated with H2O2 (Fig. 1A). Yeast cell viability was determined with the vital dye FUN1, which stains metabolically active cells red and dead cells green (plus a blue counterstain for the cell wall) (Fig. 1B). Similar results were obtained when cell death was induced by acetic acid treatment (Fig. 1A) to mimic overgrowth conditions (Madeo et al. 1997). Fis1 localizes diffusely in the mitochondrial outer membrane (Shaw and Nunnari 2002). Therefore, it was possible that deletion of any component of the outer membrane would increase susceptibility to these death stimuli. In contrast to Fis1, loss of OM45, a major component of the outer mitochondrial membrane, had no effect on cell viability following treatment with H2O2 or acetic acid (Fig. 1A). When cell death was determined by an alternative method, trypan blue exclusion, again, the same results were obtained (Fig. 1C). Also, deletion of Fis1 in a different yeast strain produced similar dose-dependent sensitivities to H2O2 and acetic acid (Fig. 1D; Jensen et al. 2000).
Figure 1.
Inhibition of programmed cell death by yeast Fis1p. (A) Cell viability was determined by FUN-1 staining at 4 h after treatment with 130 mM acetic acid (AA) or with 2.5 mM H2O2, and data are presented as mean ± se for five (wild type, Δfis1) or two (Δom45) independent experiments counting at least 200 cells per sample. One of the two experiments with Δom45 was done on BY4741 background strain; the other was on an undefined background. (B) Representative images of wild-type (WT) and Δfis1 yeast cells stained with FUN1 dye following treatment with 2.5 mM H2O2 for 4 h or untreated controls. The cell wall stains blue and viable metabolically active yeast contain a red vacuole, while nonviable yeast lack a red vesicle and stain green/yellow. (C) Viability was also determined by trypan blue exclusion. Data are presented as mean ± standard error (se) for three independent experiments counting at least 200 cells per sample. (D) Viability by FUN-1 staining was performed using yeast on RJ1365 background. Data presented are the means ± se for three independent experiments. (B–D) P < 0.001 comparing wild type (WT) and Δfis1 mutants.
To determine if Fis1 inhibits the biochemical changes characteristic of programmed cell death that are observed in both yeast and mammalian cells (Madeo et al. 1999), Δfis1 yeast were stained with annexin V to detect externalization of phosphatidylserine on the outer leaflet of the cytoplasmic membrane following treatment with a death stimulus. As expected, Δfis1 spheroplasts stained positive with annexin V under conditions where few wild-type control cells were stained (Fig. 2A,B). To verify that binding of annexin V was not due to permeabilization of the plasma membrane during spheroplast preparation, cells were costained with propidium iodide (PI). Only 40% of annexin-positive Δfis1 cells were also PI positive, confirming the specificity of annexin V staining (Fig. 2A).
Figure 2.
Fis1 inhibits phosphatidylserine exposure and caspase-dependent cell death. (A) The same field of wild-type and Δfis1 spheroplasts are shown with phase contrast (left), annexin V staining (middle, green), and propidium iodide staining (right, red) at 4 h after treatment with 2.5 mM H2O2. (B) Viability was assessed by annexin V staining of yeast spheroplasts and presented as mean ± se for five fields (>150 cells) per sample in two independent experiments. (C) Cell viability determined by FUN-1 staining of the indicated yeast mutants (BY4741) treated with 2.5 mM H2O2 or 130 mM acetic acid for 4 h is presented ± se for three independent experiments. (D) Viability of the indicated yeast mutants (BY4741) treated with 2.5 mM H2O2 or 130 mM acetic acid in the presence of DEVD-CHO or DMSO carrier control was determined by FUN-1 staining in three independent experiments. (B–D) *P < 0.001 comparing mutants to wild type. (E) Dose response assay as described for panel D.
Fis1 inhibits protease-dependent cell death
To further pursue the possibility that yeast Fis1 is an inhibitor of a programmed cell death pathway, we asked if proteases contributed to the enhanced cell death that occurs in Fis1-deficient yeast. In double-deleted yeast lacking both Fis1 and the metacaspase Yca1 (Δfis1/Δyca1), the cell death observed with Δfis1 alone was restored to wild-type levels (Fig. 2C). Interestingly, the Yca1 protease was previously reported to cleave fluorogenic peptide substrates of the sequence VEID and IETD, but not to cleave DEVD (Madeo et al. 2002b). This substrate specificity profile, together with the presence of a long pro-domain at the N terminus, suggests that Yca1 may be more analogous to the upstream mammalian caspases that subsequently activate other downstream proteases. In mammalian cells, the downstream or amplification phase caspase-3-like proteases that recognize the peptide substrate DEVD are responsible for cleaving the majority of intracellular target proteins identified to date (Earnshaw et al. 1999). Treatment of Δfis1 with the cell-permeable inhibitor DEVD provided significant (25%–30%) but partial protection against H2O2 and acetic acid treatment compared with DMSO carrier controls (Fig. 2D). Although DEVD provided dose-dependent protection, this protection was detected only in short-term assays by vital dye (Fig. 2E) and optical density (data not shown), as DEVD did not improve long-term/clonogenic survival detected by growth on plates (data not shown). This is consistent with mammalian cells where peptide inhibitors of caspases only delay death of mammalian cells (Amarante-Mendes et al. 1998; Brunet et al. 1998). Nevertheless, these findings suggest that DEVD-cleaving proteases distinct from Yca1 also contribute to yeast cell death.
Dnm1 and Mdv1/Net2 promote cell death in yeast
To further explore the possibility that yeast mitochondrial fission factors regulate programmed cell death, yeast lacking Dnm1 or Mdv1/Net2 were tested for their response to a cell death stimulus. In contrast to Fis1, but consistent with their role in promoting mitochondrial fission, the yeast knockouts Δdnm1 and Δmdv1 were significantly more resistant to death compared with wild-type and Δfis1 cells when treated with acetic acid (Fig. 3A,B). To verify that these genes also regulate the long-term survival of yeast, viability was assessed by colony formation. Although long-term survival was somewhat lower compared with the results obtained by vital dye staining at 4 h post-treatment, the same trends were observed (Fig. 3C) and were also dose-dependent (Fig. 3D). Similar to mammalian cells expressing dominant negative Drp1 (Frank et al. 2001), overexpression of the analogous dominant negative mutant of Dnm1 (K41A) also protected yeast from cell death (Fig. 3C). Consistent with previous reports (Jensen et al. 2000; Mozdy et al. 2000; Tieu and Nunnari 2000), deletion of these fission factors had no detectable effect on growth rates/survival under normal growth conditions on rich or minimal media (see legend for Fig. 4D), indicating that they affect cell survival only in the presence of a death stimulus.
Figure 3.
Fis1p and Dnm1p regulate yeast cell death. (A) Diagram of yeast protein domains; GTPase effector domain (GED), N-terminal extension (NTE), coiled coil (CC), and transmembrane (TM) were adapted from Shaw and Nunnari (2002) (© 2002, with permission from Elsevier). (B) Cell viability was also determined by viable dye FUN-1 staining as shown in Figure 1B, and data are presented as mean ± se for three independent experiments counting at least 200 cells per sample. (C) Wild-type (WT) and the mutant yeast cells (on BY4741 background strain), or cells overexpressing Dnm1(K41A) (provided by R. Jensen, Department of Cell Biology, Johns Hopkins School of Medicine, Baltimore, MD) were treated with 190 mM acetic acid for 4 h and cell viability was assessed by colony formation in three independent experiments for each assay. (B,C) The differences between all mutants and wild type (WT) are statistically significant with P < 0.001. (D) Dose response for acetic acid treatment was performed in a 96-well format for more than six independent determinations. (E) Survival of wild-type (WT) and mutant yeast (BY4741) treated with heat shock at 55°C, and the maximum heat shock survival time was determined by growth on YPD plates. Data are presented as mean ± se for five independent experiments. (F) Survival of log phase or postdiauxic yeast (BY4741) treated with heat shock at 55°C for the indicated times was determined by growth on YPD plates. A representative of three independent experiments is shown.
