Abstract
Dictyostelium mitotic kinesin Kif12 is required for cytokinesis. Myosin II localization to the cleavage furrow is severely depressed in Kif12-null (Δkif12) cells, which accounts in part for the cytokinesis failure. Myosin II-null cells, however, undergo mitosis-coupled cytokinesis when adhering to a surface, whereas the Δkif12 cells cannot. During mitosis, the rate of change of internuclear separation in Δkif12 cells is reduced compared with wild-type cells, indicating multiple roles of this molecular motor during mitosis and cytokinesis. GFP-Kif12, which rescues wild-type behavior when expressed in the Δkif12 strain, is concentrated in the nucleus in interphase cells, translocates to the cytoplasm at the onset of mitosis, appears in the centrosomes and spindle, and later is concentrated in the spindle midbody. Given these results, we hypothesize a mechanism for myosin II translocation to the furrow to set up the contractile ring.
Keywords: Kif12, cell-cycle regulation, mitosis
Cytokinesis and karyokinesis are fundamental to cell division and propagation. Karyokinesis involves nuclear segregation and requires the formation of a microtubule network on which the chromosomes are segregated with the help of kinesin-based motors. Cytokinesis involves division of cellular components of a parental cell into two daughter cells, and it is precisely timed along with nuclear division in almost all cells. Cytokinesis is generally achieved by an actin–myosin-based contractile ring that is assembled along the plane perpendicular to the axis of the spindle poles. Karyokinesis and cytokinesis are tightly synchronized to ensure high fidelity of genomic information transfer.
Classic experiments carried out on sand dollar eggs established the importance of the mitotic spindles and astral microtubules in setting up the contractile ring (1, 2). The central spindle has been shown to be important for the completion of cytokinesis in several organisms, including Dictyostelium (3). Although a number of proteins have been shown to be associated with the mitotic spindle and implicated in cytokinesis (4–6), the molecular clues for the crosstalk between the spindle and cortical midzone during mitosis remain elusive.
Members of the mitotic kinesin family have been shown to play key roles in the assembly and function of the mitotic spindle. In mammalian cells, MKLP1 is required for the formation of the midbody matrix and the completion of cytokinesis (7, 8). Similar gene disruption experiments of MKLP1 orthologs in zebrafish (9), Drosophila (10), and Caenorhabditis elegans (11) also result in a cytokinesis failure. In most cases, furrow ingression is initiated, but cytokinesis fails to complete. Thus, MKLP1 is a key player in passing information from the mitotic spindle to the cleavage furrow.
Myosin II is a major component of the cellular machinery responsible for cytokinesis (12, 13). However, little is known about how myosin is translocated to the cleavage furrow during mitosis. Using Dictyostelium as a model system, our laboratory has been studying myosin II regulation and localization during cell division. We have now identified a mitotic kinesin-like protein (Kif12), which, like MKLP1, is required for cells to undergo mitotically linked cytokinesis. Using live-cell imaging, we show that Kif12 is required for normal myosin II localization during mitosis. We show that Kif12 also is required for normal nuclear separation during late anaphase and telophase.
Materials and Methods
Cell Culture. Dictyostelium discoideum AX2 (wild-type) cells were grown in axenic HL5 medium supplemented with penicillin and streptomycin at 22°C. Kif12-null (Δkif12) cells were grown with additional 10 μg/ml blasticidin S hydrochloride. Wild-type and Δkif12 cells carrying GFP-myosin II and other plasmids were grown in medium containing penicillin, streptomycin, and 10 μg/ml G418. The cells were grown in plastic Petri dishes. Cells in suspension were grown in conical flasks on a rotating shaker at ≈150 rpm.
Identification and Cloning of the kif12 Gene from Dictyostelium. We identified a gene (kif12) in the Dictyostelium genome (www.dictybase.org) through a sequence homology search (blast, www.ncbi.nlm.nih.gov/blast) that compared mitotic kinesins from various organisms. cDNA clones for kif12 were obtained by RT-PCR from a total RNA preparation of Dictyostelium. A 4.5-kb sequence corresponding to the full length of kif12 was cloned from Dictyostelium by using the 5′ and 3′ primers ATGGATCCATGAAAGATTATATTTCATCTCC and GTTCTAGACGAGGTTTAGTCCTAAAGTTTTACGTGG, respectively. The full-length kif12 was cloned further into a pTX-based vector (14). While this work was being completed, this gene and five other mitotic kinesins were reported in the Dictyostelium genome (15).
