Summary
This study was designed to determine the molecular characteristics and antimicrobial resistance of enterococcal strains isolated from patients admitted to an Iranian Hospital. Enterococcal strains were isolated from the burn patients. All strains were screened for genes encoding resistance to aminoglycoside [aac(6')-Ie-aph(2'')-Ia, aph (3'), ant (4')], resistance to vancomycin (vanA, vanB), resistance to tetracycline (tetK, tetL, tetM, tetO), and resistance to erythromycin (ermA, ermB, ermC) by PCR and multiplex PCR-based methods. Genetic diversity was evaluated via Random Amplified Polymorphic DNA (RAPD)-PCR. All enterococcal isolates showed complete sensitivity to vancomycin with MIC ≤ 0.5μg/ml. Resistance to gentamicin, tetracycline, erythromycin, ciprofloxacin or quinopristin-dalfopristin was detected, whilst more than 96.2% of isolates were high-level gentamicinresistant (HLGR) and multiple drug resistant. The most prevalent aminoglycoside resistance gene was aac(6')-Ie-aph(2'')-Ia, that was found in 96.2% (26/27) of the isolates. The most prevalent tetracycline resistance genes were tetM, found in 85.1% (23/27) followed by tetL and tetO found in 7.4% (2/27) of the isolates. The ermA and ermB genes were detected in 33.3% (9/27) and 44.4% (12/27) of the isolates respectively. RAPD-PCR analysis yielded 17 distinct profiles among 27 investigated isolates. One cluster of isolates shared the same RAPD pattern, while 16 isolates had unique RAPD pattern. Our study showed that during the examination time period one RAPD genotype was the common type and was disseminated among patients in the burn unit. Interestingly, most of these strains had an identical or very similar antibiotic and gene resistance pattern.
Key words: Enterococcus, HLGR, RAPD-PCR
Introduction
Enterococci are Gram-positive, facultative anaerobic bacteria that are members of the normal flora in the human gastrointestinal tract but have been recognized as important pathogens worldwide [1, 2]. Enterococcus faecalis and Enterococcus faecium are the most prevalent species isolated from the urinary tract and wound infections, which affect mainly patients admitted to the medical care centers [3-6]. The disruption of the normal skin barrier, the immunocompromised state, prolonged hospitalization and antibiotic therapy put burns patients at a high risk of acquiring nosocomial infections including enterococcal infections [7].
Enterococcal infections can be difficult to treat because they have a remarkable ability of adaptation when are exposed to antibiotics; they have intrinsic resistance to several antimicrobial agents and have a tremendous capacity to acquire high levels of resistance to antibiotics [8, 9]. Vancomycin-resistant enterococci (VRE) and high-level gentamicin-resistant (HLGR) isolates have emerged as important pathogen in Iran as well as in other countries, which create serious challenges for treating the infected patients [9-11].
There are several reports on the endemicity of VRE and HLRG in Iran but there is a lack of information on enterococcal stains isolated form burn centers [3, 12]. This study was designed to determine the molecular characteristics and antimicrobial resistance of enterococcal strains isolated from patients in a burn center in Iran.
Materials and methods
BACTERIAL ISOLATES
Twenty-seven enterococcal isolates were collected from wound specimens of patients with burn injury, during April to September 2012, in a burn hospital in Tehran. Only one isolate per patient was included in the study. Identification of enterococci was performed based on a series of conventional microbiological tests including Gram reaction, catalase reaction, presence of pyrrolidonyl arylamidase (PYR), growth on bile-aesculin agar and 6.5% NaCl media, motility test, arginine decarboxylation in Moeller decarboxylase media, pyruvate utilization, and fermentation of arabinose, raffinose, mannitol and ribose [3]. To confirm the identity of the isolates as E. faecalis or E. faecium the ddlE gene was amplified by a polymerase chain reaction (PCR) method as described by Dutka-Malen et al. [13].
ANTIMICROBIAL SUSCEPTIBILITY TESTING
Antibiotic susceptibility of the strains was performed by the disk diffusion method. The antibiotics (Mast Group Ltd., Merseyside, UK) tested were ciprofloxacin (5 μg), erythromycin (15 μg), tetracycline (30 μg), and quinupristin- dalfopristin (15 μg). High-level resistance to gentamicin was also determined by disk diffusion method on Mueller– Hinton agar (Conda S.A., Madrid, Spain) using 120 μg gentamicin disk. The minimal inhibitory concentration (MIC) of vancomycin was determined by the standard agar dilution test on Brain Heart Infusion agar (Conda S.A., Madrid, Spain). All Antibiotic susceptibilities were performed and interpreted according to the criteria of the Clinical and Laboratory Standards Institute (CLSI) guidelines [14].