Figure 4.
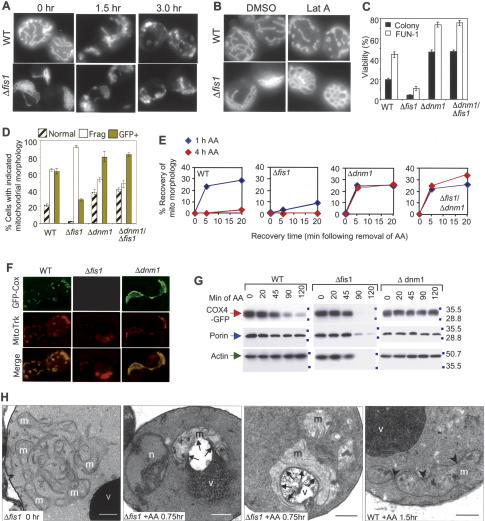
Mitochondrial fission during cell death does not require Fis1. (A) Fluorescence microscopy showing mitochondrial morphology of wild-type (WT) and Δfis1 yeast (BY4741) treated with 100 mM acetic acid for the indicated times. Mitochondria are visualized with Cox4-GFP (Sesaki and Jensen 1999). (B) Mitochondrial morphology of yeast treated with 250 μM latrunculin A (Molecular Probes) or the carrier DMSO for 1 h as described in B except without a death stimulus. (C) Wild-type (WT) and the mutant yeast cells (on BY4741 background strain) were treated with 190 mM acetic acid for 4 h and cell viability was assessed by colony formation and FUN-1 staining in three independent experiments for each assay. Data are presented as mean ± se for three independent experiments counting at least 200 cells per sample. (D) Percent of yeast cells with the indicated mitochondria morphologies (differences for all criteria between wild type [WT], Δfis1, and Δdnm1 are significant; P < 0.001). No significant differences were detected in the percent GFP-positive yeast between genotypes prior to acetic acid treatment; wild type (82%), Δfis1 (78%), Δdnm1 (76%). (C,D) P < 0.001 comparing wild type and Δfis1 mutants treated with acetic acid. (E) The indicated strains of yeast were treated for 1 h or 4 h with 130 mM acetic acid, and mitochondrial morphology (COS4-GFP) was scored at the indicated times after removal of acetic acid. (F) Fluorescence microscopy of yeast treated with 130 mM acetic acid for 20 min, resuspended in water, and stained with 0.5 μM MitoTracker. (G) Immunoblot analyses of yeast cell lysates prepared at the indicated times after treatment with 130 mM acetic acid. (H) Electron microscopy of Δfis1 and wild-type (WT) yeast treated with 100 mM acetic acid for the indicated times. (v) Vacuole; (m) mitochondrion; (arrowheads) fission sites; (n) nucleus; (arrows) association of mitochondria with vacuole. Bar, 0.5 μm.
If Dnm1 and Fis1 are part of the conserved core cell death machinery in yeast, they should modulate cell survival following treatment with multiple death stimuli. Therefore, yeast were also treated with heat shock at 55°C, a model for environmental stress (Fabrizio et al. 2001). Consistent with the acetic acid and H2O2 results, Δdnm1 was protected while Δfis1 exhibited increased sensitivity to heat shock-induced death (Fig. 3E,F). In addition, deletion of the metacaspase Yca1 provided similar protection compared with deletion of Dnm1. All the experiments described thus far were performed with log phase yeast cultures. However, Fis1 was also required for survival of postdiauxic yeast from 55°C heat shock (Fig. 3F). Taken together, the yeast fission factors appear to regulate a programmed cell death pathway in yeast.
Fis1 is not required for fragmentation of mitochondria during yeast cell death
Because mitochondrial fragmentation is linked to apoptosis of mammalian cells (Mancini et al. 1997; Nechushtan et al. 2001; Karbowski et al. 2002), we sought to determine if fragmentation of mitochondria was also a characteristic of dying yeast. The matrix-localized fluorescent protein Cox4-GFP was used to visualize mitochondria without disturbing the mitochondrial membrane (Jensen et al. 2000). Strikingly, the normal tubular mitochondria in wild-type cells became fragmented and misshapen after acetic acid treatment (Fig. 4A,D). Fis1-deficient yeast are defective for mitochondrial fission under normal growth conditions, resulting in a net-like phenotype in which the mitochondrial tubules typical of wild-type cells are fused into a single net-like structure (Fig. 4A; Shaw and Nunnari 2002). However, following acetic acid treatment, the net-like mitochondria in Δfis1 cells also fragmented. Therefore, Fis1 is apparently not required for fragmentation of mitochondria during cell death, implying that the fission machinery operating during cell death is molecularly distinct from that during normal growth.
A possible alternative explanation for the fragmentation of mitochondria during cell death is disruption of the cytoskeleton that also occurs during mammalian cell death. In fact, disruption of actin with Latrunculin A causes mitochondrial fragmentation in otherwise healthy yeast (Jensen et al. 2000). In contrast to wild-type cells, mitochondria of Latrunculin A-treated Δfis1 yeast remained profoundly fused, indicating that Fis1 is required for fission of yeast mitochondria upon actin disruption (Fig. 4B). In mammalian cells, tubulin assumes the role that actin plays in regulating yeast mitochondrial morphology. Consistent with our observations in yeast, mitochondrial fragmentation occurs prior to disruption of microtubules in at least some types of dying mammalian cells (Breckenridge et al. 2003). Our findings with Fis1 are also consistent with the requirement for Dnm1 and Mdv1/Net2 for fission in Latrunculin A-treated yeast (Jensen et al. 2000).
Fis1 protects by inhibiting Dnm1-mediated fragmentation and loss of mitochondria
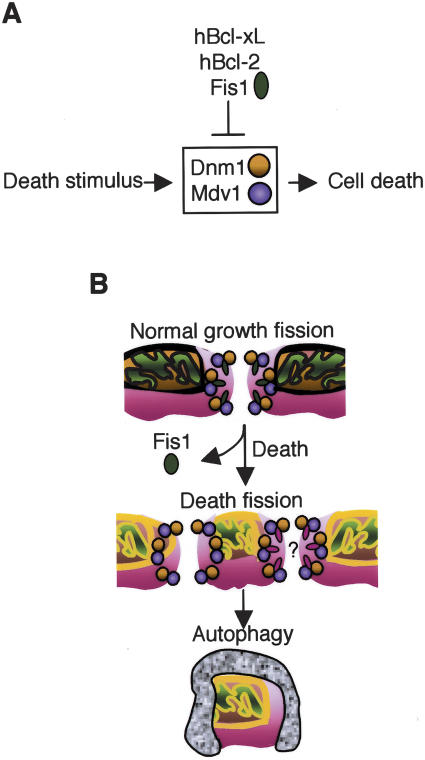
Because Fis1 is found in a complex with Dnm1, it is reasonable to propose that Fis1 protects cells by inhibiting Dnm1-mediated death. As predicted, the double Δdnm1/Δfis1 mutant behaved indistinguishably from Δdnm1 following treatment with acetic acid (Fig. 4C). Again, both colony formation and vital dye staining produced the same conclusions. These results define a genetic pathway in which Fis1 inhibits the pro-death function of Dnm1 (Fig. 7A, see below).
Figure 7.
Model of yeast cell death pathway. (A) Yeast Fis1, human Bcl-2, and Bcl-xL inhibit yeast cell death mediated by Dnm1. (B) Following a death stimulus, Fis1 may be inactivated, leading to uncontrolled/irreversible Dnm1-mediated mitochon-drial fragmentation, degradation by vacuoles, and cell death. Another possibility is that yeast Fis1 is replaced by another yet unidentified factor that mediates mitochondrial fission during cell death (red ovals).