The Knockout Construct for the Disruption of the Dictyostelium kif12 Gene. A construct for a gene knockout was created, as described in ref. 16. Two fragments of ≈500 bp each at both ends of the kif12 gene were amplified.
Fragment 1 (near the N terminus) was amplified from the kif12 gene by using the 5′ primer bam43531f, ATGGATCCGGCATATGGTGTAACAAATCTGG and the 3′ primer xba43532r, TGTCTAGACCT TGT TCA ACA ATATCACGTGCATCC. Fragment 2 (at the C terminus) was amplified by using the 5′ primer hin43532f, GTAAGCTTCCAACTAAACTTCAACCACATTCATCACC and the 3′ primer xho43532r, GTCTCGAGCGAGGTTTAGTCCTAAAGTTTTACGTGG.
The oligonucleotides used for amplification contained appropriate restriction sites to be cloned into pSP72-BSR [containing the gene conferring resistance to blasticidin (17)]. Briefly, fragment 1 of kif12 was cloned into pSP72-Bsr at the 5′ end of the blasticidin gene, and fragment 2 of kif12 was cloned into the 3′ end of the blasticidin gene, generating pSP72-fragment1-Bsr-fragment2. Through homologous recombination, nearly 70% of the gene was deleted. The deletion contained the catalytic head domain of kinesin, the neck linker, and almost the entire tail domain (except the last 200 amino acid residues), which were replaced with a blasticidin-resistance marker flanked on both sides with an actin promoter and terminator sequences.
Transformation of Dictyostelium and Selection of the Knockout Clones. The knockout construct was digested from pSP72-fragment1-BSR-fragment2 by using unique restriction sites (BamHI and XhoI) at the 5′ and 3′ ends of the construct to separate the fragment1-BSR-fragment2 from the pSP72 plasmid. The linearized knockout construct was then transformed into wild-type cells by electroporation. Transformed cells were selected on Petri plates with HL5 medium containing penicillin, streptomycin, and 10 μg/ml blasticidin. Individual clones were picked and transferred into 24-well plates. The colonies were picked and grown on 150-cm plates for obtaining genomic DNA for further analysis.
Southern Blot Analysis. Genomic DNA from each mutant cell line and wild-type parental control cells was prepared and digested with the restriction enzyme AccI and processed for transfer to a nylon membrane by using standard methods described in ref. 16. Fragment 1 and fragment 2 (Fig. 6, which is published as supporting information on the PNAS web site) were used as probes and were labeled by using the Gene Images Alkphos Direct labeling kit (Amersham Biosciences). The bound probes were detected with CDP-Star chemiluminescent detection reagent (Amersham Biosciences) after an exposure of 1 h on hyperfilm MP imaging film (Amersham Biosciences).
Expression of Protein in Dictyostelium. For the expression of GFP and Flag-tagged proteins in Dictyostelium cells, the full-length sequence encoding Kif12 was amplified by PCR from the cDNA clones and ligated into pTX-based vectors that are under the control of the actin 15 promoter (14). Transformants were selected and maintained at 7.5–10 μg/ml G418.
Immunofluorescence and Live-Cell Microscopy. D. discoideum wild-type and Δkif12 cells (HS 50) were cultivated on coverslips or in optical coverglass chambers (Lab-Tek). For immunostaining, the cells were fixed with 2–4% formaldehyde for 5 min and then incubated in methanol for 5 min at –10°C. Subsequently, the cells were stained with 4′,6-diamidino-2-phenylindole (Molecular Probes) to show nuclei.
For live-cell imaging, the cells were cultivated at 22°C in optical coverglass chambers and the nutrient medium was replaced by imaging buffer (20 mM Mes buffer, pH 6.8, containing 0.2 mM CaCl2 and 2 mM MgSO4).
