Abstract
Chondrosarcoma (CS) is the second most common primary malignant bone tumor. Unlike other bone tumors, CS is highly resistant to conventional chemotherapy and radiotherapy, thus resulting in poor patient outcomes. There is an urgent need to establish alternative therapies for CS. However, the etiology and pathogenesis of CS still remain elusive. Recently, DNA methylation-associated epigenetic changes have been found to play a pivotal role in the initiation and development of human cancers, including CS, by regulating target gene expression in different cellular pathways. Elucidating the mechanisms of DNA methylation alteration may provide biomarkers for diagnosis and prognosis, as well as novel treatment options for CS. We have conducted a critical review to summarize the evidence regarding aberrant DNA methylation patterns as diagnostic biomarkers, predictors of progression and potential treatment strategies in CS.
Keywords: : 5-azacytidine, 5-aza-2′-deoxycytidine, chondrosarcoma, DNA methylation, epigenetics
Chondrosarcoma (CS), a heterogeneous subtype of malignant tumor derived from transformed cartilage cells, ranks as the second most common primary bone cancer next to osteosarcoma [1–4]. CS accounts for more than 20% of primary bone neoplasms [5]. Unlike other primary bone sarcomas, which mostly occur in young patients, CS can affect people of any age, most often affecting the hip and thigh bones. According to clinical data, low-grade CSs rarely metastasize and can be managed with surgery alone. However, high-grade CSs often metastasize and are lethal in most cases. CS is notoriously resistant to both chemotherapy and radiation treatment [1,5]. In this regard, 5-year survival of patients with CS ranges from 10 to 25%. There has been no progress in the treatment of this disease over the last several decades [6,7]. Therefore, there is an urgent need to identify novel strategies for improving the treatment of CS. Recently, epigenetic alterations, especially DNA methylation, have been found to play an important role in detection and possible treatment, which may affect the prognosis of CS.
Epigenetics commonly consist of three main mechanisms: DNA methylation, histone modifications and post-transcriptional gene regulation by non-coding RNA [8]. DNA methylation is an epigenetic modification that occurs by the addition of a methyl group (-CH3) to a CpG dinucleotide (cytosine–guanosine base pair) in the DNA sequence that regulates gene transcription. Aberrant DNA methylations have been observed and may function as biomarkers in cancer diagnosis and prognosis [9]. Moreover, two DNA methyltransferase (DNMT) inhibitors, 5-azacytidine (azacytidine) and 5-aza-2′-deoxycytidine (decitabine), have been approved by the US FDA for the treatment of some hematological malignancies. The success observed in cutaneous lymphomas hints that a similar outcome may be expected in solid tumors, including CS [10]. In this review, we discuss aberrant DNA methylations in CS. Insight into DNA methylation-associated epigenetic changes can offer opportunities for the detection of molecular biomarkers that can be useful for diagnosis, prognosis and antimethylation drugs used for novel potential treatment strategies in CS.
DNA methylation abnormalities in CS
DNA methylation comprises the covalent addition of a methyl group to the fifth position of a certain nucleotide in DNA sequences with the assistance of DNMTs (DNMT1, DNMT3a and DNMT3b) [11,12]. It is vital to a number of cellular processes, including, but not limited to, embryonic development, X-chromosome inactivation, genomic imprinting, gene suppression, tumorigenesis and chromosome stability. Specifically, this epigenetic modification could regulate gene expression through directly interfering with the binding of specific transcription factors to DNA and/or altering the chromatin structure by recruiting methylcytosine binding proteins [13–15]. DNA methylation in CS can be classified into two methylation levels, hypomethylation and hypermethylation, both of which are significant in the study of CS tumorigenesis and progression.