To determine if Fis1 protects from cell death by inhibiting the fission function of Dnm1, mitochondrial morphology was quantified by mitochondrial-GFP staining at early times after acetic acid treatment. Fragmentation was reduced in the double-deficient Δdnm1/Δfis1 (and Δdnm1-only) yeast compared with Δfis1, indicating that Fis1 inhibits Dnm1-mediated fission (Fig. 4D, open bars). However, deletion of Dnm1 only partially inhibited mitochondrial fragmentation relative to wild-type cells, in contrast to the more obvious protection observed in viability assays (Fig. 4C,D). This raises the possibility that Dnm1 may have an additional role in promoting yeast cell death. Consistent with this idea, time course experiments revealed that deletion of Dnm1 (or both Dnm1 and Fis1) only delayed mitochondrial fragmentation, as all mitochondria eventually fragmented (Fig. 4E, time 0). We found that Dnm1-deficient cells recovered normal/fused mitochondrial morphology under these conditions almost completely within 5 min, while wild-type cells required more time to achieve only partial recovery, and cells lacking Fis1 had very poor recovery rates. Thus, Dnm1 inhibits the recovery of normal/fused mitochondrial morphology that closely correlates with survival. Deletion of Dnm1 also protected yeast from acetic acid-induced loss of mitochondrial-GFP staining and from loss of membrane potential detected by Mitotracker staining (Fig. 4D,F; data not shown). However, rare Mitotracker-stained Δfis1 cells were observed as shown in Figure 4F. Taken together, these findings suggest that fragmentation of mitochondria is not sufficient for cell death. Instead, loss of mitochondrial markers COX4-GFP and MitoTracker staining correlates with loss of cell viability suggesting that loss of functional mitochondria marks the commitment point to cell death in Δfis1.
To pursue the fate of yeast mitochondria during cell death, the levels of mitochondrial markers were determined by immunoblot analysis of cell lysates that were collected at increasing times following acetic acid treatment. The matrix marker COX4-GFP and the outer mitochondrial membrane protein porin rapidly disappeared in Δfis1 cells concomitant with the loss of cell integrity indicated by actin levels (Fig. 4G). The levels of both mitochondrial markers were stable in Δdnm1 cells, but progressively decreased relative to actin levels in wild-type cells consistent with mitochondrial disfunction prior to death. These results are consistent with the evidence above suggesting that Fis1 prevents the loss of functional mitochondria and that loss of mitochondrial function represents a commitment to cell death.
To determine the ultimate fate of mitochondria in the Δfis1 mutant, electron microscopy was performed on acetic acid-treated yeast. Evidence of mitochondrial constrictions and fragmentation was detected in wild-type cells (arrowheads), but otherwise most mitochondria appeared to have normal morphology 1.5 h post-treatment (Fig. 4H, right panel). In contrast, the netted mitochondria of healthy Δfis1 cells were dramatically altered by 45 min, the earliest time examined following acetic acid treatment. Mitochondria of treated Δfis1 were frequently fragmented, but large globular mitochondria that were partially or completely invaded by a vacuole were also prominent features (Fig. 4H, arrows). Although a small vacuole protrusion was occasionally observed in the direction of a mitochondrion in acetic acid-treated wild-type cells (data not shown), the tight association of treated Δfis1 mitochondria with vacuoles was not encountered in treated wild-type cells. The reduced electron density of mitochondria in treated Δfis1 cells is also indicative of damaged mitochondria. Condensed chromatin characteristic of apoptotic mammalian cells was also observed in nuclei (n) of treated Δfis1 but not in wild-type cells (Fig. 4H). Together these findings indicate that destruction of mitochondria by an autophagy-like process, rather than fragmentation, represents commitment to cell death in yeast. A similar process may occur in mammalian cells where nerve growth factor withdrawal from sympathetic neurons leads to loss of mitochondria (Xue et al. 2001).
Yeast Fis1 porates lipid vesicles similar to human Bcl-xL
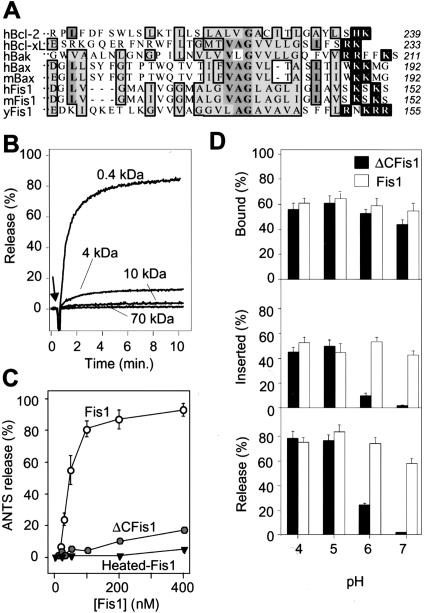
Like Bcl-2, yeast Fis1 is oriented in the outer membrane with its N terminus in the cytosol (Mozdy et al. 2000), and is composed of six α helices plus a C-terminal hydrophobic domain (Fig. 5A; Shaw and Nunnari 2002; Dohm et al. 2004). However, Fis1 is not a Bcl-2 family member based on its three-dimensional structure (Dohm et al. 2004). Nevertheless, we investigated the possibility that yeast Fis1 may share some biophysical properties of Bcl-2/Bcl-xL. Bcl-xL forms pores in synthetic lipid membranes and induces channels on intracellular mitochondria that correlate with its effects on synaptic activity (Basañez et al. 2001; Vander Heiden et al. 2001; Jonas et al. 2003, 2004). Similar to human Bcl-xL, we found that recombinant yeast Fis1 protein efficiently released >80% of 0.4 kDa and 10% of 4 kDa but not larger molecules in a dose-dependent manner from pure lipid vesicles (large unilammelar vesicles, LUV) (Fig. 5B). This release activity also required the hydrophobic C-terminal 27 amino acids of Fis1 (ΔCFis1, Fig. 5A) and the N-terminal 20 amino acids (ΔN-hFis1; data not shown). Similarly, this deletion also abolished its anti-death function (data not shown) and its fission function in healthy cells (Shaw and Nunnari 2002). Heat inactivation/denaturation also abolished pore formation, arguing against nonspecific release activity (Fig. 5C). Like Bcl-xL lacking its C-terminal hydrophobic domain (Basañez et al. 2001), ΔCFis1 became active only when the pH was lowered from pH 7 to pH 5 (Fig. 5D, bottom). The failure of ΔCFis1 to induce pores at neutral pH was not due to inefficient binding to liposomes (Fig. 5D, top), but failure to insert into the membrane, measured as resistance to alkali extraction (Fig. 5D, middle). Reduced pH presumably allows remodeling of both Fis1 and Bcl-xL proteins to facilitate membrane insertion. These findings suggest that yeast Fis1 may have two types of membrane interactions, one mediated by the tail and a second that requires other domains of the protein.
Figure 5.
Yeast Fis-1 releases small molecules from synthetic lipid vesicles. (A) Amino acid sequences of the C-terminal ends of Fis1 and Bcl-2 proteins from the indicated organisms: human (h), murine (m), and S. cerevisiae (y). Bold letters/medium shade indicate identity, light shade indicates similarity in >50% of entries; white letters mark basic residues on the C-terminal side of the hydrophobic region. Amino acid number for last residue of each protein is shown at right. (B) Release of the dye ANTS/DPX, (∼0.4 kDa), or FITC-labeled dextrans of the indicated molecular weights from synthetic lipid vesicles (25 μM lipid [1/250 mol protein/mol lipid]) at pH 7 was determined in three independent experiments on a fluorimeter following addition of 100 nM recombinant yeast Fis1 protein purified from Escherichia coli. (C) Release of ANTS/DPX at pH 7 from synthetic vesicles treated with the indicated concentrations of recombinant yeast Fis1 or ΔCFis1 (lacking the C-terminal 27 amino acids) and presented are mean ± se for four independent experiments. The fraction of recombinant yeast Fis1 proteins bound to lipid vesicles at the indicated pH conditions was determined before (D, top) or after (D, middle) alkali extraction. Release of ANTS/DPX was determined as described for A at the indicated pH conditions. Data presented are mean ± se for three to five independent experiments.
