Abstract
Among the methods used to unravel protein interaction surfaces, chemical cross-linking followed by identification of the cross-linked peptides by mass spectrometry has proven especially useful in dynamic and complex systems. During the signal recognition particle (SRP)-dependent targeting of proteins to the bacterial plasma membrane, the specific interaction between Ffh (the protein component of SRP) and FtsY (the SRP receptor) is known to be essential for the efficiency and fidelity of this process. In this work, we studied the Escherichia coli and Thermus aquaticus Ffh·FtsY complexes by using chemical cross-linking and tandem mass spectrometry to identify nine intermolecular cross-linked peptides. This information was used in conjunction with a previously undescribed model-building approach that combines geometric restraint optimization with macromolecular docking. The resulting model of the Ffh·FtsY complex is in good agreement with the crystal structure solved shortly thereafter. Intriguingly, four of the cross-linked pairs involve the M domain of Ffh, which is absent from the crystal structure, providing previously undocumented experimental evidence that the M domain is positioned in close proximity to the Ffh·FtsY interface in the complex.
Keywords: computational modeling, protein–protein interactions, protein targeting, GTPases
Cotranslational protein targeting is highly conserved in all three kingdoms of life with varying degrees of complexity (1–3). As the signal sequence of a nascent membrane or pre-secretory protein emerges from the ribosome, it is recognized by a cytosolic ribonucleoprotein complex, the signal recognition particle (SRP), to form a cytosolic targeting complex. The targeting complex then is directed to the translocation apparatus, embedded either in the endoplasmic reticulum membrane in eukaryotes or the plasma membrane in prokaryotes, through the recognition between the SRP and the SRP receptor (SR) (1, 4). Bacteria have the simplest version of SRP and SR machinery. Ffh, a bacterial homologue of mammalian SRP54, together with 4.5S RNA forms the minimal SRP, and its specific interaction with FtsY, the bacterial SR (5), ensures the efficiency and fidelity of protein translocation. Both Ffh and FtsY are GTPases, and their GTPase cycles are coupled tightly to their functional cycles to regulate the timing and effect the unidirectionality of the targeting process. Ffh and FtsY associate in a GTP-dependent manner to deliver the nascent protein to the membrane translocon; subsequent GTP hydrolysis leads to the dissociation of Ffh from FtsY, allowing the recycling of the SRP and SR (6).
Ffh and FtsY are highly homologous proteins and constitute a unique SRP GTPase subfamily (7). Structural studies on Ffh and FtsY have revealed several unusual features (8, 9). The GTPase catalytic core comprises a G domain that shares homology with the Ras GTPase fold (10). An N-terminal four-helix bundle (N domain), unique to the SRP GTPases, forms a structural and functional unit with the G domain, called collectively the “NG domain” (8, 9). The N domain contributes to the regulation of GTPase activity (11–13) and the specific interaction between Ffh and FtsY (14–16) through its highly conserved ALLEADV motif. Another unique feature of the SRP GTPases is the insertion box domain, which forms part of the nucleotidebinding pocket. The insertion box domain loop has been implicated as essential in both nucleotide exchange and the interactions between Ffh and FtsY (8, 17).
Besides the NG domain, Ffh has a C-terminal methionine-rich domain, called the M domain (7). The M domain provides the binding module for 4.5S RNA (18, 19), which facilitates Ffh·FtsY complex formation (20, 21). It is widely accepted that the M domain also contains the primary binding site for signal sequences (7, 22), although recent data implies that the NG domain may contribute to signal sequence binding as well (23). The positioning of the M domain relative to the NG domain has long been ambiguous (22) because of the highly flexible peptide linkage between the M and NG domains. Recent crystal structures of archaeal Sulfolobus solfataricus full-length SRP54 both alone and with RNA suggest that the M domain is juxtaposed to the N domain, forming direct hydrophobic contacts (24).
The regulatory mechanism of the SRP GTPase family appears to be unique as well. Most GTPases function through the “GTPase switch” mechanism, cycling between kinetically stable GTP-bound and GDP-bound states. Switch between these states is regulated by two sets of proteins, guanine nucleotide exchange factors, which facilitate the release of GDP in exchange for GTP, and GTPase-activating proteins, which facilitate the hydrolysis of GTP to GDP. Unlike other subfamilies of GTPases, Ffh and FtsY possess only weak nucleotide affinities (25); thus, in this case, there is no obvious need for external guanine nucleotide exchange factors. This notion is consistent with the crystal structures of the apoproteins Ffh and FtsY, which show empty nucleotide-binding sites stabilized by extensive hydrogen bonds (8, 9). In contrast, upon Ffh·FtsY complex formation, GTP is bound much more stably, and both proteins act as reciprocal GTPase-activating proteins for each other (26). The recently solved crystal structures of the Ffh·FtsY NG complex (15, 16) reveal the remarkable twinning of substrates and the recruitment of several essential residues into the catalytic chamber through conformational changes, shedding light on the basis for the nucleotide affinity increase and reciprocal activation during the complex formation. However, it remains to be understood how the SRP GTPase cycles are coupled to the protein targeting reaction.