The cells were observed with a 64×/1.3 numerical aperture objective on an Axiovert microscope (Zeiss). Images were collected by using metamorph software (Universal Imaging, Downingtown, PA), and the images were processed with photoshop (Adobe Systems, San Jose, CA) and imagej (National Institutes of Health). Live-cell imaging was performed by collecting images at 30-s intervals.
Results
Identification of kif12 from Dictyostelium. We identified and cloned the kif12 gene from D. discoideum (Fig. 7, which is published as supporting information on the PNAS web site). The Dictyostelium genomic database was screened by using protein sequence similarity to the MKLP1 mitotic kinesin family from other organisms. The sequence of the kif12 gene encodes a protein with a molecular mass of 168 kDa that consists of a kinesin motor domain near the N terminus, with an N-terminal extension of ≈100 residues (Fig. 6). The motor domain is followed by a central stalk and then a C-terminal tail region.
The motor domain of Kif12 shows the highest degree of sequence homology with the head domain of the mitotic kinesin MKLP1. The full-length sequence of Dictyostelium Kif12 is available through GenBank (accession no. AY484465), and the motor domain has sequence similarity to human (50%), C. elegans (56%), and zebrafish (56%) MKLP1. However, outside the motor domain, Kif12 does not show significant homology to MKLP1, any other mitotic kinesin-related proteins, or conventional kinesins.
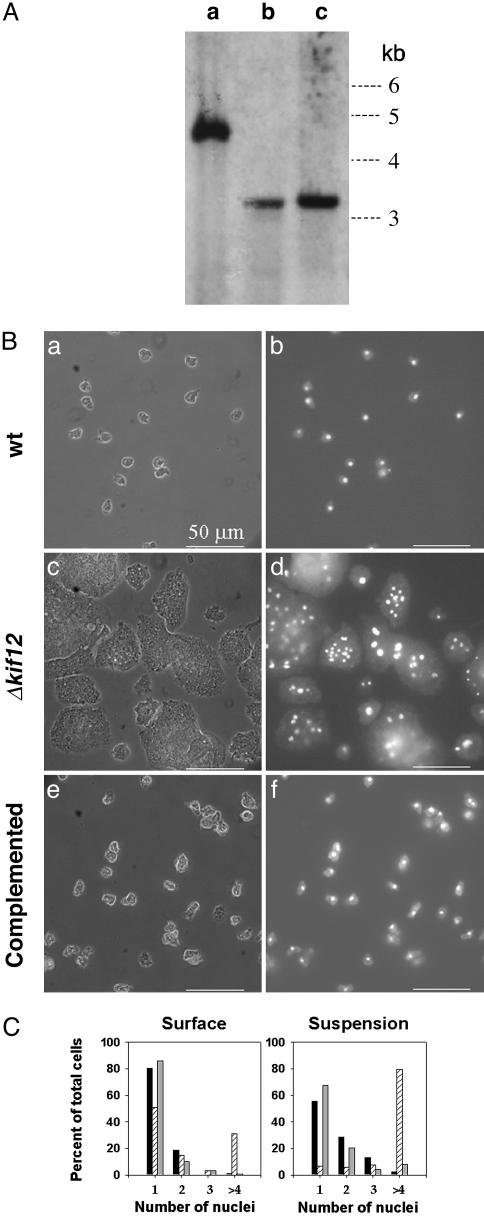
Disruption of the kif12 Gene. To examine the cellular function of Kif12, we deleted the region of the kif12 gene that codes for the motor domain, neck linker, and most of the tail domain (Fig. 6) by homologous recombination using a knockout construct (16, 17). Briefly, a gene conferring resistance to blasticidin was inserted as a marker between two regions of the kif12 gene, one near the N terminus and the other at the C terminus. This construct was transformed into wild-type Dictyostelium (AX2) cells, and the resulting colonies were selected for blasticidin resistance. Deletion of kif12 was confirmed by Southern blot analysis (Fig. 1A). By using the same construct, kif12 also was deleted in an independent strain of Dictyostelium (Orf+).
Fig. 1.