DNA hypomethylation in CS
DNA hypomethylation refers to a decreased/low methylation level and was the first discovered epigenetic alteration [16]. It can occur at normally methylated DNA sequences as repetitive DNA sequences and gene regulatory regions, and can be classified into two subtypes: global hypomethylation (loss of total DNA methylation content) and individual gene hypomethylation (e.g., hypomethylation of an oncogene) [17]. Since repetitive DNA sequences, such as satellite 2 and LINE-1, make up approximately 50% of the genome, their hypomethylation always acts as a marker for evaluating global hypomethylation that is a frequent event in human malignancies, including Swarm rat CS (SRC), which has been shown to resemble the human disease [17–20]. In a 2009 study, global hypomethylation was induced by using decitabine in SRC, a rat model of human CS [18,20–23]. This study used rat-specific pyrosequencing assays to compare the methylation status of satellite 1 and LINE-1 between SRC cells with decitabine treatment and without treatment [18]. Results showed that both repetitive DNA sequences were hypomethylated in decitabine-treated SRC cells. To further assess individual gene methylation status, growth factor MDK and pluripotent transcription factor Sox2 were shown to be overexpressed, and there was a notable decrease in methylation levels in the promoter region of both genes following decitabine treatment. These studies demonstrated increased invasiveness of the SRC cells with decitabine in vitro and tumor growth with decitabine in vivo [18]. It may be plausible that hypomethylation of the individual genes, MDK and Sox2, functions in CS development. However, drug-induced global hypomethylation has complex impacts on the whole genome, affecting the progression of CS. There is a lack of available data on the hypomethylation of individual genes in CS in part possibly because hypomethylation of individual genes is rather an infrequent event in cancers [17,24]. Thus, further investigations are required.
A correlation between changes in the microenvironment and DNA methylation alteration was evaluated in 2010 [20]. SRC tumors were transplanted in different locations of Sprague-Dawley rats, including subcutaneous tissue and the tibia. Rat-specific pyrosequencing detected the methylation status of satellite 1 in SRC tumor tissues from various locations versus that in rat normal articular cartilage, which were obtained from the femoral heads of healthy 37–40-day-old male Sprague-Dawley rats. Results showed that the SRC tumor tissues exhibited a lower methylation level than normal cartilage. Specifically, statistically significant differences of methylation levels were revealed among SRC tumors tissues in different transplantation sites [20]. These findings indicated that DNA methylation may be regulated by microenvironment changes, providing insight into the influence of environmental factors on DNA methylation alterations in CS.
DNA hypermethylation & abnormalities in CS
Another form of abnormal DNA methylation, hypermethylation of CpG islands in promoters of tumor-related genes, refers to increased/high methylation level. The silencing of tumor-related genes induced by hypermethylation has been observed to have a significant influence on tumorigenesis in CS.
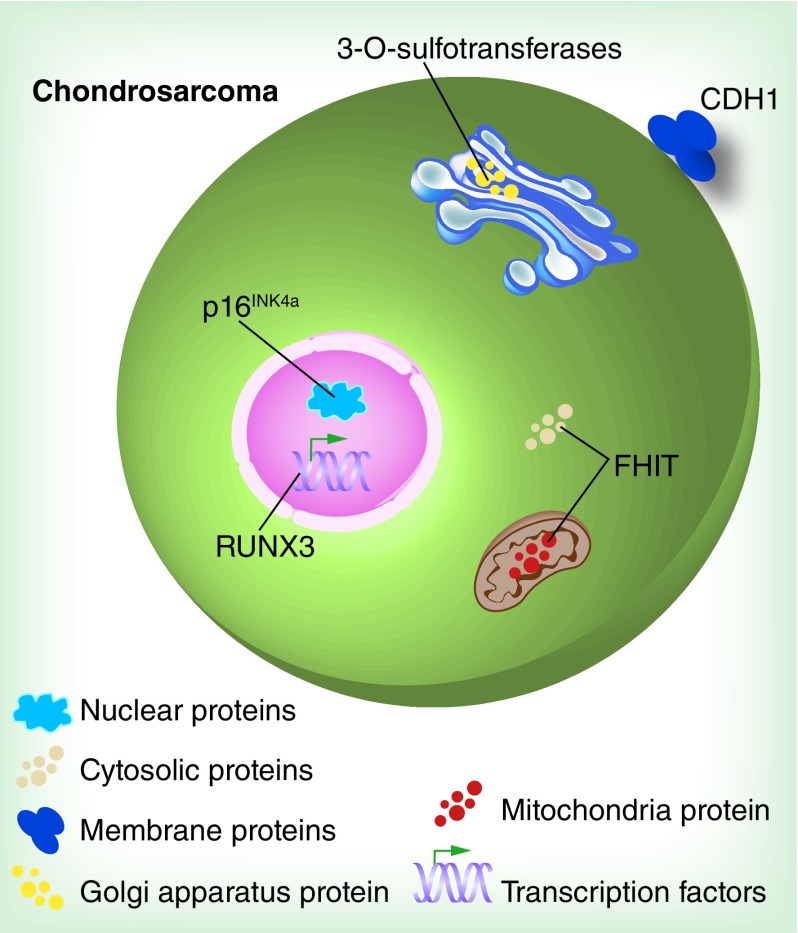