Chemical cross-linking followed by proteolytic digestion and identification of the cross-linked species by tandem mass spectrometry (MS) constitutes a valuable complementary strategy to probe interactions between macromolecules (27, 28). Determination of the cross-linked amino acid residues provides valuable spatial constraints that can be used to elucidate structural information on folding of the proteins (29, 30), the connectivity and positioning of secondary structures (31, 32), and protein interaction surfaces (33–37). Chemical cross-linking has proven especially useful in providing structural information on dynamic and complex systems. Such insight becomes critical in cases when crystallization of the protein complex is difficult to achieve. In the present work, we studied the Ffh·FtsY complexes from two species, Escherichia coli and Thermus aquaticus. Because of the reasonable distribution of lysines on these protein surfaces, we used two homofunctional, amine-reactive N-hydroxysuccinimide ester cross-linking reagents, which react with protein N-termini and lysine ε-amino groups. During these studies, nine intermolecular cross-linked peptides were detected, and the cross-linked residues were identified unambiguously with high-performance tandem MS. Spatial restraints, derived from the maximal distance that the two reactive groups of the cross-linkers could span, were imposed simultaneously on the cross-linked residues to generate low-resolution models of the complex. Subsequently, macromolecular docking and energy minimization were used to build a refined model of the Ffh·FtsY NG domain complex.
Materials and Methods
Materials. Disuccinimidyl suberate (DSS) was purchased from Pierce. Bis(sulfosuccinimidyl) adipate (BSSA) was a generous gift from R. Kiplin Guy (University of California, San Francisco). Modified trypsin was obtained from Promega, and 5′-guanylylimidodiphosphate was obtained from Sigma. The E. coli Ffh and FtsY were overexpressed from the pDMF6 plasmid in BL21(DE3)-pLysE cells (Stratagene), as described in ref. 21. FtsY (47–497), a truncated form of E. coli FtsY, was used for this study. E. coli Ffh·FtsY complex was formed at 25°C for 1 h in assay buffer [50 mM Hepes/150 mM KOAc/2 mM Mg(OAc)2/0.01% Nikkol (Anatrace, Maumee, OH), pH 7.5] in the presence of 100 μM purified 5′-guanylylimidodiphosphate. T. aquaticus Ffh and FtsY were subcloned in the pET28b vector as N-terminal hexahistidine fusion proteins and expressed by using the E. coli BL21(DE3)-Rosetta strain (Novagen) (15). The histidine tags were removed by thrombin cleavage, and the Ffh·FtsY complex was formed by incubation and purified by gel filtration in 50 mM KCl, 20 mM Hepes, 5 mM GDP, 0.5 mM BeSO4, and 5 mM NaF.
Cross-Linking Reactions and In-Solution Tryptic Digestion. Stock solutions of the cross-linking reagents were prepared freshly at a concentration of 25 mM in DMSO for DSS and in H2O for BSSA. The cross-linking reactions were performed in a final volume of 70 μl, with the final concentration of 5 μM Ffh·FtsY complex and 0.5 mM cross-linking reagents. DSS was used for the E. coli Ffh·FtsY complex, and BSSA was used for the T. aquaticus Ffh·FtsY complex. The reactions were allowed to proceed for 1 h at room temperature and were quenched with ammonium hydroxide. For the controls, Ffh and FtsY were treated separately with DSS or BSSA at the same protein and cross-linking reagent concentrations. Ffh and FtsY were not combined until the reactions were quenched with ammonium hydroxide. The proteins were digested by 2% trypsin (wt/wt) at 37°C for 4 h.
On-Line Capillary Liquid Chromatography (LC)-MS and LC-Tandem MS (LC-MS/MS) Analysis of Cross-Linked Peptides. A1-μl aliquot of the digestion mixture was injected into an UltiMate capillary LC system (LC Packings, Sunnyvale, CA) by means of a FAMOS autosampler (LC Packings) and separated by a 75-μm × 15-cm reverse-phase capillary column at a flow rate of ≈300 nl/min. The HPLC eluent was connected directly to the microion electrospray source of a QSTAR Pulsar QqTOF mass spectrometer (Applied Biosystems). Typical performance characteristics were >8,000 resolution with 30 ppm mass measurement accuracy in both MS and collision-induced dissociation mode. LC-MS data were acquired by using the analyst qs software (Applied Biosystems). The centroided LC-MS data then were deisotoped and reconstructed to generate a list of peptides detected during each LC-MS run. The peptide lists from the cross-linked Ffh·FtsY complex and the control were compared thoroughly to differentiate two related LC-MS runs and indicate the peptides unique to the digest of the cross-linked complex. For those unique peptides in the digest of the cross-linked complexes, all possible cross-linking combinations were predicted by ms-bridge, a program in the University of California, San Francisco proteinprospector package (http://prospector.ucsf.edu).