DNA EXTRACTION AND GENE DETECTION
DNA extraction was performed as described previously and this DNA was used as a template for PCR analysis [11]. All strains were screened for genes encoding resistance to aminoglycoside [aac(6')-Ie-aph(2'')-Ia, aph (3'), ant (4')], vancomycin (vanA, vanB), tetracycline (tetK, tetL, tetM, tetO), and resistance to erythromycin (ermA, ermB, ermC) by PCR and multiplex PCR-based methods, using specific primers [3, 11,12, 15, 16].
RANDOM AMPLIFIED POLYMORPHIC DNA (RAPD)-PCR
The RAPD-PCR assay was carried out in 25 μl reaction volumes containing 0.5 μM of each primer (5′-AGCGGGCCAA-3′ and 5′- ACGGCCGACC-3′), 12.5 μl of PCR master (Sinaclone Inc, Iran) and 5 μl of DNA template. Cycling conditions were as follow: initial denaturation at 95°C for 5 min, followed by 30 cycles of 95°C for 1 min, 38.5°C for 1 min, and 72°C for 2 min, and final extension at 72°C for 10 min. PCR products were then separated by electrophoresis in 1.4% agarose gel with 0.5X TBE buffer. DNA bands were observed by staining with KBC power load dye (Kawsar Biotech Co. Iran) and photographed under UV illumination. RAPD patterns were analyzed by visual inspection. Only the obvious, prominent and reproducible bands from repeated experiments (at least twice) were considered and any pattern differing by one or more bands was classified as a distinct RAPD type.
Results
Distribution of species according to conventional microbiological tests and PCR method included 19 (70.3%) E. faecalis and 8 (29.7%) E. faecium. The prevalence of antimicrobial resistance pattern and RAPD types have been summarized in Table I. In the present study, no VRE was recovered and all enterococcal isolates showed complete sensitivity to vancomycin with MIC ≤ 0.5 μg/ml.
Tab. I.
The distribution of antimicrobial resistance genes, virulence genes and biofilm production among enterococcal species.
| Isolate | Date of Isolation | Species | Resistance pattern | Resistance genes | RAPD type |
|---|---|---|---|---|---|
| 1 | 20/4/2012 | E. faecalis | E*,GM, TET,SYN,CIP | tetM,ermA, aac(6')-Ie-aph(2'')-Ia, ant(4') | A |
| 2 | 22/4/2012 | E. faecium | GM, TET, SYN | tetM, aac(6')-Ie-aph(2'')-Ia, ant(4') | B |
| 3 | 24/4/2012 | E. faecium | E,GM, TET,SYN,CIP | tetM, aac(6')-Ie-aph(2'')-Ia | H |
| 4 | 26/4/2012 | E. faecium | E,GM, TET | tetL, tetM, ermB, aac(6')-Ie-aph(2'')-Ia, ant(4') | G |
| 5 | 26/4/2012 | E. faecalis | E,GM, TET | tetM,ermB, aac(6')-Ie-aph(2'')-Ia | P |
| 6 | 1/5/2012 | E. faecalis | E,GM, TET,SYN,CIP | tetM, ermB, aac(6')-Ie-aph(2'')-Ia | A |
| 7 | 4/5/2012 | E. faecium | E,GM, TET | tetL, tetM, ermB, aac(6')-Ie-aph(2'')- Ia, ant(4') | J |
| 8 | 8/5/2012 | E. faecalis | E,GM, TET,SYN,CIP | ermB, aac(6')-Ie-aph(2'')-Ia | A |
| 9 | 15/5/2012 | E. faecalis | E,GM, TET,SYN,CIP | tetM, aac(6')-Ie-aph(2'')-Ia | N |