DNA hypermethylation contributes to the development of CS via various cell pathways, including cell cycle, apoptosis, cell adherence and cell-to-cell interaction [25–28]. For example, P16INK4a, a well-known tumor suppressor gene, was found to be hypermethylated in the promoter region in CS [26]. P16INK4a is located on chromosome 9p21 and encodes an inhibitor of cyclin-dependent kinase which is involved in the control of G1 progression and arrests the growth of deregulated tumor cells [29]. Five high-grade CS tissues (dedifferentiated, central grade II and grade III tumors) were found to be partially methylated by methylation-specific PCR (MSP) across 22 CSs [26]. The methylation levels of eight candidate tumor suppressor genes, p21WAF1, p16INK4a, p14ARF, DAPK, FHIT, hMLH1, p73 and E-cadherin (CDH1), in dedifferentiated CS tissue were measured by MSP analysis [27]. The samples analyzed consisted of two different areas: a low-grade chondroid site and a highly malignant osteosarcomatous site. The results showed p16INK4a and E-cadherin (cell-adhesion-related gene) was methylated in both dedifferentiated CS sites. However, methylation of FHIT (an apoptosis and cell cycle control-related gene) was only detected in the highly malignant osteosarcomatous site [27]. Furthermore, gene silencing induced by DNA hypermethylation is involved in cell-to-cell interaction in CS. Heparan sulfate (HS) proteoglycan is a core protein linked by long linear glycosaminoglycan HS located on the surface of almost every animal cell, and interacts with numerous biological molecules, such as growth factors and cytokines [28,30]. Thereby, HS proteoglycans regulate a number of biological processes, including cell proliferation, migration and adhesion. Abnormal promoter DNA hypermethylation of one HS biosynthetic enzyme, 3-OST, was examined in human CS cell line HEMC compared with that in normal human blood [28]. Semiquantitative real-time PCR analysis of 3-OST mRNA showed significantly elevated expression in the decitabine-treated HEMC cell line as compared with untreated cells, suggesting a downregulation of transcriptional activity by methylation modification. Further assessment showed the effect of the restoration of 3-OST expression on CS cell activities. Decitabine treatment of HEMC cells or transfection of 3-OST cDNA increased cell adhesion and reduced cell proliferation and migration versus untreated cells or untransfected cells [28]. These findings indicate that hypermethylation of 3-OST contributes to invasive phenotypes in CS in vitro. Additionally, hypermethylation influences the gene expression of upstream regulators. In CS patient specimens, the hypermethylation of RUNX3, an encoding transcription factor regulating cell cycle arrest and apoptosis, acting as a tumor suppressor was examined by MSP [25]. Western blot and real-time PCR for mRNA exhibited a reduction of RUNX3 protein levels as well as a low transcription level in CS tissues relative to normal tissues [25]. Furthermore, a correlation was observed between RUNX3 promoter hypermethylation and downregulated expression, implicating hypermethylation of RUNX3 as a mechanism of inactivating gene expression. Colony formation assays were performed to examine the antitumor activities of RUNX3 in CS cell line SW1353 and results showed lower proliferation of cDNA-transfected CS cells relative to untransfected cells. A high rate of apoptosis was also confirmed in cDNA-transfected cells versus untransfected cells. Collectively, hypermethylation of RUNX3 correlated with increasing proliferation and reducing apoptosis in CS cells in vitro [25]. Hypermethylation of tumor-related genes in human CS is shown in Figure 1.
Figure 1. . Hypermethylation of tumor-related genes in human chondrosarcoma.
Five tumor-related genes are shown to be hypermethylation in human chondrosarcoma: 3-OST, RUNX3, p16INK4a, CDH1 and FHIT.