Bcl-2 and Bcl-xL functionally replace anti-death activity of yeast Fis1
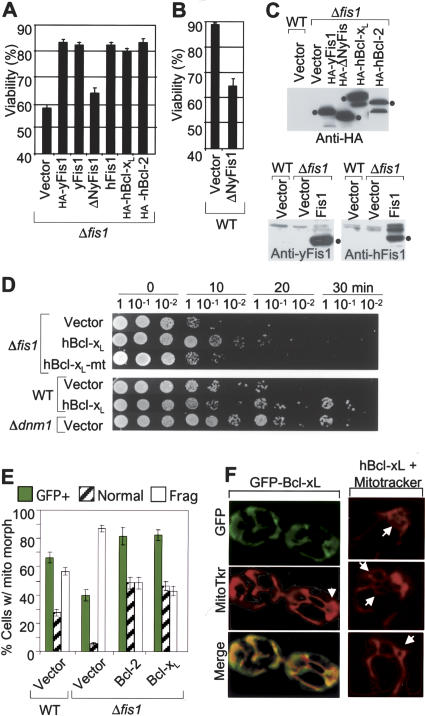
If yeast Fis1 can function in part like mammalian Bcl-2 proteins, then human Bcl-2 may substitute for yeast Fis1 in the Δfis1 mutant. Indeed, overexpression of human Bcl-2 or Bcl-xL (HA-tagged) from a GAL1 expression plasmid restored viability of acetic acid-treated Δfis1 as efficiently as overexpressed yeast Fis1 (±N-terminal HA-tag) (Fig. 6A). Human Fis1 also functionally substituted for the endogenous yeast homolog. Cleavage by caspases or engineered N-terminal deletions can convert Bcl-2 and Bcl-xL into Bax-like killer proteins (Cheng et al. 1997; Clem et al. 1998). Although ΔNFis1 (lacking amino acids 2–30) had no deleterious effect in the absence of a death stimulus (data not shown), this deletion abolished its protective activity (Fig. 6A). Furthermore, ΔNFis1 enhanced acetic acid-induced cell death of wild-type yeast (Fig. 6B). This finding suggests the possibility that ΔNFis1 behaves as a dominant negative inhibitor of the anti-apoptotic function of endogenous Fis1. Immunoblot analyses (Fig. 6C) verified the expression of all constructs in yeast except untagged ΔNFis1, which was confirmed only by sequencing because it lacks the antibody epitope. To extend this analysis, human Bcl-xL (expressed from a constitutive promoter) was tested for the ability to protect Δfis1 cells from heat shock-induced death. Human Bcl-xL, but not a BH1 domain point mutant, functionally substituted for yeast Fis1 in this assay (Fig. 6D, top) and further enhanced the survival of the wild-type yeast similar to the effect of deleting Dnm1 (Fig. 6D, lower).
Figure 6.
Human Bcl-2 and Bcl-xL rescue the cell death phenotype of Δfis1 mutant yeast. (A,B) Viability of Δfis1 and wild-type yeast (RJ1365) expressing the indicated N-terminal HA-tagged or untagged human (h) or yeast (y) proteins from GAL1 expression vectors was determined by trypan blue staining 6 h after treatment with 100 mM acetic acid. Data are presented as ±se for seven independent experiments; P < 0.001. (C) Immunoblot analyses of the yeast strains shown in panels A and B using anti-Fis1 rabbit sera or anti-HA antibody. (D) Yeast strains constitutively expressing the indicated proteins via a CEN plasmid were analyzed for survival following heat shock treatment as described in Figure 3E, except that samples were plated on minimal medium for selection, resulting in smaller viability differences between strains. A representative of three to five experiments is shown. All samples shown were from the same experiment, but the lanes were rearranged for presentation. (E) Percent of yeast cells transformed with the indicated plasmids with the indicated mitochondria morphology and protein expression was induced with galactose. Data presented are for three to five independent experiments; P < 0.01 for Bcl-xL, and P = 0.03 for Bcl-2 compared with wild type. (F) Yeast stably expressing GFP-Bcl-xL via a CEN plasmid were stained with MitoTracker and examined by fluorescence microscopy.
Because we found that human Bcl-2 and Bcl-xL can substitute for yeast Fis1 in cell viability assays, these mammalian factors may also inhibit Dnm1-induced mitochondrial fission and degradation observed in the Δfis1 mutant. Strikingly, human Bcl-2 and Bcl-xL significantly inhibited mitochondrial fragmentation that occurs in the absence of Fis1. Furthermore, Bcl-2 and Bcl-xL inhibited the loss of GFP-positive mitochondria in acetic acid-treated Δfis1 yeast, providing somewhat greater protection relative to endogenous Fis1 observed in wild-type yeast (Fig. 6E). Furthermore, overexpressed Bcl-xL localized to mitochondria and induced a partially fused mitochondrial phenotype in wild-type cells (Fig. 6F). However, Bcl-2 and Bcl-xL failed to replace the fission function of yeast Fis1 in healthy log phase cells. Together, these results suggest that like yeast Fis1, mammalian Bcl-2 proteins inhibit both mitochondrial fragmentation and cell death in yeast.
Discussion
We found that yeast mitochondria undergo an active fragmentation/degradation process during cell death requiring the function of Dnm1. Like Dnm1, Fis1 was genetically and biochemically defined as an essential component of the normal mitochondrial fission machinery (Shaw and Nunnari 2002; Cerveny and Jensen 2003). However, we found that Fis1 is not required for mitochondrial fission during cell death. To the contrary, Fis1 limits mitochondrial fission and death, apparently by blocking an irreversible step mediated by Dnm1 that leads to loss of functional mitochondria (Fig. 7A). Thus, in the absence of Fis1, mitochondria fail to regain their ability to fuse following a death stimulus, subsequently leading to loss of mitochondria that is concomitant with cell death. These findings in yeast may parallel those in mammalian cells where mitochondrial fragmentation/destruction appears to play a causal role in programmed death (Frank et al. 2001; Tolkovsky et al. 2002; Breckenridge et al. 2003). This loss of mitochondria in neurons deprived of growth factor is inhibited by the anti-apoptotic protein Bcl-2 (Tolkovsky et al. 2002). Similarly, the ability of yeast Fis1 to inhibit mitochondrial fission and degradation during yeast cell death can be replaced by the human anti-apoptotic proteins Bcl-2 and Bcl-xL.
Our observation that yeast Fis1 limits mitochondrial fission mediated by Dnm1 during cell death raises the possibility that Fis1 may also serve to limit mitochondrial fission during normal growth conditions. Consistent with this idea, the effect of ectopically overexpressed yeast Fis1 on mitochondrial morphology is dependent on its expression level. Reintroducing low levels of Fis1 protein into Δfis1 cells restored mitochondrial fission, but overexpression of yeast Fis1 in healthy yeast inhibited mitochondrial fission in growing cells, leading to the appearance of partially netted (and globular) mitochondria rather than fission of mitochondria (data not shown). This dual function of yeast Fis1 in promoting but also limiting fission likely explains the unexpected protective effects of Fis1 during yeast cell death. Our results are also consistent with a previous report showing that overexpression of yeast Fis1 in yeast cells displayed no enhanced mitochondrial fragmentation (Mozdy et al. 2000). However, overexpression of human Fis1 in mammalian cells was reported to induce mitochondrial fragmentation and cytochrome c release (James et al. 2003), though others have described alternative human Fis1-induced morphologies in mammalian cells (Yoon et al. 2003; Stojanovski et al. 2004).