| 10 | 2/6/2012 | E. faecalis | E,GM, TET,SYN,CIP | tetM,ermB, aac(6')-Ie-aph(2'')-Ia | A |
| 11 | 12/7/2012 | E. faecalis | E,GM, TET,SYN,CIP | tetM,ermA, aac(6')-Ie-aph(2'')-Ia, ant(4') | A |
| 12 | 14/7/2012 | E. faecalis | E,GM, TET,SYN,CIP | tetM,ermA, aac(6')-Ie-aph(2'')-Ia, ant(4') | A |
| 13 | 14/7/2012 | E. faecalis | E,GM, TET,SYN,CIP | tetM, ermA, aac(6')-Ie-aph(2'')-Ia, ant(4') | A |
| 14 | 21/7/2012 | E. faecalis | E,GM, TET,SYN,CIP | tetM, ermA, aac(6')-Ie-aph(2'')-Ia, ant(4') | A |
| 15 | 23/7/2012 | E. faecium | TET | tetM | M |
| 16 | 25/7/2012 | E. faecium | E, GM,TET,SYN,CIP | tetM,ermB, aac(6')-Ie-aph(2'')-Ia | F |
| 17 | 7/8/2012 | E. faecalis | E, GM,TET,SYN,CIP | tetM, ermA, aac(6')-Ie-aph(2'')-Ia, ant(4') | A |
| 18 | 7/8/2012 | E. faecalis | GM,SYN,CIP | tetM,ermB, aac(6')-Ie-aph(2'')-Ia | E |
| 19 | 14/8/2012 | E. faecalis | E, GM,TET,SYN,CIP | ermA, aac(6')-Ie-aph(2'')-Ia, ant(4') | D |
| 20 | 14/8/2012 | E. faecalis | E, GM,TET,SYN,CIP | tetM,ermB, aac(6')-Ie-aph(2'')-Ia, ant(4') | I |
| 21 | 21/8/2012 | E. faecalis | E, GM,TET,SYN,CIP | tetM,ermA, aac(6')-Ie-aph(2'')-Ia, ant(4') | A |
| 22 | 25/8/2012 | E. faecalis | E, GM,TET,SYN,CIP | tetM,ermA aac(6')-Ie-aph(2'')-Ia, ant(4') | A |
| 23 | 11/9/2012 | E. faecalis | E, GM,TET,SYN,CIP | tetM,tetO, ermB, aac(6')-Ie-aph(2'')-Ia, ant(4') | K |
| 24 | 11/9/2012 | E. faecium | E, GM | aac(6')-Ie-aph(2'')-Ia, ant(4') | Q |
| 25 | 16/9/2012 | E. faecalis | E, GM,TET,SYN,CIP | tetM, ermB, aac(6')-Ie-aph(2'')-Ia | C |
| 26 | 16/9/2012 | E. faecalis | E, GM,TET,SYN,CIP | tetM aac(6')-Ie-aph(2'')-Ia, ant(4') | L |
| 27 | 16/9/2012 | E. faecium | E, GM,TET,SYN,CIP | tetM,tetO, ermB, aac(6')-Ie-aph(2'')-Ia, ant(4') | O |
E, Erythromycin; G, Gentamicin; SYN, Quinupristin-dalfopristin; TET, Tetracycline; CIP, Ciprofloxacin
All isolates at least exhibited resistance to one antibiotic and 6 different profiles were observed on the basis of their antibiotic resistance patterns. High-level resistance to gentamicin and multiple drug resistance were observed in 96.2% of isolates. The most frequent profile was resistance to erythromycin, gentamicin, tetracycline, quinupristin-dalfopristin and ciprofloxacin that was observed in 74% of isolates.
The most prevalent aminoglycoside resistance gene was aac(6')-Ie-aph(2'')-Ia, found in 96.2% (26/27) of the isolates. These genes were detected in all HLGR isolates. The ant (4') gene was detected in 62.9% (17/27) of isolates. The most prevalent tetracycline resistance gene was tetM, found in 85.1% (23/27) of the isolates followed by tetL and tetO found in 7.4% (2/27) of the isolates. The ermA and ermB genes were detected in 33.3% (9/27) and 44.4% (12/27) of the isolates respectively. The aph (3'), tetK, ermC, vanA, and vanB genes were not detected in any of the enterococcal isolates in this study.