DNA methylation alterations induced by genetic changes in CS
Disordered DNA methylation patterns led by IDH gene mutation prove that DNA methylation can be regulated by genetic modification. IDH mutations (IDH1 and IDH2 mutations) are prevalent in more than 50% of patients with CS [31]. Mutant IDH in CS produces elevated 2-hydroxyglutarate compared with normal tissues [32]. 2-hydroxyglutarate is an inhibitor of TET proteins that participate in DNA demethylation. Thus, increasing 2-hydroxyglutarate produced by mutant IDH results in genome-wide hypermethylation [31,32]. Collectively, IDH mutations can maintain appropriate DNA methylation of genes associated with regulation of cells differentiation and ultimately contribute to tumorigenesis [31,32]. In murine 10T1/2 mesenchymal progenitor cells, the expression of mutant IDH2 led to genome-wide DNA hypermethylation and impairment in the differentiation of mesenchymal cells, which could be reversed by treatment with azacytidine [31–33]. Therefore, further investigation into the genetic impact on DNA methylation in CS and the interaction between genetics and epigenetics is necessary for the understanding of initiation and progression of this malignancy.
DNA methylation profiles in CS
Sox family genes code proteins that are involved in cell processes, such as cell development, homeostasis and regeneration [34]. Dysregulations of these genes have been found in sarcoma, including CS. Besides Sox2 mentioned in Table 1, another member of the Sox family, Sox9, which acts as a transcriptional factor and plays a significant role in chondrogenesis, has been found to be upregulated in three CS types (conventional, mesenchymal and clear cell), but downregulated in dedifferentiated CS [34–38]. Consequently, Sox family DNA methylation may exist in CS.
Table 1. . DNA methylation profiles in chondrosarcoma.
| Gene | Sample | Function | Methylation status | Ref. |
|---|---|---|---|---|
|
MDK |
Rat CS cells |
Growth factor |
Induced hypomethylation† |
[18] |
|
Sox2 |
Rat CS cells |
Pluripotent transcription factor |
Induced hypomethylation† |
[18] |
|
Satellite 1 |
Rat CS cells, rat CS tissues |
Repetitive DNA elements |
Global hypomethylation |
[18,20] |
|
LINE-1 |
Rat CS cells |
Repetitive DNA elements |
Global hypomethylation |
[18] |
|
Maspin |
Human CS cell lines |
Epithelial-specific marker |
Hypomethylation |
[39] |
|
14-3-3σ |
Human CS cell lines |
Epithelial-specific marker |
Hypomethylation |
[39] |
|
p16INK4a (CDKN2A) |
Human CS tissues |
Inhibitor of cyclin-dependent kinase |
Hypermethylation |
[26,27] |
|
FHIT |
Human CS tissues |
Induce apoptosis and in cell-cycle control |
Hypermethylation |
[27] |
|
E-cadherin (CDH1) |
Human CS tissues |
Cell adhesion |
Hypermethylation |
[27] |
|
3-OST |
Human CS cell lines |
HS biosynthetic enzymes |
Hypermethylation |
[28] |
| RUNX3 | Human CS tissues | Induce cell cycle arrest and apoptosis | Hypermethylation | [25] |
†Induced hypomethylation denotes certain gene is detected as hypomethylated after using DNA methyltransferase inhibitor but unknown methylation status in CS.
CS: Chondrosarcoma; HS: Heparan sulfate.
CS may be characterized by epithelial features of cancer. CS cells were observed to have acquired the expression of four epithelial markers: E-cadherin, Desmocollin 3, Maspin and 14-3-3σ in vitro [39]. Furthermore, hypomethylation of two of the four markers, Maspin and 14-3-3σ, were examined. There is a progression transitioning from mesenchymal to epithelial during CS development. DNA methylation modification is involved in this progression. Importantly, substantial DNA methylation profiles of cancers could be utilized for exploring unclaimed DNA methylation in CS.
Gene silencing induced by DNA methylation occurs not only in protein-coding genes, but also in non-coding RNA-coding genes, such as miRNAs [40]. MiRNAs are one of the small non-coding RNAs and are approximately 22 nucleotides long, functioning as post-transcriptional regulators of target genes expression. MiRNAs participate in cellular processes, such as proliferation, apoptosis, differentiation, oncogenesis and drug resistance, by means of the function of its target genes in a large number of malignancies, including CS [41,42]. For example, miRNA-145 is involved in chondrogenic differentiation through downregulating the expression of Sox9. The observed overexpression of Sox9 in CS implies the declining expression of miRNA-145 [43]. MiRNA-494, which inhibits cell proliferation and invasion of CS, was reported to be downregulated in CS tissues and human CS cell line SW1353 [42]. However, the combination of DNA methylation and miRNA in CS has not been studied. Accordingly, further investigation on DNA methylation of CS-related miRNA may yield promising results.
A summary of DNA methylation profiles is presented in Table 1.