Establishing the kif12 knockout in Dictyostelium. (A) Genomic DNA from the parental strain and two independent knockout strains was subjected to restriction digestion by AccI and used for Southern blot analysis. A 500-bp DNA fragment from the N terminus of the gene was used as a probe (indicated in Fig. 6). Lane a shows a 4.6-kb band, expected for the parental strain. Lanes b and c, containing DNA from strains KO1 (HS50) and KO2 (HS51), respectively, show a 3.4-kb band, which is expected from homologous recombination with the knockout construct shown in Fig. 6. (B) Morphology of Dictyostelium cells. The phase-contrast image is shown in a, c, and e; b, d, and f show nuclear staining (with 4′,6-diamidino-2-phenylindole) for the parental wild-type (wt) strain (a and b), the Δkif12 strain (c and d), and the Δkif12 strain rescued with GFP-Kif12 (e and f). (C) Effect of deletion of kif12 on cytokinesis of Dictyostelium. Left shows the comparison of the number of nuclei per cell when grown on a surface for wild-type (black bars) and Δkif12 (hatched bars) cells and Δkif12 cells complemented with kif12 (gray bars). In each case, n = 200. Right shows the comparison of the number of nuclei per cell when grown in suspension.
Δkif12 Cells Fail to Undergo Cytokinesis. Compared with wild-type cells, which predominantly were mononucleated, Δkif12 cells generally were large and multinucleated when grown on a surface (Fig. 1B). The Δkif12 cells were unable to divide in a cell-cycle-dependent manner and remained multinucleated during the vegetative cycle. When grown in suspension, the Δkif12 cells showed a severe cytokinesis defect (Fig. 1C), and the cells did not survive for more than a couple of days in suspended culture. The Δkif12 cells lacked the ability to maintain a relatively homogeneous cell volume (Fig. 8, which is published as supporting information on the PNAS web site).
Myosin II is required for division of Dictyostelium cells in suspension (cytokinesis A), but Dictyostelium cells lacking the myosin heavy chain gene (mhcA–) can undergo mitosis-coupled division in the absence of myosin II on a surface by an adhesion-dependent process (cytokinesis B) (18–20). The Δkif12 cells failed to divide by using cell-cycle-dependent cytokinesis both on a surface and in suspension, suggesting that this gene is common to both cytokinesis A and cytokinesis B in Dictyostelium. On a surface, the Δkif12 cells could pull themselves apart into fragments and undergo mitosis-independent, traction-mediated cytofission (21). When cells divided in this way, they typically divided asymmetrically, forming smaller cells that survived as long as they captured at least one nucleus. The Δkif12 cells that were mononucleated showed various defects during mitosis. Some Δkif12 cells got arrested at metaphase, and the cells remained rounded for a very long time. Other Δkif12 cells showed some furrow ingression during late mitosis, but the furrow ingression did not progress to completion, and the cells failed to undergo complete cytokinesis (Movie 1, which is published as supporting information on the PNAS web site). The furrow ingression frequently was asymmetric. Occasionally, when the cleavage furrow formation went nearly to completion, the two daughter cells retracted and fused back to form a single parent cell. The same deletion phenotype was observed with both the AX2 and Orf+ strains.
Δkif12 Cells Are Complemented by Extrachromosomal Expression of Kif12. Two constructs were made to express N-terminal fusion proteins with full-length Kif12 in Dictyostelium. Flag-kif12 and GFP-kif12 were made and transformed into Δkif12 cells. Both kif12 constructs fully rescued wild-type behavior in Δkif12 cells (Fig. 1 B and C).
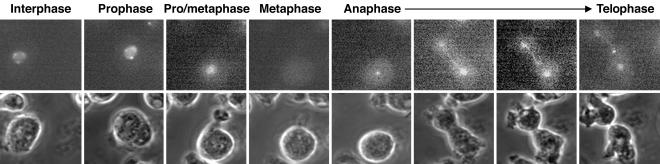
Localization of GFP-Kif12 Is Cell-Cycle Regulated. Δkif12 cells transformed with GFP-kif12 were monitored for cellular localization of GFP-Kif12. During interphase, most of the GFP-Kif12 was localized in punctate spots associated with nuclei (Fig. 2). As the cells entered metaphase, GFP-Kif12 appeared to completely leave the nucleus and was found diffuse in the cytoplasm. Upon the onset of anaphase, GFP-Kif12 localized to the centrosomes and along the spindle fibers. After anaphase, the GFP-Kif12 showed some concentration in the midbody (Movies 2 and 3, which are published as supporting information on the PNAS web site).