Regulating mitochondrial fission in dying cells
Mitochondrial fragmentation has been observed in many apoptotic cell death paradigms from flies to humans (Igaki et al. 2000; Frank et al. 2001; Karbowski et al. 2002; Vieira et al. 2002; Breckenridge et al. 2003). Ectopic expression of the dominant-negative mutant of Drp1, Drp1K38A/E, inhibits not only mitochondrial fragmentation, but also cytochrome c release and caspase activation, suggesting that fragmentation of mitochondria or other functions of Drp1 are required for efficient programmed cell death (Frank et al. 2001; Breckenridge et al. 2003). This is supported by our observation that deletion of yeast DNM1 protects yeast cells from death stimuli.
Perhaps Dnm1/Drp1-induced fission activity is elevated above tolerable rates in dying cells, consistent with the global translocation of Drp1 from the cytosol to mitochondria in mammalian cells following a death stimulus (Frank et al. 2001). Given the importance of Dnm1/Drp1 in mitochondrial fission, we anticipated that deletion of Dnm1 would abolish fission in Δdnm1 during cell death. Instead, fission was only delayed and removal of the death stimulus caused Δdnm1 mitochondria to rapidly refuse. However, we cannot rule out the possibility that the mitochondrial fragments in Δdnm1 mutant cells remain linked in some manner due to incomplete severing of mitochondrial tubules. Interestingly, time-lapse monitoring of mitochondrial fission in Δdnm1 also failed to detect fission events that were not rapidly rejoined, unlike Δfis1 mutant yeast (Jakobs et al. 2003).
Mitochondrial fission in healthy versus dying cells
While Fis1 is required for normal levels of fission in healthy cells, fission occurs robustly during yeast cell death in the absence of Fis1. Thus, mitochondrial fission during cell death is molecularly and functionally distinguishable from mitochondrial fission that occurs every 2 min or so in healthy yeast cells apparently without deleterious effects (Nunnari et al. 1997). This is consistent with the variable morphology of fragmented mitochondria during death compared with that induced by actin depolymerization in the absence of cell death. In addition, the accelerated loss of mitochondria that is so prominently associated with excessive fission during cell death is not observed in healthy cells. This implies that either the increased frequency of fission or additional damaging events also distinguish fission during cell death from that during normal growth. We favor the idea that loss of mitochondria following a death stimulus is caused by more than fission alone, supported by the observation that Dnm1-deficiency protects from cell death more efficiently than from mitochondrial fragmentation. Consistent with this idea, other examples of enhanced mitochondrial fission or impaired mitochondrial fusion do not result in cell death. Mitochondrial fission apparently proceeds normally while fusion is impaired in healthy mammalian cells with reduced mitochondrial membrane potential (Smaili et al. 2001; Legros et al. 2002). Embryonic fibroblasts from knockout mice lacking mitofusin 1 (Mfn1) or Mfn2, dynamin GTPases that promote mitochondrial fusion, have profoundly fragmented mitochondria (Chen et al. 2003). Nevertheless, a portion of Mfn-deficient mitochondria has reduced membrane potential suggesting that fusion may be required for proper maintenance of membrane potential. Consistent with this observation, we found that yeast lacking its Mfn1 homolog, Δfzo1, were not more susceptible to acetic acid-induced death even though their mitochondria are profoundly fragmented (data not shown). Furthermore, the anti-apoptotic factor vMIA encoded by cytomegalovirus induces mitochondrial fragmentation but inhibits cytochrome c release and cell death (Goldmacher 2002). Thus, excessive mitochondrial fission per se is not sufficient to induce cell death.
Ancient origins of programmed cell death
The origins of programmed death in single-cell organisms may reflect survival mechanisms in response to pathogens such as viruses, or to environmental stresses such as nutrient deprivation that triggers sporulation, or to other adaptations (Skulachev 2002). The existence of mammalian caspases that are largely specific to inflammatory responses in the immune system (e.g., caspase-1) or to the cell death pathway (e.g., caspase-9) could conceivably have a common origin represented by the metacaspase or other protease in yeast.
Recent evidence for a “programmed cell death” pathway in the bacterium Bacillus subtilis provides compelling support that these processes are conserved in unicellular organisms (Gonzalez-Pastor et al. 2003). When death of the entire population is eminent due to nutrient deprivation, a cell that is destined to survive produces a factor, SdpC, which up-regulates its own survival strategies while compromising the survival strategies of neighboring cells that are destined to die. SdpC produced by the surviving cell promotes death in neighboring cells by triggering the susceptible cell to down-regulate gene expression of a protective factor (Gonzalez-Pastor et al. 2003). In this manner, it appears that the dying cell contributes to its own demise. In addition, SdpC also delays sporulation in the surviving cell by turning on expression of ATP synthase and lipid catabolism enzymes, arguably a protective mechanism to take advantage of the new nutrient source supplied by dying cell corpses.
Do yeast encode a Bcl-2-like function?
Human Bcl-2 and Bcl-xL could potentially function independently of all other yeast proteins to explain their protective effects in yeast, but it is also possible that some aspects of the mammalian Bcl-2-dependent pathway are conserved in yeast. Although both Fis1 and Bcl-2/Bcl-xL consist of a series of approximately six α helices plus a hydrophobic tail, Fis1 folds like a TPR repeat protein and lacks both structural and sequence similarity to Bcl-2 (Suzuki et al. 2003; Dohm et al. 2004). Nevertheless, the evidence presented suggests that Fis1may have Bcl-2-like anti-death activity in yeast, and several lines of evidence suggest that yeast Fis1 and human Bcl-2/Bcl-xL act in the same biochemical pathway. The subcellular localization, topology, and some biophysical properties of Fis1 are consistent with this possibility. Like Bcl-2/Bcl-xL in mammals, Fis1 also protects yeast from multiple cell death stimuli and inhibits a metacaspase-dependent death pathway, but is not important for growth of cells in the absence of a death stimulus. Furthermore, human Bcl-2 and Bcl-xL functionally replace the anti-death activity of Fis1 in yeast. In addition, Bcl-xL acts parallel to/downstream of Fis1, based on the observation that Bcl-xL inhibits fission/fragmentation in Δfis1 yeast. Finally, we suggest that Bcl-xL inhibits Dnm1-mediated mitochondrial fragmentation, based on the observation that Bcl-xL inhibits fission/fragmentation in Δfis1 (and wild-type) yeast cells, a process mediated by Dnm1. However, we cannot formally eliminate the possibility that Bcl-2/Bcl-xL inhibits an additional/alternative parallel pathway. Nevertheless, the ability of Fis1 as well as Bcl-2 proteins to protect yeast cells from death by inhibiting mitochondrial fission/destruction supports and extends the evidence for analogous pathways in mammalian cells (Fig. 7).
Our structure of human Fis1 suggests that homodimers form through the binding of the hinged N-terminal α helix of one molecule into the cleft of its binding partner (Dohm et al. 2004). Although the role of dimerization between Bcl-2 family proteins remains controversial, and any role for Fis1 dimers is unknown, this N-terminal dimerization domain of Fis1 is required for inhibition of cell death. It is also possible that yeast Fis1 encompasses both the cell death regulatory function of Bcl-2 and the fission function of mammalian Fis1 that were subsequently split into separate proteins in higher organisms. Alternatively, Fis1 may interact with or share a common target with pro- and anti-death Bcl-2 family proteins. While Fis1 is required for normal levels of fission in healthy cells, fission occurs robustly during yeast cell death in the complete absence of Fis1. It is possible that Fis1 is inactivated or displaced by another factor in the death fission “machine”, perhaps a factor analogous to Bax (Fig. 7B). This is consistent with colocalization of Bax (and Bak) with Drp1 at mitochondrial constriction sites and tubule ends in apoptotic mammalian cells (Karbowski et al. 2002). In contrast, human Fis1 and Bcl-xL are diffusely localized on mitochondria and do not appear to localize specifically with Drp1 at mitochondrial constriction sites in apoptotic mammalian cells (Karbowski et al. 2002). However, it is interesting to note that in yeast cells lacking the Fis1 protein, Dnm1 and Mdv1 form fewer but larger clusters on mitochondria (Shaw and Nunnari 2002), consistent with the possibility that Fis1 restricts their coalescence as well as their propensity to induce irreversible mitochondrial damage and cell death.