Potential biomarkers for DNA methylation in CS
DNA methylation analysis could be used to identify biomarkers for diagnosis of CS. RUNX3 expressions induced by different DNA methylation levels have shown a significant association with CS pathological types through statistical analysis [25]. Thus, evaluating the methylation level of RUNX3 may serve as a biomarker to distinguish the particular histological features associated with clinical diagnosis for CS [25]. At present, the main diagnostic methods of CS are imaging and pathological biopsy assessment. The previous studies cited in this review suggest that a feasible application of detection of disordered DNA methylation could be used as tumor markers in aiding the diagnosis of CS. In order to build a library of aberrant DNA methylation profiles associated with clinical parameters in CS patients, more hypermethylation and hypomethylation biomarkers relating to CS development and progression need to be explored.
DNA methylation of particular tumor-related genes could be considered as putative biomarkers of prognosis in patients with CS. In the case of RUNX3 as previously mentioned, methylation of RUNX3 was also associated with patient survival in clinic [25]. Positive expression of RUNX3 revealed a more favorable outcome as compared with negative expression of RUNX3 in 63 patients with CS, and it was demonstrated that hypermethylation was responsible for decreased RUNX3 expression [25]. Accordingly, monitoring of the dynamic changes in prognosis-related DNA methylation after tumor resections could help evaluate the effect of therapy, as well as anticipate the prognosis of CS patients.
Additionally, the measurement of DNA markers has several unique merits. As compared with protein, another frequently used indicator of tumor progression and diagnosis in clinic, DNA is stable and easy to isolate from different kinds of material, whereas proteins are more difficult to harvest in amounts necessary for analysis [24]. Moreover, the detection of DNA methylation requires less sophisticated methods over, for example, mutation of traditional genetic alteration which has been recognized as valuable in clinical diagnosis [44]. DNA methylation locating at CpG dinucleotides was previously well defined, unlike mutations, which affect sites distributed sparsely throughout the genome sequence, and hence requires more complicated analyses [24]. Nevertheless, the application of DNA methylation methods in the clinic faces challenges as well. Studies on DNA methylation biomarkers in CS are either scarce or employ too few number of samples for broad interpretation. Further application in clinical practice will require rapid, cost-effective techniques, objective criteria for selection of the candidate gene markers as well as the establishment of false-positive and false-negative rates [9].
Prospective therapy for using DNMT inhibitors in CS
DNMT inhibitors, which consist of decitabine, azacytidine and hydralazine, mainly interfere with DNA methylation through disrupting DNMTs’ activity and blocking subsequent DNA methylation [10,45]. Re-expressing silenced tumor suppressor genes, such as p16INK4a, RASSF1A and DAPK, consequently impedes tumors progression. In the CS cell line HEMC, decitabine-mediated demethylation restored the expression of tumor suppressor 3-OST and resulted in reduced proliferative and invasive properties, as well as increased adhesion of CS cells [28]. Accordingly, the strategy of using DNMT inhibitors can attenuate CS progression induced by appropriate hypermethylation of tumor suppressor gene in vitro. However, the nonspecific effects of DNA demethylation agents do not always yield the expected outcomes in antitumor studies. In SRC, global hypomethylation, induced by decitabine, contributed to CS progression both in vitro and in vivo. The invasiveness of rat CS cells dramatically decreased compared with control after withdrawing decitabine in vitro [18]. Moreover, a specific DNA methylation pattern often is restored after the removal of the demethylation agents and the toxic effects of demethylation agents exist, such as neutropenia caused by high doses of azacytidine [10,46]. Thus, exploring specific demethylation therapy and management of demethylation drugs in clinical practice are challenges that remain to be solved.
Conclusion
Evidence regarding DNA methylation, especially hypomethylation, is quite limited so far. It is necessary, as well, to accumulate more data on methylation modification in regard to clinical parameters for those patients with CS. However, recent developments in investigations have provided valuable insights into the role of these modifications in several tumor-related genes and disruption of normal biological process leading to CS tumorigenesis and progression. These new insights hold potential applications in diagnosis, prognosis and treatment of CS.
Future perspective
CSs are specific resistant to chemotherapy and the prognosis of patients with unresectable or metastatic disease remains poor. As many different genetic and epigenetic alterations in CS have been identified, it is important to identify druggable targets that may improve the prognosis and the treatment of CS patients. There is still much to explore regarding the aberrant methylation of CS-related genes; how they contribute to oncogenic process; and what factors affect them in CS. Future characterization of DNA methylation pathways involved in the pathogenesis of CS may offer the improved treatment of patients with CS.