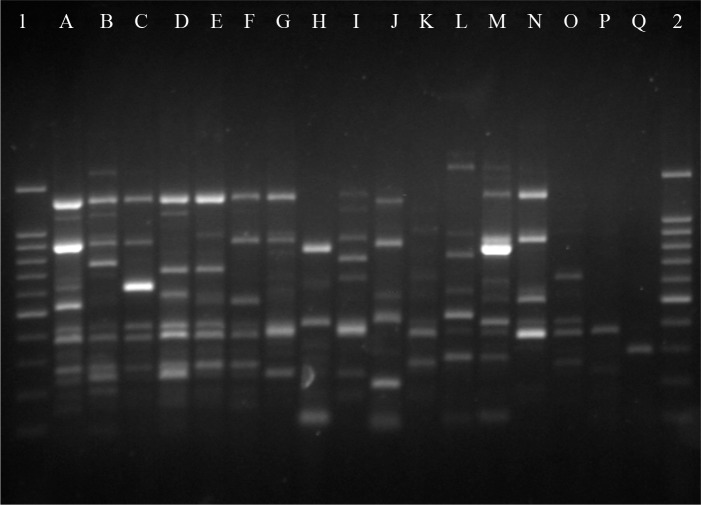
To determine the degree of clonality among enterococcal isolates, the RAPD typing was used. RAPD analysis yielded 17 distinct profiles among 27 investigated isolates (Fig. 1). One clusters of isolates shared the same RAPD patterns, while 16 isolates had unique RAPD patterns. A dominant RAPD type designated as A, that consisted 11 isolates of E. faecalis and were HLGR.
Fig. 1.
RAPD-PCR profiles of Enterococcus faecalis and E. faecium isolates. Lanes 1and 2; DNA size marker (100bp), Lanes A-Q; Enterococcal isolates with different RAPD patterns.
Discussion
Among Enterococci isolates, E. faecalis and E. faecium are the most common species caused enterococcal nosocomial infections [1]. In our study, the most prevalent species was E. faecalis (70.3%) followed by E. faecium (29.7%) isolated from burn wound infections. Similar results were reported for clinical isolates by other studies [3, 11, 16, 17].
In the treatment of enterococcal infections, the combination of gentamicin with a beta lactam antibiotic or a glycopeptide is used to obtain a synergistic bactericidal effect. However, strains that are highly resistant to gentamicin are no longer susceptible to the combination therapy [18]. Several studies have demonstrated that VRE have caused both outbreaks and endemic infections in burn units [5, 6], However, in this study, there were no VRE isolates and more than 96% of isolates were HLGR and 74% of them were simultaneously resistant to erythromycin, tetracycline, quinupristin-dalfopristin and ciprofloxacin. Our study is the first investigation on enterococcal resistance among burn patients in Iran and the findings indicated that vancomycin keeps its therapeutic effects against enterococcal infections.
Resistance against aminoglycosides among enterococci is commonly due to enzymatic modification. Genes encoding aminoglycoside-modifying enzymes (AMEs) are often carried on transposable elements. One of the most prevalent AMEs genes among enterococci is aac(6')-Ie-aph(2'')-Ia encoding a bifunctional enzyme Aac(6')-Ie-Aph(2'')-Ia which confers resistance to virtually all of the clinically available aminoglycosides including gentamicin [11, 19].
In the current study, as in many other reports, the aac(6')- Ie-aph(2'')-Ia gene was the most prevalent AME gene, encountered in 96.2% of isolates followed by the ant (4') gene and was detected in 62.9% of isolates [16, 20].
Acquired resistance to tetracyclines and macrolides in enterococci is often by mobile genetic elements [20]. In the present study, as in many previous reports, the most common tetracycline and erythromycin resistance mechanism was mediated by the tetM and ermB genes, respectively [21-25]. To take preventive measures and apply infection control, identification of the source of infection and nature of outbreaks are required. RAPD-PCR has become a reliable tool for the differentiation and identification of enterococci from clinical origin [26-28]. In this study, RAPD-PCR profiles showed that eleven E. faecalis isolates shared identical banding patterns. In addition, the results of RAPD typing corresponded with antibiogram findings and the resistance genes patterns for eight of these eleven isolates. This could be explained by the predominance or an outbreak of a particular clonal group of E. faecalis with high-level gentamicin resistance in the assessed burn unit.
In conclusion, we found that over the examination time period one RAPD genotype was the common type and was disseminated among patients in the burn unit. Interestingly, most of these strains had an identical or very similar antibiotic and gene resistance pattern.
Acknowledgments
This research has been supported by Iran University of Medical Sciences & Health Services grant number: 17989/91-03-13 and Tehran University of Medical Sciences & Health Services grant 95-02-30-32393. Authors deny any existing or potential conflicts of interest of a financial, personal or any other nature that could affect or bias their research.