Materials and methods
Yeast strains and plasmids
Wild-type and FIS1 disruption were analyzed on two different background strains of yeast, Boeke yeast BY4741 (MATa fis1::kanMX4 his3Δ1 leu2Δ0 met15Δ0 ura3Δ0), and the Winston strain RJ1365 (MATa fis1::HIS3 leu2Δ0 ura3Δ0 trp1Δ0 lys2Δ0) (Cerveny et al. 2001). Yeast strains disrupted in YCA1 (Y02453) (MATa YCA1::kanMX4 his3Δ1 leu2Δ0 met15Δ0 ura3Δ0), Net2/MDV1 (RJ1253 [Cerveny et al. 2001]), and FZO1 (RJ1232 [Sesaki and Jensen 1999]) were derived from BY4741. The DNM1/FZO1 double deletion was derived from BY4734 (Sesaki and Jensen 1999) (MATa dnm1::HIS3 fzo1::kanMX4 his3Δ200 leu2Δ0 met15Δ0 trp1Δ63 ura3Δ0). To disrupt FIS1 in YCA1 deletion strain, the HIS3-cassette was PCR amplified from genomic DNA isolated from RJ1365 lacking FIS1 and transformed into the YCA1 deletion strain. Overexpression plasmids were generated by inserting PCR amplified coding sequences for yFis1, ΔNyFis1, hFis1, and hBcl-xL (with or without N-terminal HA-tag) into SalI and EcoRI of pRS314GU, a GAL1-Trp plasmid, and transformed into RJ1365 wild-type or Δfis1.
Cell viability assays
For H2O2 and acetic acid treatment, yeast was cultured in liquid medium (2% dextrose, 1% yeast extract, and 2% peptone) and incubated on a mechanical rotator at 30°C. After reaching an OD600 of 0.5, cells were exposed to 1.5–4.5 mM of H2O2 or 90 mM 190 mM of acetic acid for 4 h by direct addition of undiluted reagents. Heat stress assays were performed on cells grown to either OD600 of 0.5 or for 1 d (postdiauxic) in liquid medium and then incubated at 55°C. For each time point cells were diluted and spotted onto plates and incubated 2 d at 30°C. For FUN 1 staining, yeast cells were washed in staining buffer (2% glucose and 10 mM Na-HEPES at pH 7.2), resuspended in staining buffer with 5 μM FUN-1 and 25 μM Calcofluor White M2R (Molecular Probes), and incubated at 30°C in the dark for 30 min. For trypan blue staining, cells were resuspended in PBS with 0.4% trypan blue for 20 min at 25°C. For survival platings, yeast cells were counted with a hemocytometer, and 200 cells were plated on plates (2% agar) for 2 d at 30°C. Phosphatidylserine exposure was detected by an FITC-coupled annexin V reaction using ApoAlert (Clontech Lab., Inc) as reported (Madeo et al. 1997). To determine viability of yeast transformed with GAL1 expression plasmids, cells were cultured in liquid medium containing 0.67% yeast nitrogen base with ammonium sulfate and 30 mg/L amino acids except tryptophan with 2% raffinose. After reaching an OD600 of 0.4–0.5, cells were induced with 2% galactose and treated simultaneously with 100 mM acetic acid for 6 h at 30°C with shaking. For inhibition of caspase-like proteases, DEVD-CHO (Sigma) from a 20-mM stock in DMSO was added to a final concentration of 20 μM and incubated for 1 h prior to and during H2O2 or acetic acid treatment.
For viability determination in a 96-well colony formation assay, wild type, Δfis1 and Δdnm1 were seeded as 24 replicates from a fresh culture on Omnitray (Nunc, catalog no. 242811) plates into a 96-well plate with fresh liquid media. Cells were grown to saturation (48 h), diluted into new 96 well plates containing 200 μL of media and allowed to grow to 0.5 OD600/mL in a HiGro shaker (Gene Machine). Yeast cultures were transferred with a multichannel pipetter into premade acetic acid containing 96-well plates for 4 h with shaking. Cultures were diluted and 10 μL were plated onto solid media with a 96-well replicator (Fogger, V&P Scientific, Inc.). Plates were incubated for 2 d and colonies were counted.
Recovery of mitochondrial morphology
Yeast strains expressing the matrix-targeted domain of cytochrome oxidase subunit IV (Cox4) fused to GFP (Sesaki and Jensen 1999) to label mitochondria were grown to 0.5 OD600/mL and treated with 130 mM of acetic acid for 1–4 h. Cells were fixed as described (http://www.ciwemb.edu/labs/koshland/Protocols/MICROSCOPY/gfpfix.html) or allowed to recover for 5 or 20 min at RT in fresh media prior to fixation. Mitochondrial morphology was quantified by fluorescence microscopy.
Immunoblot analysis
Yeast cells (OD600 0.4) grown in selection medium were harvested at 3 h after galactose induction, and cytosolic extracts were prepared by mixing with equal volumes of lysis buffer (0.06 M Tris-HCl at pH 6.8; 10% [v/v] glycerol; 2% [w/v] SDS; 5% [v/v] 2-mercaptoehanol; 0.1 M PMSF; 0.5 M benzamidine; and 0.0025% [w/v] bromophenol blue) and glass beads for 4 min on a Vortex mixer. Cell extracts were analyzed by SDS-PAGE and immunoblotted with antibodies against HA-epitope (12CA5, Roche) or rabbit antisera (1:1000) raised against a peptide of human Fis1 (amino acids 12–28, EDLLKFEKKFQSEKAAG) or amino acids 1–127 of yeast Fis1 purified from E. coli. Yeast porin was detected with an antibody from Molecular Probes (A-6449).
Lipid vesicles
LUVs composed of DOPC/DOPG (6/4) at a lipid concentration of 25 μM were prepared in 100 mM KCl, 10 mM Hepes (pH 7.0), and 0.2 mM EDTA, and release of LUV-entrapped fluorescence markers was previously described (Basañez et al. 2001). To measure total amount of protein bound to vesicles, LUVs were prepared with D20 instead of H20, facilitating separation of LUV from protein by ultracentrifugation (2 h, 100,000g). Protein content in lipid-associated and lipid-free fractions was determined by fluorescence intensity at λex 280 nm and λem 345 nm, after addition of the detergent dodecyl octaethyleneglycol mono ether (Ostolaza and Goni 1995). To discriminate between protein inserted into and adsorbed to the membrane, the same protocol was followed except that a second incubation was performed at pH 11.5 for 30 min, the alkaline pH being maintained during sample centrifugation.
Electron microscopy
Briefly, the cells are fixed in 3% glutaraldehyde contained in 0.1 M Na cacodylate (pH 7.4), 5 mM CaCl2, 5 mM MgCl2, and 2.5% sucrose for 1 h at 25°C with gentle agitation; spheroplasted; embedded in 2% ultra low temperature agarose (prepared in water); cooled; and subsequently cut into small pieces (∼1 mm3) (Rieder et al. 1996). The cells are then post-fixed in 1% OsO4/1% potassium ferrocyanide contained in 0.1 M cacodylate/5 mM CaCl2 (pH 7.4) for 30 min at room temperature. The blocks are washed thoroughly four times with ddH20, 10 min total; transferred to 1% thiocarbohydrazide at room temperature for 3 min; washed in ddH2O (four times, 1 min each); and transferred to 1% OsO4/1% potassium ferrocyanide in cacodylate buffer (pH 7.4) for an additional 3 min at room temp. The cells are then washed four times with ddH2O (15 min total), en bloc stained in Kellenberger's uranyl acetate for 2 h to overnight, dehydrated through a graded series of ethanol, and subsequently embedded in Spurr resin. Sections were cut on a Reichert Ultracut T ultramicrotome, post-stained with UA and lead citrate, and observed on a Philips TEM 420 at 80 kV. Images were recorded with a Soft Imaging System Megaview III digital camera and figures were assembled in Adobe Photoshop 7.0.