Computational Modeling of Low-Resolution NG Domain Complex by Using modeller and Cross-Linking Results. NG domains of T. aquaticus Ffh and FtsY were treated as “pseudo” rigid bodies with the intraatomic distances and dihedral angels restrained to their crystallographic values (8). An upper bound distance restraint of 30 Å was imposed on the α-carbons of the cross-linked pairs. Nonbonded interactions were modeled with a Lennard–Jones potential by using the charmm (Accelrys, San Diego) force-field parameters and a distance cut-off of 9 Å. We optimized 1,000 Ffh·FtsY NG-domain complex models with random positions and orientations to obtain an ensemble of models that maximally satisfy all of the input restraints. The procedure for optimizing a single configuration consisted of 1,000 steps of molecular dynamics with simulated annealing followed by a final conjugate gradient minimization.
Refinement of Ffh·FtsY NG Domain Complex Model by Using dock and multidock. The apo NG domains of Ffh and FtsY were docked against each other as rigid bodies by using the program dock (version 4.01) (38). The program dms (www.cgl.ucsf.edu/Overview/software.html) was used to generate molecular surfaces over the Ffh and FtsY structures. The dock accessory program sphgen was used with default parameters to build a set of receptor spheres over FtsY and a set of ligand spheres over Ffh. Orientations were scored with the dock contact score, by using default values for the cutoff distance, the clash overlap, and the clash penalty parameters of 4.5, 0.75, and 50, respectively. We applied 100 steps of simplex minimization to each orientation by using default values for the initial translation, the initial rotation, the contact convergence, and the maximum cycles parameters of 1, 0.1, 0.1, and 1, respectively. The top-scoring 250 orientations were selected. The Ffh·FtsY docked models were scored, optimized, and ranked with the multidock program (39). multidock was run with default parameters, with FtsY selected as the imobile_mol, and Ffh selected as the mobile_mol. The Total Energy score of each complex was used to rank order the complexes.
Results and Discussion
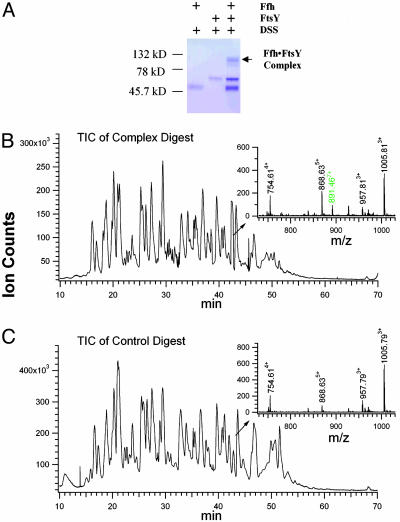
Cross-Linking and LC-MS, LC-MS/MS Analysis of E. coli Ffh·FtsY Complex. By using homobifunctional cross-linker DSS, we investigated various cross-linking conditions and optimized the cross-linking yield of the Ffh·FtsY complex while minimizing any nonspecific cross-links between homodimeric Ffh or FtsY (Fig. 1A). As expected, the appearance of the cross-linked gel band was strictly dependent on the presence of both nucleotide triphosphate and Mg2+ (data not shown), which are required for complex formation (25). In addition, the extent of FtsY tryptophan fluorescence change in the DSS-treated E. coli Ffh·FtsY complex was indistinguishable from that of the native complex. This fluorescence change monitors a relatively late conformational change during the formation of active Ffh·FtsY complex. These results therefore indicate that the integrity of the complex was sustained during the cross-linking reaction.
Fig. 1.
Cross-linking of E. coli Ffh·FtsY complex. (A) SDS/PAGE analysis of the cross-linking reaction. (B and C) Total ion chromatograms (TIC) of the tryptic digestion mixture of the cross-linked Ffh·FtsY complex (B) and the control (C). (Insets) The mass spectra of the peptides eluted at ≈42.5 min.