References
- 1.Cetinkaya Y, Falk P, Mayhall CG. Vancomycin-resistant enterococci. Clin Microbiol Rev. 2000;13:686–707. doi: 10.1128/cmr.13.4.686-707.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klare I, Konstabel C, Mueller-Bertling S, Werner G, Strommenger B, Kettlitz C, Borgmann S, Schulte B, Jonas D, Serr A, et al. Spread of ampicillin/vancomycin- resistant Enterococcus faecium of the epidemic-virulent clonal complex-17 carrying the genes esp and hyl in German hospitals. Eur J Clin Microbiol Infect Dis. 2005;24:815–825. doi: 10.1007/s10096-005-0056-0. [DOI] [PubMed] [Google Scholar]
- 3.Fatholahzadeh B, Hashemi FB, Emaneini M, Aligholi M, Nakhjavani FA, Kazemi B. Detection of Vancomycin Resistant Enterococci (VRE) isolated from urinary tract infections (UTI) in Tehran, Iran. Daru. 2006;14:141–145. [Google Scholar]
- 4.Titze-de-Almeida R, Belkum A, Felipe MS, Zanella RC, Top J, Willems RJ. Multilocus sequence typing of hospital-associated Enterococcus faecium from Brazil reveals their unique evolutionary history. Microb Drug Resist. 2006;12:121–121. doi: 10.1089/mdr.2006.12.121. [DOI] [PubMed] [Google Scholar]
- 5.Wibbenmeyer L, Williams I, Ward M, Xiao X, Light T, Latenser B, Lewis R, Kealey GP, Herwaldt L. Risk factors for acquiring vancomycin-resistant Enterococcus and methicillin-resistant Staphylococcus aureus on a burn surgery step-down unit. J Burn Care Res. 2010;31:269–279. doi: 10.1097/BCR.0b013e3181d0f479. [DOI] [PubMed] [Google Scholar]
- 6.Altoparlak U, Koca O, Ozkurt Z, Akcay MN. Incidence and risk factors of vancomycin-resistant enterococcus colonization in burn unit patients. Burns. 2011;37:49–53. doi: 10.1016/j.burns.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 7.Branski LK, Al-Mousawi A, Rivero H, Jeschke MG, Sanford AP, Herndon DN. Emerging infections in burns. Surg Infect (Larchmt) 2009;10:389–397. doi: 10.1089/sur.2009.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaye KS, Kaye D. Multidrug-resistant pathogens: mechanisms of resistance and epidemiology. Curr Infect Dis Rep. 2000;2:391–398. doi: 10.1007/s11908-000-0065-1. [DOI] [PubMed] [Google Scholar]
- 9.Nishimoto Y, Kobayashi N, Alam MM, Ishino M, Uehara N, Watanabe N. Analysis of the prevalence of tetracycline resistance genes in clinical isolates of Enterococcus faecalis and Enterococcus faecium in a Japanese hospital. Microb Drug Resist. 2005;11:146–153. doi: 10.1089/mdr.2005.11.146. [DOI] [PubMed] [Google Scholar]
- 10.Hryniewicz W, Zareba T, Kawalec M. Susceptibility patterns of Enterococcus spp. isolated in Poland during 1996. Int J Antimicrob Agents. 1998;10:303–307. doi: 10.1016/s0924-8579(98)00058-2. [DOI] [PubMed] [Google Scholar]
- 11.Emaneini M, Aligholi M, Aminshahi M. Characterization of glycopeptides, aminoglycosides and macrolide resistance among Enterococcus faecalis and Enterococcus faecium isolates from hospitals in Tehran. Pol J Microbiol. 2008;57:173–178. [PubMed] [Google Scholar]
- 12.Emaneini M, Hashemi FB, Aligholi M, Fatholahzadeh B, Kazemi B, Sadeghi F. Detection of vanB genotype enterococci in Iran. Int J Antimicrob Agents. 2005;26:98–99. doi: 10.1016/j.ijantimicag.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 13.Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33:1434–1434. doi: 10.1128/jcm.33.5.1434-1434.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; twenty-third informational supplement. M100-S23, 2013.