Executive summary.
DNA methylation abnormalities in chondrosarcoma
DNA global hypomethylation induced by DNA methyltransferase (DNMT) inhibitor increases chondrosarcoma (CS) invasiveness in vitro and tumor growth in vivo.
DNA methylation may be regulated by microenvironment change.
DNA hypermethylation contributes to CS progression via dysregulating various tumor-related genes.
IDH mutations are involved in modulating inappropriate DNA methylation in CS.
DNA methylation profiles in CS
Sox family DNA methylation may exist in CS.
DNA methylation profiles of cancers could be utilized for exploring unclaimed DNA methylation in CS.
DNA methylation of CS-related miRNA may yield promising results.
Prospective biomarkers for DNA methylation in CS
DNA methylation of RUNX3 could be used as biomarkers for diagnosis and prognosis in patients with CS.
Measuring of DNA methylation has several unique merits, but the application of it in the clinic faces challenges as well.
Potential therapy for using DNMT inhibitors in CS
DNMTs may work in curing CS, but there is a challenge because of their nonspecific effects and toxic effects.
Footnotes
Financial & competing interests disclosure
This work was supported, in part, by the Gattegno and Wechsler funds. Z Duan is supported, in part, through a grant from Sarcoma Foundation of America (SFA), a grant from National Cancer Institute (NCI)/NIH, UO1, CA 151452-01, a pilot grant from Sarcoma SPORE/NIH, and a grant from an Academic Enrichment Fund of MGH Orthopaedics. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Jiao G, Guo W, Ren T, et al. BMPR2 inhibition induced apoptosis and autophagy via destabilization of XIAP in human chondrosarcoma cells. Cell Death Dis. 2014;5:e1571. doi: 10.1038/cddis.2014.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Neal LW, Ackerman LV. Chondrosarcoma of bone. Cancer. 1952;5(3):551–577. doi: 10.1002/1097-0142(195205)5:3<551::aid-cncr2820050317>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 3.Bauer HC, Brosjo O, Kreicbergs A, Lindholm J. Low risk of recurrence of enchondroma and low-grade chondrosarcoma in extremities. 80 patients followed for 2–25 years. Acta Orthop. Scand. 1995;66(3):283–288. doi: 10.3109/17453679508995543. [DOI] [PubMed] [Google Scholar]
- 4.Eriksson AI, Schiller A, Mankin HJ. The management of chondrosarcoma of bone. Clin. Orthop. Relat. Res. 1980;153:44–66. [PubMed] [Google Scholar]
- 5.Totoki Y, Yoshida A, Hosoda F, et al. Unique mutation portraits and frequent COL2A1 gene alteration in chondrosarcoma. Genome Res. 2014;24(9):1411–1420. doi: 10.1101/gr.160598.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soderstrom M, Ekfors TO, Bohling TO, Teppo LH, Vuorio EI, Aro HT. No improvement in the overall survival of 194 patients with chondrosarcoma in Finland in 1971–1990. Acta Orthop. Scand. 2003;74(3):344–350. doi: 10.1080/00016470310014292. [DOI] [PubMed] [Google Scholar]
- 7.Giuffrida AY, Burgueno JE, Koniaris LG, Gutierrez JC, Duncan R, Scully SP. Chondrosarcoma in the United States (1973 to 2003): an analysis of 2890 cases from the SEER database. J. Bone Joint Surg. Am. 2009;91(5):1063–1072. doi: 10.2106/JBJS.H.00416. [DOI] [PubMed] [Google Scholar]
- 8.Yu X, Li Z. Epigenetic deregulations in chordoma. Cell Prolif. 2015;48(5):497–502. doi: 10.1111/cpr.12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esteller M. Epigenetics in cancer. N. Engl. J. Med. 2008;358(11):1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]; •• Aberrant DNA methylations have been observed and may function as biomarkers in cancer diagnosis and prognosis.