The total ion chromatograms of the tryptic digestion peptide mixtures from the cross-linked complex (Fig. 1B) and the control (Fig. 1C) resembled each other, yet closer inspection revealed significant differences. For example, among the peptides that elute at ≈42.5 min from the reverse-phase capillary column, a peptide at m/z 891.467+ was present only in the digest of the cross-linked Ffh·FtsY complex (Fig. 1 B Inset and C Inset). Its monoisotopic mass value and charge state were assigned without ambiguity (Fig. 2 Inset). Depending on the accuracy of peptide mass measurement, there are many possible cross-linking combinations that could give rise to this species (e.g., assuming a mass measurement window of ± 20 ppm yielded >900 possibilities). In retrospect, even exact mass measurement would not lead to any conclusive assignment, allowing four isobaric cross-linked species with different peptide combinations as possible solutions. Therefore, sequence information was necessary to identify unambiguously the actual cross-linked peptide moieties. Interpretation of the low-energy collision-induced dissociation spectrum (Fig. 2) obtained for this component revealed that it was composed of three cross-linked peptide moieties, FtsY Lys-406–Lys-447, Ffh Met-1–Arg-8, and FtsY Lys-399–Arg-402. The mass values of the N-terminal sequence ion series of the FtsY Lys-406–Lys-447 moiety, annotated as B2–B11, contained the mass values anticipated for FtsY Lys-399–Arg-402 moiety and the cross-linking bridge, thus establishing an intramolecular cross-link between Lys-399FtsY and Lys-406FtsY. Similarly, the mass values of some C-terminal sequence ions of the FtsY Lys-406–Lys-447 moiety, annotated as Y19–Y24, contained the mass value of Ffh Met-1–Arg-8 and the cross-linking bridge. In contrast, the C-terminal sequence ion series of the FtsY Lys-406–Lys-447 moiety up to Leu-431FtsY, i.e., y1–y15, appears unmodified. These results establish that the α-amino group of Ffh N terminus is cross-linked to the ε-amino of FtsY Lys-432. This particular cross-linked species, therefore, contains both an intramolecular and an intermolecular cross-link.
Fig. 2.
Structure and low-energy collision-induced dissociation spectrum of the cross-linked species with a m/z value of 891.467+ from E. coli complex. (Inset) The MS spectrum of this species. (Lower) Structure indicates the residue numbers of the cross-linked sites. C-terminal sequence ions that did not contain the cross-linker moiety are labeled in lowercase, e.g., yi# (red), y′i (brown), and yi (blue) ions. N-terminal sequence ions of FtsY K406–K447 that contained cross-linked FtsY K399–R402 peptide moiety are annotated as Bi (green). C-terminal sequence ions of FtsY K406–K447 that contained the cross-linked Ffh M1–R8 peptide moiety are annotated as Yi (magenta).
The identity and structure of another intermolecular cross-linked species observed with a m/z of 817.753+ was determined in the same way (Table 1). This species corresponds to a cross-link between the N-terminal Met of E. coli Ffh and Lys-247 of E. coli FtsY.
Table 1. Intermolecular cross-linked species identified by tandem MS.
| Exp. m/z | Δ, ppm | Cross-linked residues | Peptide sequences |
|---|---|---|---|
| 891.467+ | -10 | Ffh M1-FtsY K432 | Ffh M1-R8 |
| (Tfh M1-TtsY K241)* | FtsY K406-K447 | ||
| FtsY K399-R402 | |||
| 817.753+ | 0 | Ffh M1-FtsY K247 | Ffh M1-R8 |
| (Tfh M1-TtsY E51)* | FtsY K247-R258 | ||
| 529.314+ | 19 | Tfh K28-TtsY K13 | Tfh I22-R32 |
| TtsY A9-R15 | |||
| 613.554+ | 0 | Tfh K28-TtsY G(-3)† | Tfh I22-R32 |
| TtsY G(-3)-R6 | |||
| 685.614+ | 18 | Tfh K236-TtsY G(-3)† | Tfh A232-K246 |
| TtsY G(-3)-R6 | |||
| 577.034+ | 17 | Tfh K404-TtsY G(-3)† | Tfh F402-K410 |
| TtsY G(-3)-R6 | |||
| 652.834+ | 20 | Tfh K390-TtsY G(-3)† | Tfh I388-R401 |
| TtsY G(-3)-R6 | |||
| 489.243+ | 12 | Tfh K386-TtsY G(-3)† | Tfh K386-R387 |
| TtsY G(-3)-R6 | |||
| 515.304+ | 25 | Tfh K390-TtsY K62 | Tfh I388-R401 |
| TtsY K62-K65 |
The cross-linked species, namely 891.467+ and 817.753+, were obtained from E. coli Ffh·FtsY complex, and other cross-linked species were observed in T. aquaticus Ffh·FtsY complex. T. aquaticus Ffh and FtsY were annotated as Tfh and TtsY, respectively, to distinguish from E. coli Ffh and FtsY. A high proportion of the cross-links involve the N termini of the proteins. This characteristic is presumably due to the lower pKa value of the α-amino of proteins (pKa ≈ 8) than that of the ε-amino of lysines (pKa ≈ 10). In addition, the higher mobility of the protein terminal tails may contribute to their higher cross-linking frequency. Exp. m/z is experimentally measured mass to charge ratio.