Acknowledgments
We thank Rob Jensen, Kara Cerveny, and Jef Boeke for yeast strains and helpful discussions, and Clare Poynton for technical assistance. This work was supported by the National Institutes of Health NS37402 (J.M.H.), the National Science Foundation DBI 0099706 (J.M.M.), and the Ministerio de Ciencia y Techologia, Spain, BMC 2002-00784 (G.B.).
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1247904.
References
- Amarante-Mendes G.P., Finucane, D.M., Martin, S.J., Cotter, T.G., Salvesen, G.S., and Green, D.R. 1998. Anti-apoptotic oncogenes prevent caspase-dependent and independent commitment for cell death. Cell Death Differ. 5: 298-306. [DOI] [PubMed] [Google Scholar]
- Basañez G., Zhang, J., Chau, B.N., Maksaev, G.I., Frolov, V., Brandt, T.A., Burch, J., Hardwick, J.M., and Zimmerberg, J. 2001. Pro-apoptotic cleavage products of Bcl-xL form cytochrome c-conducting pores in pure lipid bilayers. J. Biol. Chem. 276: 31083-31091. [DOI] [PubMed] [Google Scholar]
- Breckenridge D.G., Stojanovic, M., Marcellus, R.C., and Shore, G.C. 2003. Caspase cleavage product of BAP31 induces mitochondrial fission through endoplasmic reticulum calcium signals, enhancing cytochrome c release to the cytosol. J. Cell Biol. 160: 1115-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet C.L., Gunby, R.H., Benson, R.S., Hickman, J.A., Watson, A.J., and Brady, G. 1998. Commitment to cell death measured by loss of clonogenicity is separable from the appearance of apoptotic markers. Cell Death Differ. 5: 107-115. [DOI] [PubMed] [Google Scholar]
- Cerveny K.L. and Jensen, R.E. 2003. The WD-repeats of Net2p interact with Dnm1p and Fis1p to regulate division of mitochondria. Mol. Biol. Cell 14: 4126-4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerveny K.L., McCaffery, J.M., and Jensen, R.E. 2001. Division of mitochondria requires a novel DMN1-interacting protein, Net2p. Mol. Biol. Cell 12: 309-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Detmer, S.A., Ewald, A.J., Griffin, E.E., Fraser, S.E., and Chan, D.C. 2003. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 160: 189-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng E.H.Y., Kirsch, D.G., Clem, R.J., Ravi, R., Kastan, M.B., Bedi, A., Ueno, K., and Hardwick, J.M. 1997. Conversion of Bcl-2 to a Bax-like death effector by caspases. Science 278: 1966-1968. [DOI] [PubMed] [Google Scholar]
- Clem R.J., Cheng, E.H.Y., Karp, C.L., Kirsch, D.G., Ueno, K., Takahashi, A., Kastan, M.B., Griffin, D.E., Earnshaw, W.C., Veliuona, M.A., et al. 1998. Modulation of cell death by Bcl-xL through caspase interaction. Proc. Natl. Acad. Sci. 95: 554-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohm J.A., Lee, S.J., Hardwick, J.M., Hill, R.B., and Gittis, A.G. 2004. Cytosolic domain of the human mitochondrial fission protein fis1 adopts a TPR fold. Proteins 54: 153-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W.C., Martins, L.M., and Kaufmann, S.H. 1999. Mammalian caspases: Structure, activation, substrates, and functions during apoptosis. Annu. Rev. Biochem. 68: 383-424. [DOI] [PubMed] [Google Scholar]
- Fabrizio P., Pozza, F., Pletcher, S.D., Gendron, C.M., and Longo, V.D. 2001. Regulation of longevity and stress resistance by Sch9 in yeast. Science 292: 288-290. [DOI] [PubMed] [Google Scholar]
- Frank S., Gaume, B., Bergmann, L.E.S., Leitner, W.W., Robert, E.G., Catez, F., Smith, C.L., and Youle, R.J. 2001. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev. Cell 1: 515-525. [DOI] [PubMed] [Google Scholar]
- Goldmacher V.S. 2002. vMIA, a viral inhibitor of apoptosis targeting mitochondria. Biochimie 84: 177-185. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Pastor J.E., Hobbs, E.C., and Losick, R. 2003. Cannibalism by sporulating bacteria. Science 301: 510-513. [DOI] [PubMed] [Google Scholar]
- Hanada M., Aimé-Sempé, C., Sato, T., and Reed, J.C. 1995. Structure-function analysis of Bcl-2 protein. J. Biol. Chem. 270: 11962-11969. [DOI] [PubMed] [Google Scholar]
- Hockenbery D.M., Oltvai, Z.N., Yin, X.-M., Milliman, C.L., and Korsmeyer, S.J. 1993. Bcl-2 functions in an antioxidant pathway to prevent apoptosis. Cell 75: 241-251. [DOI] [PubMed] [Google Scholar]
- Hoeberichts F.A., ten Have, A., and Woltering, E.J. 2003. A tomato metacaspase gene is upregulated during programmed cell death in Botrytis cinerea-infected leaves. Planta 217: 517-522. [DOI] [PubMed] [Google Scholar]
- Igaki T., Kanuka, H., Inohara, N., Sawamoto, K., Nunez, G., Okano, H., and Miura, M. 2000. Drob-1, a Drosophila member of the Bcl-2/CED-9 family that promotes cell death. Proc. Natl. Acad. Sci. 97: 662-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda S. and Kawasaki, N. 2001. Isolation and characterization of the Schizosaccharomyces pombe cDNA encoding the mitochondrial endonuclease(1). Biochim. Biophys. Acta 1519: 111-116. [DOI] [PubMed] [Google Scholar]
- Jakobs S., Martini, N., Schauss, A.C., Egner, A., Westermann, B., and Hell, S.W. 2003. Spatial and temporal dynamics of budding yeast mitochondria lacking the division component Fis1p. J. Cell Sci. 116: 2005-2014. [DOI] [PubMed] [Google Scholar]
- James D.I., Parone, P.A., Mattenberger, Y., and Martinou, J.C. 2003. hFis1, a novel component of the mammalian mitochondrial fission machinery. J. Biol. Chem. 278: 36373-36379. [DOI] [PubMed] [Google Scholar]
- Jensen R.E., Hobbs, A.E., Cerveny, K.L., and Sesaki, H. 2000. Yeast mitochondrial dynamics: Fusion, division, segregation, and shape. Microsc. Res. Tech. 51: 573-583. [DOI] [PubMed] [Google Scholar]
- Jonas E.A., Hoit, D., Hickman, J.A., Brandt, T.A., Polster, B.M., Fannjiang, Y., McCarthy, E., Hardwick, J.M., and Kaczmarek, L.K. 2003. Modulation of synaptic transmission by the BCL-2 family protein BCL-XL. J. Neurosci. 23: 8423-8431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas E.A., Hickman, J.A., Chachar, M., Polster, B.M., Brandt, T.A., Fannjiang Y., Ivanovska, I., Basañez, G., Kinnally, K.W., Zimmerberg, J., et al. 2004. Proapoptotic N-truncated BCL-xL protein activates endogenous mitochondrial channels in living synaptic terminals. Proc. Natl. Acad. Sci. 101: 13590-13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M., Lee, Y.J., Gaume, B., Jeong, S.Y., Frank, S., Nechushtan, A., Santel, A., Fuller, M., Smith, C.L., and Youle, R.J. 2002. Spatial and temporal association of Bax with mitochondrial fission sites, Drp1, and Mfn2 during apoptosis. J. Cell Biol. 159: 931-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M., Arnoult, D., Chen, H., Chan, D.C., Smith, C.L., and Youle, R.J. 2004. Quantitation of mitochondrial dynamics by photolabeling of individual organelles shows that mitochondrial fusion is blocked during the Bax activation phase of apoptosis. J. Cell Biol. 164: 493-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann T., Schlipf, S., Sanz, J., Neubert, K., Stein, R., and Borner, C. 2003. Characterization of the signal that directs Bcl-x(L), but not Bcl-2, to the mitochondrial outer membrane. J. Cell Biol. 160: 53-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legros F., Lombes, A., Frachon, P., and Rojo, M. 2002. Mitochondrial fusion in human cells is efficient, requires the inner membrane potential and is mediated by mitofusins. Mol. Biol. Cell 13: 4343-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L.Y., Luo, X., and Wang, X. 2001. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature 412: 95-99. [DOI] [PubMed] [Google Scholar]