- 15.Emaneini M, Bigverdi R, Kalantar D, Soroush S, Jabalameli F, Noorazar Khoshgnab B, Asadollahi P, Taherikalani M. Distribution of genes encoding tetracycline resistance and aminoglycoside modifying enzymes in Staphylococcus aureus strains isolated from a burn center. Ann Burns Fire Disasters. 2013;30:76–80. 26. [PMC free article] [PubMed] [Google Scholar]
- 16.Udo EE, Al-Sweih N, Phillips OA, Chugh TD. Species prevalence and antibacterial resistance of enterococci isolated in Kuwait hospitals. J Med Microbiol. 2003;52:163–168. doi: 10.1099/jmm.0.04949-0. [DOI] [PubMed] [Google Scholar]
- 17.Titze-de-Almeida R, Rollo Filho M, Nogueira CA, Rodrigues IP, Eudes Filho J, Nascimento RS, Ferreira RF, 2nd, Moraes LM, Boelens H, Belkum A, et al. Molecular epidemiology and antimicrobial susceptibility of Enterococci recovered from Brazilian intensive care units. Braz J Infect Dis. 2004;8:197–205. doi: 10.1590/s1413-86702004000300002. [DOI] [PubMed] [Google Scholar]
- 18.Kaçmaz B, Aksoy A. Antimicrobial resistance of enterococci in Turkey. Int J Antimicrob Agents. 2005;25:535–538. doi: 10.1016/j.ijantimicag.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 19.Chow JW. Aminoglycoside resistance in enterococci. Clin Infect Dis. 2000;31:586–589. doi: 10.1086/313949. [DOI] [PubMed] [Google Scholar]
- 20.Hegstad K, Mikalsen T, Coque TM, Werner G, Sundsfjord A. Mobile genetic elements and their contribution to the emergence of antimicrobial resistant Enterococcus faecalis and Enterococcus faecium. Clin Microbiol Infect. 2010;16:541–554. doi: 10.1111/j.1469-0691.2010.03226.x. [DOI] [PubMed] [Google Scholar]
- 21.Vignaroli C, Zandri G, Aquilanti L, Pasquaroli S, Biavasco F. Multidrug-resistant enterococci in animal meat and faeces and co-transfer of resistance from an Enterococcus durans to a human Enterococcus faecium. Curr Microbiol. 2011;62:1438–1447. doi: 10.1007/s00284-011-9880-x. [DOI] [PubMed] [Google Scholar]
- 22.Aarestrup FM, Agerso Y, Gerner-Smidt P, Madsen M, Jensen LB. Comparison of antimicrobial resistance phenotypes and resistance genes in Enterococcus faecalis and Enterococcus faecium from humans in the community, broilers, and pigs in Denmark. Diagn Microbiol Infect Dis. 2000;37:127–137. doi: 10.1016/s0732-8893(00)00130-9. [DOI] [PubMed] [Google Scholar]
- 23.De Leener E, Martel A, Decostere A, Haesebrouck F. Distribution of the erm (B) gene, tetracycline resistance genes, and Tn1545- like transposons in macrolide- and lincosamide-resistant enterococci from pigs and humans. Microb Drug Resist. 2004;10:341–345. doi: 10.1089/mdr.2004.10.341. [DOI] [PubMed] [Google Scholar]
- 24.Nishimoto Y, Kobayashi N, Alam MM, Ishino M, Uehara N, Watanabe N. Analysis of the prevalence of tetracycline resistance genes in clinical isolates of Enterococcus faecalis and Enterococcus faecium in a Japanese hospital. Microb Drug Resist. 2005;11:146–153. doi: 10.1089/mdr.2005.11.146. [DOI] [PubMed] [Google Scholar]
- 25.Reyes J, Hidalgo M, Díaz L, Rincón S, Moreno J, Vanegas N, Castañeda E, Arias CA. Characterization of macrolide resistance in Gram-positive cocci from Colombian hospitals: a countrywide surveillance. Int J Infect Dis. 2007;11:329–336. doi: 10.1016/j.ijid.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 26.Braak N, Power E, Anthony R, Endtz HP, Verbrugh HA, Belkum A. Random amplification of polymorphic DNA versus pulsed field gel electrophoresis of SmaI DNA macrorestriction fragments for typing strains of vancomycin-resistant enterococci. FEMS Microbiol Lett. 2000;192:45–52. doi: 10.1016/s0378-1097(00)00407-9. [DOI] [PubMed] [Google Scholar]
- 27.Monstein HJ, Quednau M, Samuelsson A, Ahrné S, Isaksson B, Jonasson J. Division of the genus Enterococcus into species groups using PCR-based molecular typing methods. Microbiology. 1998;144:1171–1179. doi: 10.1099/00221287-144-5-1171. [DOI] [PubMed] [Google Scholar]
- 28.Gzyl A, Augustynowicz E. Technical aspects of random amplified polymorphic DNA (RAPD) technique in genotyping of bacterial strains. Acta Microbiol Pol. 1999;48:243–259. [PubMed] [Google Scholar]