The intermolecular cross-linked residues in E. coli complex were mapped back onto the corresponding T. aquaticus proteins as indicated in parentheses.
Gly(-3)FtsY corresponds to the N terminus of the recombinant FtsY used in this study, which contained a three-amino acid extension derived from the expression tag (15).
Cross-Linking and LC-MS, LC-MS/MS Analysis of T. aquaticus Ffh·FtsY Complex. To further constrain the Ffh·FtsY interface, we treated the Ffh·FtsY complex purified from T. aquaticus with a different cross-linker BSSA. This complex from a different species permitted us to explore a set of differently positioned Lys residues as cross-linking targets. Seven pairs of intermolecular cross-links from T. aquaticus Ffh·FtsY complex were found, and their identity and structure were established by tandem MS (Table 1).
Generating Ffh·FtsY NG Domain Complex Models Based on the Cross-Linking Results. Distance constraints generated from chemical cross-linking studies have been exploited as filters to select the best-fit models (29). Nevertheless, conformational rearrangements upon complex formation and the motion of terminal tails and hinge regions between domains can significantly alter the distance of some residues on the apo structures. To accommodate these kinds of dynamic properties in computational modeling of protein complexes, we developed a multistep strategy of using cross-linking distance information to determine the binding interface and relative orientation of the complex, followed by rigorous docking and energy minimization in the restricted search space corresponding to the putative low-resolution interface.
To this end, we first calculated an ensemble of FtsY·Ffh NG domain complex structures that maximally satisfied all distance restraints derived from the cross-linking experiments. We optimized 1,000 FtsY·Ffh NG domain complex models with randomly generated orientations to simultaneously minimize violations of the spatial restraints. Remarkably, the selected top 10% ranking models were highly clustered, sharing a tight binding interface and a single interacting orientation. A discrete patch of surface on each protein was defined readily as the putative interaction surface by a cluster of residues that were in direct contact with its partner protein in >75% of the selected models.
Within the boundaries of this putative binding interface and orientation, we used macromolecular docking to further refine our computational models. The docking program used here sampled the protein–protein interface in a much more fine-grained manner than the restraints optimization program used in the first step. The apo NG domains of Ffh and FtsY were treated as rigid bodies, and 1,000 models with varying orientations of Ffh to FtsY were generated. The models were optimized and ranked by their steric complementarity, and the best 250 were selected for further refinement. Any minimized orientations that had moved significantly from the predicted binding interface were discarded to restrict the sampling to the interface defined by the cross-linking geometry. The remaining docked models then were energy-minimized over both the rigid body and side-chain rotamer degrees of freedom to optimize the binding interface of the two proteins. The minimized energies were used to score and sort the computational models.
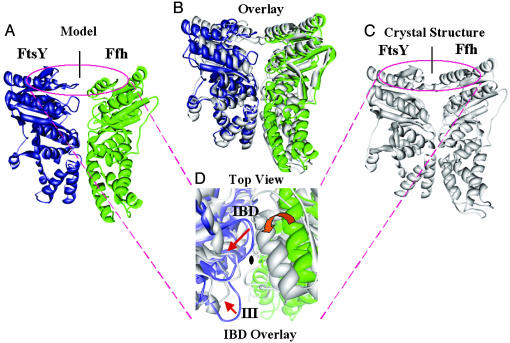
The best-scoring model was superimposed onto the crystal structure of the Ffh·FtsY NG domain complex (Fig. 3). The heavy-atom and backbone rms deviations between the model and the crystal structure were 4.19 and 3.80 Å, respectively. Among the 11 residues involved in the cross-linking reactions, 4 residues with cross-links in the NG domain were present on the crystal structure of the T. aquaticus Ffh·FtsY NG domain complex, namely Lys-241FtsY, Glu-51FtsY, Lys-28Ffh, and Lys-236Ffh. Their solvent accessibility and close proximity to the Ffh·FtsY complex interface further indicates that our cross-linking results are a good reflection of Ffh·FtsY complex conformation in solution. In addition, among the intramolecular cross-links we identified by tandem MS, four cross-linked pairs have distance information available from the crystal structure of the NG complex (Table 2). The length of DSS linker bridge is 11.4 Å, although the distance of two cross-linked α-carbons can span up to 24 Å if the mobility of Lys side-chains is considered (29). The satisfaction of the distance restraint of these cross-linked pairs indicates that the use of the cross-linking reagents did not induce global conformational change in the proteins.
Fig. 3.
Modeling of the Ffh·FtsY NG domain complex. (A–C) Side view of the computational model (A), crystal structure (C), and their overlay (B). (D) Top view of the overlaid structures. Red arrows indicate the rearrangement of motif II (IBD, insertion box domain loop) and motif III on FtsY, which account for the roughly 12° rotation of Ffh between the model and the crystal structure (red block arrow).