Fig. 2.
Localization of GFP-Kif12 in live Δkif12 cells undergoing cytokinesis. Upper shows the GFP fluorescence during various stages of mitosis, and Lower shows the corresponding phase-contrast images.
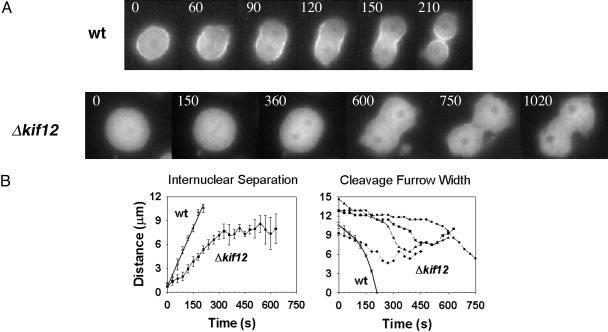
Localization of Myosin II in Δkif12 Cells. Δkif12 cells were transformed with GFP-myosin II, and the cells were grown on a surface. These cells were monitored, and their attempts at cell division were compared with the wild-type cell-division process by using GFP-myosin II localization as a marker for cytokinesis, as shown in Fig. 3A.
Fig. 3.
Characterization of Δkif12 cells expressing GFP-myosin II. (A) Time-lapse images show the localization of GFP-myosin II in wild-type (wt) and Δkif12 cells. (Upper) Wild-type cells undergoing cytokinesis. GFP-myosin II is localized strongly at the cleavage furrow (representative of 20 cells imaged). (Lower) Δkif12 cells expressing GFP-myosin II at a stage similar to that of the wild-type cells. No distinct localization of GFP-myosin II is observable at the cleavage furrow in the Δkif12 cells (representative of 30 cells imaged). Time is indicated in seconds. (B) Internuclear distance and width of the cleavage furrow with time in wild-type and Δkif12 cells. Left shows the internuclear separation, and Right shows the width of the cleavage furrow. Five cells were averaged to obtain the wild-type nuclear separation data and the wild-type cytokinesis data. The width of the cleavage furrow is shown for representative Δkif12 cells because there was a large variation in the cleavage furrow width, unlike the situation for wild-type cells, for which the average of six cells is shown.
The nuclear position was easily measured because it appears relatively dark because of exclusion of the GFP-myosin II from the nucleus. Wild-type cells completed cell division (Movie 4, which is published as supporting information on the PNAS web site) <200 s after anaphase, whereas the Δkif12 cells attempted division 3–6 times longer after anaphase before failing to divide (Fig. 3). The internuclear distance and the width of the cleavage furrow were measured as a function of time. Nuclear separation occurred in the Δkif12 cells, but the rate was slowed compared with wild-type cells, and the separation was no longer linear with time (22) (Fig. 3B). Thus, Kif12 may work in concert with other mitotic kinesins to provide the normal rate of karyokinesis. For example, KLP61F, a Drosophila bipolar kinesin, and Ncd, a minus-end-directed kinesin, have been implicated in a similar anaphase role (23). Dynein also may play a role in this process by pulling on astral microtubules at the poles of the cell (24, 25).
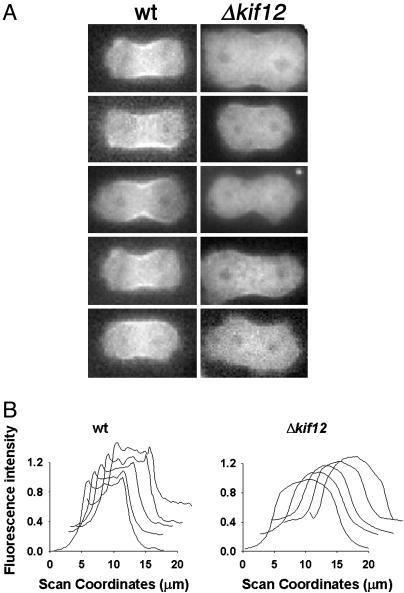
Unlike wild-type cells, Δkif12 cells expressing GFP-myosin II failed to localize GFP-myosin II to the cleavage furrow (Fig. 4A and Movie 5, which is published as supporting information on the PNAS web site). The line scan of fluorescence intensity of GFP-myosin across the width of the cleavage furrow indicated the lack of accumulation of myosin II in the furrow cortex (Fig. 4B).