- 10.Nervi C, De Marinis E, Codacci-Pisanelli G. Epigenetic treatment of solid tumours: a review of clinical trials. Clin. Epigenetics. 2015;7:127. doi: 10.1186/s13148-015-0157-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ouadid-Ahidouch H, Rodat-Despoix L, Matifat F, Morin G, Ahidouch A. DNA methylation of channel-related genes in cancers. Biochim. Biophys. Acta. 2015;1848(10 Pt B):2621–2628. doi: 10.1016/j.bbamem.2015.02.015. [DOI] [PubMed] [Google Scholar]
- 12.Bestor TH. Gene silencing as a threat to the success of gene therapy. J. Clin. Invest. 2000;105(4):409–411. doi: 10.1172/JCI9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mann DA. Epigenetics in liver disease. Hepatology. 2014;60(4):1418–1425. doi: 10.1002/hep.27131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bergman Y, Cedar H. DNA methylation dynamics in health and disease. Nat. Struct. Mol. Biol. 2013;20(3):274–281. doi: 10.1038/nsmb.2518. [DOI] [PubMed] [Google Scholar]
- 15.Cedar H, Bergman Y. Programming of DNA methylation patterns. Annu. Rev. Biochem. 2012;81:97–117. doi: 10.1146/annurev-biochem-052610-091920. [DOI] [PubMed] [Google Scholar]
- 16.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301(5895):89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 17.Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics. 2009;1(2):239–259. doi: 10.2217/epi.09.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamm CA, Xie H, Costa FF, et al. Global demethylation of rat chondrosarcoma cells after treatment with 5-aza-2′-deoxycytidine results in increased tumorigenicity. PLoS ONE. 2009;4(12):e8340. doi: 10.1371/journal.pone.0008340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32(3):e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamm CA, Stevens JW, Xie H, et al. Microenvironment alters epigenetic and gene expression profiles in Swarm rat chondrosarcoma tumors. BMC Cancer. 2010;10:471. doi: 10.1186/1471-2407-10-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Breitkreutz D, Diaz de Leon L, Paglia L, Gay S, Swarm RL, Stern R. Histological and biochemical studies of a transplantable rat chondrosarcoma. Cancer Res. 1979;39(12):5093–5100. [PubMed] [Google Scholar]
- 22.Kenan S, Steiner GC. Experimental transplantation of the Swarm rat chondrosarcoma into bone: radiological and pathological studies. J. Orthop. Res. 1991;9(3):445–451. doi: 10.1002/jor.1100090317. [DOI] [PubMed] [Google Scholar]
- 23.Grimaud E, Damiens C, Rousselle AV, Passuti N, Heymann D, Gouin F. Bone remodelling and tumour grade modifications induced by interactions between bone and swarm rat chondrosarcoma. Histol. Histopathol. 2002;17(4):1103–1111. doi: 10.14670/HH-17.1103. [DOI] [PubMed] [Google Scholar]
- 24.Kulis M, Esteller M. DNA methylation and cancer. Adv. Genet. 2010;70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2. [DOI] [PubMed] [Google Scholar]; • Demostrates that DNA methylation can be divided into hypermethylation and hypomethylation, genes that acquire promoter hypermethylation are involved in a variety of important cellular pathways.
- 25.Jin Z, Han YX, Han XR. Loss of RUNX3 expression may contribute to poor prognosis in patients with chondrosarcoma. J. Mol. Histol. 2013;44(6):645–652. doi: 10.1007/s10735-013-9511-x. [DOI] [PubMed] [Google Scholar]
- 26.Asp J, Sangiorgi L, Inerot SE, et al. Changes of the p16 gene but not the p53 gene in human chondrosarcoma tissues. Int. J. Cancer. 2000;85(6):782–786. doi: 10.1002/(sici)1097-0215(20000315)85:6<782::aid-ijc7>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 27.Ropke M, Boltze C, Neumann HW, Roessner A, Schneider-Stock R. Genetic and epigenetic alterations in tumor progression in a dedifferentiated chondrosarcoma. Pathol. Res. Pract. 2003;199(6):437–444. doi: 10.1078/0344-0338-00443. [DOI] [PubMed] [Google Scholar]
- 28.Bui C, Ouzzine M, Talhaoui I, et al. Epigenetics: methylation-associated repression of heparan sulfate 3-O-sulfotransferase gene expression contributes to the invasive phenotype of H-EMC-SS chondrosarcoma cells. FASEB J. 2010;24(2):436–450. doi: 10.1096/fj.09-136291. [DOI] [PubMed] [Google Scholar]