Table 2. Intramolecular cross-linked species identified by tandem MS.
| Exp. m/z | Δ, ppm | Peptide sequence | Cross-linked residues | Distances, Å |
|---|---|---|---|---|
| 816.143+ | 30 | Ffh 59-79 | Ffh K66-K76 (Tfh E66-A76)* | 15.7 |
| 583.335+ | 28 | Ffh 120-125, 272-289 | Ffh K122-K278 (Tfh L120-K275)* | 9.2 |
| 681.924+ | 23 | TftsY 87-101, 272-278 | TftsY K87-K274 | 19.7 |
| 581.594+ | 24 | TftsY 70-81, 272-278 | TftsY K71-K274 | 15.3 |
T. aquaticus Ffh and FtsY are annotated as Tfh and TftsY, respectively to distinguish from E. coli Ffh and FtsY. Exp. m/z is experimentally measured mass to charge ratio.
The cross-linked residues in E. coli complex were mapped back onto the corresponding T. aquaticus proteins as indicated in parentheses.
The largest difference between the model and the crystal structure is situated at the interface region, where induced-fit conformational changes in the small loop regions allow Ffh and FtsY to pack more closely together in the crystal structure. As shown in Fig. 3D, the highly conserved motif II (insertion box domain loop) and motif III of FtsY move away from the complex interface to seal the upper face and lateral entrance of the catalytic chamber and, at the same time, to assume the correct position for catalysis of GTP hydrolysis (15, 16). Thus, it is most likely that discrepancies between the model and the crystal structure arose because such secondary structure displacements were not allowed during the modeling, treating Ffh and FtsY as rigid bodies.
Implications from Ffh·FtsY Complex Model and Ffh M Domain Position. We have established that four of the cross-linked species detected in the T. aquaticus Ffh·FtsY complex involve three residues, Lys-386Ffh, Lys-390Ffh, and Lys-404Ffh, on the Ffh M domain and two residues, Gly(-3)FtsY and Lys-62FtsY, on the FtsY N domain. These cross-links therefore suggest the proximity of the M domain toward the FtsY N domain (Fig. 4B). This finding is in contrast to the domain arrangements in the recently solved crystal structure of SRP54 from archaea S. solfataricus (24) (Fig. 4 A and B, M domain shown in gray) and that proposed from a low-resolution, single-particle cryoelectron microscopic structure of eukaryotic SRP bound to the ribosome (data not shown) (40). Both studies position the M domain at the side, facing away from the FtsY N terminus (Fig. 4A). In this arrangement, Lys-386, Lys-390, and Lys-404 of T. aquaticus Ffh, which correspond to Arg-401, Glu-405, and Glu-419 of S. solfataricus SRP54, respectively, are positioned too far from the N domain of FtsY to be plausible cross-linking targets. Although Gly(-3)FtsY, to which Lys-386Ffh, Lys-390Ffh, and Lys-404Ffh were cross-linked, is absent in the complex crystal structure, its location is well confined around the Ffh·FtsY interface by the position of the other three residues to which Gly(-3)FtsY cross-linked in good stoichiometry, namely Lys-28Ffh, Lys-236Ffh, and Lys-62FtsY(Fig. 4B). Therefore, our cross-linking studies suggest that the M domain assumes a different position in the Ffh·FtsY complex from those suggested in the two previous studies. However, the Ffh domain organization proposed here is consistent with the model of Ffh NG and M domain arrangement, cited to be based on fluorescence data (41).
Fig. 4.
A different M domain position in the Ffh·FtsY complex. (A) Mapping of T. aquaticus Lys-386Ffh, Lys-390Ffh, and Lys-404Ffh on the structure of S. solfataricus SRP54. S. solfataricus SRP54 (gray) was superimposed with Ffh (green) of T. aquaticus Ffh·FtsY NG domain complex (green, Ffh; blue, FtsY). Residues that cross-linked to Gly(-3)Ffh are shown as sticks in magenta. (B) A close proximity of the M domain toward Ffh·FtsY complex interface, suggested by the cross-linking data. Magenta asterisks, relevant cross-linked residues; lines, cross-links; gray cylinders, different positions of the M domain.
One possible explanation for this difference in the M domain positioning is that FtsY (SRP receptor) was not present in the crystal and electron microscopy studies. The M domain, being loosely tethered to the surface of Ffh, might assume different positions depending on the step of the targeting reaction. Upon interacting with FtsY, the M domain might undergo a dynamic rearrangement, either by means of rearrangements in the N domain that is transmitted to the M domain or additional interactions formed between the M domain and FstY. This rearrangement could be further modulated by other components of the targeting reaction, such as the SRP RNA and the signal sequence, such that loading and unloading of signal sequences to SRP may be appropriately controlled in space and time.