Fig. 4.
Observation of early furrowing in cells expressing GFP-myosin II. (A) Left shows five live wild-type (wt) cells undergoing cytokinesis. GFP-myosin II localization is observed prominently in the cleavage furrow. Right shows attempts of cytokinesis in live Δkif12 cells. The cells fail to localize GFP-myosin II to the cleavage furrow. (B) Line-scan analysis of fluorescence intensity of GFP-myosin II across the width of the cleavage furrow for each cell shown in A. The y axis shows the normalized fluorescence, and the x axis shows the scanning coordinate in micrometers.
Discussion
In this study, we successfully cloned the kif12 gene and showed that Kif12 plays a critical role in cytokinesis in Dictyostelium. Localization of Kif12 in interphase cells occurs predominantly in regions associated with the nucleus. Kif12 has two nuclear localization signals (Fig. 6).
In budding yeast, nucleoli serve as storage depots for regulatory factors, including Mdm2, Pch2, Sir2, and Cdc14, which regulate the onset of anaphase and telophase. These factors are known collectively as the mitotic exit network (26). It has been shown in vitro that cdk1/cyclin B phosphorylates the motor domain of ZEN-4, a C. elegans MKLP1 ortholog, and thus reduces its affinity for microtubules (27). However, a constitutively dephosphorylated form of ZEN-4 localizes to the central spindle during mitosis (27). Phosphorylation by cyclin B seems to account for the release of mitotic exit activator cdc14 from the nucleolus (28). It is possible that cdc14 dephosphorylates MKLP1, causing it to bind to microtubules, which results in localization of mitotic machinery (including INCENP-Aurora B) to the spindle (29).
Previous studies showed that the localization of Dictyostelium myosin II to the cleavage furrow region is independent of its interaction with actin (30, 31). In wild-type cells, after metaphase, myosin II localizes to the cell cortex and then is depleted at the poles by the action of myosin heavy chain kinases (MHCKs) (32). Bipolar thick filaments of myosin tails carrying the regulatory light chain but having the essential light chain and catalytic domain replaced by a GFP molecule were found to accumulate in the cytoplasmic domain of the cleavage furrow region, rather than in a circumferential cortical ring in the furrow (30). These observations would be consistent with myosin II bipolar thick filaments being pulled to the center of the cell along spindle microtubules by way of the myosin tail domain to accumulate in the cytoplasmic region of the furrow. Activation of the myosin II motor domains then would allow the thick filaments to interact with actin filaments attached to the cortical membrane, resulting in pulling the filaments into the cortex in the furrow region. The activity of MHCK A and MHCK C in the polar regions of the dividing cell could release myosin II from the poles (33), thus biasing myosin II localization to the central cortex of the dividing cell.
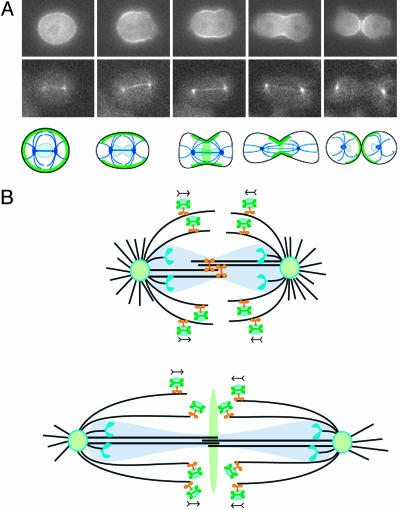
In Dictyostelium, the nuclear membrane does not completely break down during mitosis. The centrosomes are positioned on the outside of the nuclear membrane, and a central spindle of overlapping microtubules is intranuclear throughout mitosis (Fig. 5A and ref. 3). The role of Kif12 in myosin localization could be relatively indirect and may be the result of complex structural, signaling, and regulatory mechanisms. Kif12, for example, may play a role in the cellular distribution of mechanoregulatory proteins, which collectively ensure faithful cell division coupled to mitosis. However, it is worth considering the simplest hypothesis: that Kif12 may be transporting myosin II along mitotic extranuclear microtubules toward the center of the cell (Fig. 5B). Kinesins are known to transport vesicular cargo, and roles of membrane insertion during cytokinesis and deposition of special lipids in the region of the cleavage furrow have been documented recently (34, 35). Thus, this simplest model of Kif12-driven myosin translocation could be vesicle-mediated, with the vesicles being inserted into the cleavage furrow membrane during furrow formation (36).