- Liang X.H., Jackson, S., Seaman, M., Brown, K., Kempkes, B., Hibshoosh, H., and Levine, B. 1999. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402: 672-676. [DOI] [PubMed] [Google Scholar]
- Longo V.D., Ellerby, L.M., Bredesen, D.E., Valentine, J.S., and Gralla, E.B. 1997. Human Bcl-2 reverses survival defects in yeast lacking superoxide dismutase and delays death of wild-type yeast. J. Cell Biol. 137: 1581-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F., Frohlich, E., and Frohlich, K.U. 1997. A yeast mutant showing diagnostic markers of early and late apoptosis. J. Cell Biol. 139: 729-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F., Frohlich, E., Ligr, M., Grey, M., Sigrist, S.J., Wolf, D.H., and Frohlich, K.U. 1999. Oxygen stress: A regulator of apoptosis in yeast. J. Cell Biol. 145: 757-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F., Engelhardt, S., Herker, E., Lehmann, N., Maldener, C., Proksch, A., Wissing, S., and Frohlich, K.U. 2002a. Apoptosis in yeast: A new model system with applications in cell biology and medicine. Curr. Genet. 41: 208-216. [DOI] [PubMed] [Google Scholar]
- Madeo F., Herker, E., Maldener, C., Wissing, S., Lachelt, S., Herlan, M., Fehr, M., Lauber, K., Sigrist, S.J., Wesselborg, S., et al. 2002b. A caspase-related protease regulates apoptosis in yeast. Mol. Cell 9: 911-917. [DOI] [PubMed] [Google Scholar]
- Mancini M., Anderson, B.O., Caldwell, E., Sedghinasab, M., Paty, P.B., and Hockenbery, D.M. 1997. Mitochondrial proliferation and paradoxical membrane depolarization during terminal differentiation and apoptosis in a human colon carcinoma cell line. J. Cell Biol. 138: 449-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdy A.D., McCaffery, J.M., and Shaw, J.M. 2000. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 151: 367-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechushtan A., Smith, C.L., Lamensdorf, I., Yoon, S.H., and Youle, R.J. 2001. Bax and Bak coalesce into novel mitochondria-associated clusters during apoptosis. J. Cell Biol. 153: 1265-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmeyer D.D. and Ferguson-Miller, S. 2003. Mitochondria: Releasing power for life and unleashing the machineries of death. Cell 112: 481-490. [DOI] [PubMed] [Google Scholar]
- Nunnari J., Marshall, W.F., Straight, A., Murray, A., Sedat, J.W., and Walter, P. 1997. Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol. Biol. Cell 8: 1233-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostolaza H. and Goni, F.M. 1995. Interaction of the bacterial protein toxin α-haemolysin with model membranes: Protein binding does not always lead to lytic activity. FEBS Lett. 371: 303-306. [DOI] [PubMed] [Google Scholar]
- Parrish J., Li, L., Klotz, K., Ledwich, D., Wang, X., and Xue, D. 2001. Mitochondrial endonuclease G is important for apoptosis in C. elegans. Nature 412: 90-94. [DOI] [PubMed] [Google Scholar]
- Rieder S.E., Banta, L.M., Koher, K., McCaffery, J.M., and Emr, S.D. 1996. Multilamellar endosome-like compartment accumulates in the yeast vps28 vacuolar protein sorting mutant. Mol. Biol. Cell 7: 985-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesaki H. and Jensen, R.E. 1999. Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J. Cell Biol. 147: 699-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J.M. and Nunnari, J. 2002. Mitochondrial dynamics and division in budding yeast. Trends Cell Biol. 12: 178-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skulachev V.P. 2002. Programmed death in yeast as adaptation? FEBS Lett. 528: 23-26. [DOI] [PubMed] [Google Scholar]
- Smaili S.S., Hsu, Y.T., Sanders, K.M., Russell, J.T., and Youle, R.J. 2001. Bax translocation to mitochondria subsequent to a rapid loss of mitochondrial membrane potential. Cell Death Differ. 8: 909-920. [DOI] [PubMed] [Google Scholar]
- Smirnova E., Griparic, L., Shurland, D.L., and van der Bliek, A.M. 2001. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell 12: 2245-2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovski D., Koutsopoulos, O.S., Okamoto, K., and Ryan, M.T. 2004. Levels of human Fis1 at the mitochondrial outer membrane regulate mitochondrial morphology. J. Cell Sci. 117: 1201-1210. [DOI] [PubMed] [Google Scholar]
- Suarez M.F., Filonova, L.H., Smertenko, A., Savenkov, E.I., Clapham, D.H., von Arnold, S., Zhivotovsky, B., and Bozhkov, P.V. 2004. Metacaspase-dependent programmed cell death is essential for plant embryogenesis. Curr. Biol. 14: R339-R340. [DOI] [PubMed] [Google Scholar]
- Suzuki M., Jeong, S.Y., Karbowski, M., Youle, R.J., and Tjandra, N. 2003. The solution structure of human mitochondria fission protein Fis1 reveals a novel TPR-like helix bundle. J. Mol. Biol. 334: 445-458. [DOI] [PubMed] [Google Scholar]
- Tieu Q. and Nunnari, J. 2000. Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J. Cell Biol. 151: 353-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieu Q., Okreglak, V., Naylor, K., and Nunnari, J. 2002. The WD repeat protein, Mdv1p, functions as a molecular adaptor by interacting with Dnm1p and Fis1p during mitochondrial fission. J. Cell Biol. 158: 445-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolkovsky A.M., Xue, L., Fletcher, G.C., and Borutaite, V. 2002. Mitochondrial disappearance from cells: A clue to the role of autophagy in programmed cell death and disease? Biochimie 84: 233-240. [DOI] [PubMed] [Google Scholar]
- Vander Heiden M.G., Plas, D.R., Rathmell, J.C., Fox, C.J., Harris, M.H., and Thompson, C.B. 2001. Growth factors can influence cell growth and survival through effects on glucose metabolism. Mol. Cell. Biol. 21: 5889-5912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira H.L., Boya, P., Cohen, I.E.l., Hamel, C., Haouzi, D., Druillenec, S., Belzacq, A.S., Brenner, C., Roques, B., and Kroemer, G. 2002. Cell permeable BH3-peptides overcome the cytoprotective effect of Bcl-2 and Bcl-X(L). Oncogene 21: 1963-1977. [DOI] [PubMed] [Google Scholar]
- Wang X., Yang, C., Chai, J., Shi, Y., and Xue, D. 2002. Mechanisms of AIF-mediated apoptotic DNA degradation in Caenorhabditis elegans. Science 298: 1587-1592. [DOI] [PubMed] [Google Scholar]
- Xue L., Fletcher, G.C., and Tolkovsky, A.M. 2001. Mitochondria are selectively eliminated from eukaryotic cells after blockade of caspases during apoptosis. Curr. Biol. 11: 361-365. [DOI] [PubMed] [Google Scholar]
- Yoon Y., Krueger, E.W., Oswald, B.J., and McNiven, M.A. 2003. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol. Cell. Biol. 23: 5409-5420. [DOI] [PMC free article] [PubMed] [Google Scholar]