Acknowledgments
We thank David Maltby and Drs. Lan Huang and Katalin M. Medzihradszky for helpful discussions on MS experiments. This work was supported by National Institutes of Health Grants RR 01614 (to A.L.B.), GM 32384 (to P.W.), GM 60641 (to R.M.S.), RR 12961 (to A.L.B.), and RR 15804 (to A.L.B.). F.C. was a Eugene Cota-Robles graduate student fellow. S.o.S. started as a Damon Runyon–Walter Winchell Cancer Research fellow and now is a Burroughs Wellcome Fund fellow. P.W. is an Investigator of the Howard Hughes Medical Institute.
Author contributions: F.C., S.-o.S., D.T.M., F.A., P.F.E., R.M.S., P.W., and A.L.B. designed research; F.C., S.-o.S., D.T.M., F.A., and P.F.E. performed research; F.C., S.-o.S., D.T.M., F.A., and P.F.E. contributed new reagents/analytic tools; F.C., D.T.M., F.A., S.-o.S., and P.F.E. analyzed data; and F.C., S.-o.S., D.T.M., F.A., P.F.E., P.W., R.M.S., and A.L.B. wrote the paper.
Abbreviations: SRP, signal recognition particle; SR, SRP receptor; LC-MS, liquid chromatography MS; LC-MS/MS, LC-tandem MS; DSS, disuccinimidyl suberate; BSSA, bis(sulfosuccinimidyl) adipate.
Note. Shortly after the completion of this work, the crystal structures of the complex of the NG GTPase domains of Ffh and FtsY were reported (15, 16). These x-ray structures are in good agreement with the computational model of this complex presented here. In addition, four of the cross-linked pairs provide new insight regarding the position of Ffh M domain relative to the rest of the Ffh·FtsY complex that was not revealed by crystallographic studies.
References
- 1.Walter, P. & Johnson, A. E. (1994) Annu. Rev. Cell Biol. 10, 87–119. [DOI] [PubMed] [Google Scholar]
- 2.Koch, H. G., Moser, M. & Muller, M. (2003) Rev. Physiol. Biochem. Pharmacol. 146, 55–94. [DOI] [PubMed] [Google Scholar]
- 3.Keenan, R. J., Freymann, D. M., Stroud, R. M. & Walter, P. (2001) Annu. Rev. Biochem. 70, 755–775. [DOI] [PubMed] [Google Scholar]
- 4.Stroud, R. M. & Walter, P. (1999) Curr. Opin. Struct. Biol. 9, 754–759. [DOI] [PubMed] [Google Scholar]
- 5.Gilmore, R., Blobel, G. & Walter, P. (1982) J. Cell Biol. 95, 463–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller, J. D., Wilhelm, H., Gierasch, L., Gilmore, R. & Walter, P. (1993) Nature 366, 351–354. [DOI] [PubMed] [Google Scholar]
- 7.Bernstein, H. D., Poritz, M. A., Strub, K., Hoben, P. J., Brenner, S. & Walter, P. (1989) Nature 340, 482–486. [DOI] [PubMed] [Google Scholar]
- 8.Freymann, D. M., Keenan, R. J., Stroud, R. M. & Walter, P. (1997) Nature 385, 361–364. [DOI] [PubMed] [Google Scholar]
- 9.Montoya, G., Svensson, C., Luirink, J. & Sinning, I. (1997) Nature 385, 365–368. [DOI] [PubMed] [Google Scholar]
- 10.Pai, E. F., Kabsch, W., Krengel, U., Holmes, K. C., John, J. & Wittinghofer, A. (1989) Nature 341, 209–214. [DOI] [PubMed] [Google Scholar]
- 11.Padmanabhan, S. & Freymann, D. M. (2001) Structure (London) 9, 859–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shepotinovskaya, I. V. & Freymann, D. M. (2002) Biochim. Biophys. Acta 1597, 107–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freymann, D. M., Keenan, R. J., Stroud, R. M. & Walter, P. (1999) Nat. Struct. Biol. 6, 793–801. [DOI] [PubMed] [Google Scholar]
- 14.Lu, Y., Qi, H. Y., Hyndman, J. B., Ulbrandt, N. D., Teplyakov, A., Tomasevic, N. & Bernstein, H. D. (2001) EMBO J. 20, 6724–6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egea, P. F., Shan, S. O., Napetschnig, J., Savage, D. F., Walter, P. & Stroud, R. M. (2004) Nature 427, 215–221. [DOI] [PubMed] [Google Scholar]