- 29.Liggett WH, Jr, Sidransky D. Role of the p16 tumor suppressor gene in cancer. J. Clin. Oncol. 1998;16(3):1197–1206. doi: 10.1200/JCO.1998.16.3.1197. [DOI] [PubMed] [Google Scholar]
- 30.Poulain FE, Yost HJ. Heparan sulfate proteoglycans: a sugar code for vertebrate development? Development. 2015;142(20):3456–3467. doi: 10.1242/dev.098178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guilhamon P, Eskandarpour M, Halai D, et al. Meta-analysis of IDH-mutant cancers identifies EBF1 as an interaction partner for TET2. Nat. Commun. 2013;4:2166. doi: 10.1038/ncomms3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Azzi G, Velez M, Mathias-Machado MC. Isocitrate dehydrogenase mutations in chondrosarcoma: the crossroads between cellular metabolism and oncogenesis. Curr. Opin. Oncol. 2014;26(4):403–407. doi: 10.1097/CCO.0000000000000092. [DOI] [PubMed] [Google Scholar]
- 33.Lu C, Venneti S, Akalin A, et al. Induction of sarcomas by mutant IDH2 . Genes Dev. 2013;27(18):1986–1998. doi: 10.1101/gad.226753.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J, Shen J, Wang K, Hornicek F, Duan Z. The roles of Sox family genes in sarcoma. Curr. Drug Targets. 2016 doi: 10.2174/1389450117666160502145311. PMID:27138764. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 35.Tang X, Lu X, Guo W, et al. Different expression of Sox9 and Runx2 between chondrosarcoma and dedifferentiated chondrosarcoma cell line. Eur. J. Cancer Prev. 2010;19(6):466–471. doi: 10.1097/CEJ.0b013e32833d942f. [DOI] [PubMed] [Google Scholar]
- 36.Fanburg-Smith JC, Auerbach A, Marwaha JS, Wang Z, Rushing EJ. Reappraisal of mesenchymal chondrosarcoma: novel morphologic observations of the hyaline cartilage and endochondral ossification and beta-catenin, Sox9, and osteocalcin immunostaining of 22 cases. Hum. Pathol. 2010;41(5):653–662. doi: 10.1016/j.humpath.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 37.Soderstrom M, Bohling T, Ekfors T, Nelimarkka L, Aro HT, Vuorio E. Molecular profiling of human chondrosarcomas for matrix production and cancer markers. Int. J. Cancer. 2002;100(2):144–151. doi: 10.1002/ijc.10457. [DOI] [PubMed] [Google Scholar]
- 38.Matsuura S, Ishii T, Endo M, et al. Epithelial and cartilaginous differentiation in clear cell chondrosarcoma. Hum. Pathol. 2013;44(2):237–243. doi: 10.1016/j.humpath.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 39.Fitzgerald MP, Gourronc F, Teoh ML, et al. Human chondrosarcoma cells acquire an epithelial-like gene expression pattern via an epigenetic switch: evidence for mesenchymal-epithelial transition during sarcomagenesis. Sarcoma. 2011;2011:598218. doi: 10.1155/2011/598218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lujambio A, Ropero S, Ballestar E, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67(4):1424–1429. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- 41.Sun R, Shen JK, Choy E, Yu Z, Hornicek FJ, Duan Z. The emerging roles and therapeutic potential of microRNAs (miRs) in liposarcoma. Discov. Med. 2015;20(111):311–324. [PubMed] [Google Scholar]
- 42.Li J, Wang L, Liu Z, et al. MicroRNA-494 inhibits cell proliferation and invasion of chondrosarcoma cells in vivo and in vitro by directly targeting SOX9 . Oncotarget. 2015;6(28):26216–26229. doi: 10.18632/oncotarget.4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mak IW, Singh S, Turcotte R, Ghert M. The epigenetic regulation of SOX9 by miR-145 in human chondrosarcoma. J. Cell Biochem. 2015;116(1):37–44. doi: 10.1002/jcb.24940. [DOI] [PubMed] [Google Scholar]
- 44.Chin L, Gray JW. Translating insights from the cancer genome into clinical practice. Nature. 2008;452(7187):553–563. doi: 10.1038/nature06914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hamm CA, Costa FF. Epigenomes as therapeutic targets. Pharmacol. Ther. 2015;151:72–86. doi: 10.1016/j.pharmthera.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 46.Braiteh F, Soriano AO, Garcia-Manero G, et al. Phase I study of epigenetic modulation with 5-azacytidine and valproic acid in patients with advanced cancers. Clin. Cancer Res. 2008;14(19):6296–6301. doi: 10.1158/1078-0432.CCR-08-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]