Fig. 5.
Progression of a live cell during cytokinesis and model for spindle-pole and vesicle movement during mitosis. (A) Top shows the localization and reorganization of myosin II during cytokinesis in a wild-type cell expressing GFP-myosin. Middle shows GFP-α-tubulin expressed in a wild-type cell under-going cytokinesis. Bottom is a schematic that shows the localization of myosin (green) and tubulin (dark blue) during cytokinesis in Dictyostelium wild-type cells. The position of the nuclei is indicated in light blue. (B) Upper shows Kif12 (orange) on microtubules (black) in the spindle region of a cell during anaphase. The nuclear membrane does not completely break down during mitosis of Dictyostelium and is shown in transparent light blue. The poles (light-green ovals) are separated, in part, by the action of Kif12 on the interdigitating central spindle microtubules. Kif12 also transports vesicles (light-blue ovals) carrying mechanoregulatory factors, perhaps including myosin II (green), on the spindle fibers. Chromosomes (blue) are being segregated bound to kinetochore microtubules. Lower shows a cell in late telophase. The vesicles drop off in the center of the cell and transfer the signals for cytokinesis.
Supplementary Material
Acknowledgments
We thank Dr. Arturo De Lozanne (University of Texas, Austin) for the PSP72 plasmid construct, Dr. Gunther Gerisch (Max-Planck-Institut für Biochemie, Martinsried, Germany) for the GFP-α-tubulin construct, Natalie Dye for extensive help in collecting the immunofluorescence data, Sara Ocon for help during the course of the work, Tom Purcell for help on the model figure, and other members of J.A.S.'s laboratory for critical comments on the work. G.S.L. is supported by a Damon Runyon Cancer Research Foundation Postdoctoral Fellowship. This work was supported by National Institutes of Health Grant GM46551 (to J.A.S.).
Author contributions: G.S.L. and J.A.S. designed research; G.S.L. and H.M.W. performed research; G.S.L., H.M.W., and J.A.S. contributed new reagents/analytic tools; G.S.L. and J.A.S. analyzed data; and G.S.L. and J.A.S. wrote the paper.
References
- 1.Rappaport, R. (1961) J. Exp. Zool. 148, 81–89. [DOI] [PubMed] [Google Scholar]
- 2.Rappaport, R. (1996) Cytokinesis in Animal Cells (Cambridge Univ. Press, Cambridge, U.K.).
- 3.McIntosh, J. R., Roos, U. P., Neighbors, B. & McDonald, K. L. (1985) J. Cell Sci. 75, 93–129. [DOI] [PubMed] [Google Scholar]
- 4.Glotzer, M. (2003) Curr. Opin. Cell Biol. 15, 684–690. [DOI] [PubMed] [Google Scholar]
- 5.Straight, A. F. & Field, C. M. (2000) Curr. Biol. 10, R760–R770. [DOI] [PubMed] [Google Scholar]
- 6.Robinson, D. N. & Spudich, J. A. (2000) Trends Cell Biol. 10, 228–237. [DOI] [PubMed] [Google Scholar]
- 7.Matuliene, J. & Kuriyama, R. (2002) Mol. Biol. Cell 13, 1832–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuriyama, R., Gustus, C., Terada, Y., Uetake, Y. & Matuliene, J. (2002) J. Cell Biol. 156, 783–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, M. C., Zhou, Y. & Detrich, H. W., III (2002) Physiol. Genomics 8, 51–66. [DOI] [PubMed] [Google Scholar]