- 16.Focia, P. J., Shepotinovskaya, I. V., Seidler, J. A. & Freymann, D. M. (2004) Science 303, 373–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montoya, G., Kaat, K., Moll, R., Schafer, G. & Sinning, I. (2000) Struct. Fold Des. 8, 515–525. [DOI] [PubMed] [Google Scholar]
- 18.Zopf, D., Bernstein, H. D., Johnson, A. E. & Walter, P. (1990) EMBO J. 9, 4511–4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Batey, R. T., Rambo, R. P., Lucast, L., Rha, B. & Doudna, J. A. (2000) Science 287, 1232–1239. [DOI] [PubMed] [Google Scholar]
- 20.Peluso, P., Herschlag, D., Nock, S., Freymann, D. M., Johnson, A. E. & Walter, P. (2000) Science 288, 1640–1643. [DOI] [PubMed] [Google Scholar]
- 21.Peluso, P., Shan, S. O., Nock, S., Herschlag, D. & Walter, P. (2001) Biochemistry 40, 15224–15233. [DOI] [PubMed] [Google Scholar]
- 22.Keenan, R. J., Freymann, D. M., Walter, P. & Stroud, R. M. (1998) Cell 94, 181–191. [DOI] [PubMed] [Google Scholar]
- 23.Cleverley, R. M. & Gierasch, L. M. (2002) J. Biol. Chem. 277, 46763–46768. [DOI] [PubMed] [Google Scholar]
- 24.Rosendal, K. R., Wild, K., Montoya, G. & Sinning, I. (2003) Proc. Natl. Acad. Sci. USA 100, 14701–14706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shan, S. O. & Walter, P. (2003) Proc. Natl. Acad. Sci. USA 100, 4480–4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Powers, T. & Walter, P. (1995) Science 230, 32–36. [Google Scholar]
- 27.Back, J. W., de Jong, L., Muijsers, A. O. & de Koster, C. G. (2003) J. Mol. Biol. 331, 303–313. [DOI] [PubMed] [Google Scholar]
- 28.Sinz, A. (2003) J. Mass Spectrom. 38, 1225–1237. [DOI] [PubMed] [Google Scholar]
- 29.Young, M. M., Tang, N., Hempel, J. C., Oshiro, C. M., Taylor, E. W., Kuntz, I. D., Gibson, B. W. & Dollinger, G. (2000) Proc. Natl. Acad. Sci. USA 97, 5802–5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dihazi, G. H. & Sinz, A. (2003) Rapid Commun. Mass Spectrom. 17, 2005–2014. [DOI] [PubMed] [Google Scholar]
- 31.Doyle, D. A., Morais Cabral, J., Pfuetzner, R. A., Kuo, A., Gulbis, J. M., Cohen, S. L., Chait, B. T. & MacKinnon, R. (1998) Science 280, 69–77. [DOI] [PubMed] [Google Scholar]
- 32.Cohen, S. L. & Chait, B. T. (2001) Annu. Rev. Biophys. Biomol. Struct. 30, 67–85. [DOI] [PubMed] [Google Scholar]
- 33.Person, M. D., Brown, K. C., Mahrus, S., Craik, C. S. & Burlingame, A. L. (2001) Protein Sci. 10, 1549–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kopp, D. A., Berg, E. A., Costello, C. E. & Lippard, S. J. (2003) J. Biol. Chem. 278, 20939–20945. [DOI] [PubMed] [Google Scholar]
- 35.Lanman, J., Lam, T. T., Barnes, S., Sakalian, M., Emmett, M. R., Marshall, A. G. & Prevelige, P. E., Jr. (2003) J. Mol. Biol. 325, 759–772. [DOI] [PubMed] [Google Scholar]
- 36.D'Ambrosio, C., Talamo, F., Vitale, R. M., Amodeo, P., Tell, G., Ferrara, L. & Scaloni, A. (2003) Biochemistry 42, 4430–4443. [DOI] [PubMed] [Google Scholar]
- 37.Trester-Zedlitz, M., Kamada, K., Burley, S. K., Fenyo, D., Chait, B. T. & Muir, T. W. (2003) J. Am. Chem. Soc. 125, 2416–2425. [DOI] [PubMed] [Google Scholar]
- 38.Ewing, T. J., Makino, S., Skillman, A. G. & Kuntz, I. D. (2001) J. Comput. Aided Mol. Des. 15, 411–428. [DOI] [PubMed] [Google Scholar]
- 39.Jackson, R. M., Gabb, H. A. & Sternberg, M. J. (1998) J. Mol. Biol. 276, 265–285. [DOI] [PubMed] [Google Scholar]
- 40.Halic, M., Becker, T., Pool, M. R., Spahn, C. M., Grassucci, R. A., Frank, J. & Beckmann, R. (2004) Nature 427, 808–814. [DOI] [PubMed] [Google Scholar]
- 41.Buskiewicz, I., Deuerling, E., Gu, S. Q., Jockel, J., Rodnina, M. V., Bukau, B. & Wintermeyer, W. (2004) Proc. Natl. Acad. Sci. USA 101, 7902–7906. [DOI] [PMC free article] [PubMed] [Google Scholar]