- 10.Minestrini, G., Harley, A. S. & Glover, D. M. (2003) Mol. Biol. Cell 14, 4028–4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Powers, J., Bossinger, O., Rose, D., Strome, S. & Saxton, W. (1998) Curr. Biol. 8, 1133–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mabuchi, I. & Okuno, M. (1977) J. Cell Biol. 74, 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Satterwhite, L. L. & Pollard, T. D. (1992) Curr. Opin. Cell Biol. 4, 43–52. [DOI] [PubMed] [Google Scholar]
- 14.Levi, S., Polyakov, M. & Egelhoff, T. T. (2000) Plasmid 44, 231–238. [DOI] [PubMed] [Google Scholar]
- 15.Kollmar, M. & Glockner, G. (November 27, 2003) BMC Genomics, 10.1186/1471-2164-4-47. [DOI] [PMC free article] [PubMed]
- 16.Manstein, D. J., Titus, M. A., De Lozanne, A. & Spudich, J. A. (1989) EMBO J. 8, 923–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang, N., Wu, W. I. & De Lozanne, A. (2002) J. Cell. Biochem. 86, 561–570. [DOI] [PubMed] [Google Scholar]
- 18.Neujahr, R., Heizer, C. & Gerisch, G. (1997) J. Cell Sci. 110, 123–137. [DOI] [PubMed] [Google Scholar]
- 19.Zang, J. H., Cavet, G., Sabry, J. H., Wagner, P., Moores, S. L. & Spudich, J. A. (1997) Mol. Biol. Cell 8, 2617–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerisch, G. & Weber, I. (2000) Curr. Opin. Cell Biol. 12, 126–132. [DOI] [PubMed] [Google Scholar]
- 21.De Lozanne, A. & Spudich, J. A. (1987) Science 236, 1086–1091. [DOI] [PubMed] [Google Scholar]
- 22.Sabry, J. H., Moores, S. L., Ryan, S., Zang, J. H. & Spudich, J. A. (1997) Mol. Biol. Cell 8, 2605–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharp, D. J., Rogers, G. C. & Scholey, J. M. (2000) Nature 407, 41–47. [DOI] [PubMed] [Google Scholar]
- 24.Busson, S., Dujardin, D., Moreau, A., Dompierre, J. & De Mey, J. R. (1998) Curr. Biol. 8, 541–544. [DOI] [PubMed] [Google Scholar]
- 25.Gaetz, J. & Kapoor, T. M. (2004) J. Cell Biol. 166, 465–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Visintin, R. & Amon, A. (2000) Curr. Opin. Cell Biol. 12, 372–377, and corrigendum (2000) 12, 752. [DOI] [PubMed] [Google Scholar]
- 27.Mishima, M., Pavicic, V., Gruneberg, U., Nigg, E. A. & Glotzer, M. (2004) Nature 430, 908–913. [DOI] [PubMed] [Google Scholar]
- 28.Azzam, R., Chen, S. L., Shou, W., Mah, A. S., Alexandru, G., Nasmyth, K., Annan, R. S., Carr, S. A. & Deshaies, R. J. (2004) Science 305, 516–519. [DOI] [PubMed] [Google Scholar]
- 29.Pereira, G. & Schiebel, E. (2003) Science 302, 2120–2124. [DOI] [PubMed] [Google Scholar]
- 30.Zang, J. H. & Spudich, J. A. (1998) Proc. Natl. Acad. Sci. USA 95, 13652–13657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uyeda, T. Q., Kitayama, C. & Yumura, S. (2000) Cell Struct. Funct. 25, 1–10. [DOI] [PubMed] [Google Scholar]
- 32.Berlot, C. H., Devreotes, P. N. & Spudich, J. A. (1987) J. Biol. Chem. 262, 3918–3926. [PubMed] [Google Scholar]
- 33.Liang, W., Licate, L., Warrick, H., Spudich, J. & Egelhoff, T. (July 24, 2002) BMC Cell Biol., 10.1186/1471-2121-3-19. [DOI] [PMC free article] [PubMed]
- 34.Shuster, C. B. & Burgess, D. R. (2002) Proc. Natl. Acad. Sci. USA 99, 3633–3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finger, F. P. & White, J. G. (2002) Cell 108, 727–730. [DOI] [PubMed] [Google Scholar]
- 36.Musch, A., Cohen, D. & Rodriguez-Boulan, E. (1997) J. Cell Biol. 138, 291–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.