Abstract
Diversity generally protects communities from unstable environmental conditions. This principle, known as the “insurance hypothesis,” has been tested in many different ecosystems. Here we show that the opportunistic pathogen Pseudomonas aeruginosa undergoes extensive genetic diversification during short-term growth in biofilm communities. The induced genetic changes are produced by a recA-dependent mechanism and affect multiple traits, including the behavior of the bacteria in biofilms. Some biofilm-derived variants exhibit an increased ability to disseminate, whereas others manifest accelerated biofilm formation. Furthermore, the presence of these functionally diverse bacteria increases the ability of biofilms to resist an environmental stress. These findings suggest that self-generated diversity in biofilms provides a form of biological insurance that can safeguard the community in the face of adverse conditions.
Keywords: genetic diversity, Pseudomonas aeruginosa, insurance hypothesis, recA
Many bacterial species are capable of living in structures known as biofilms. In biofilms, bacteria live clustered together in matrix-encased groups attached to some surface (1, 2). Biofilms are thought to be the predominant growth mode for bacteria in natural environments, and increasing evidence implicates them as a cause of human infections (2–4). Biofilms also contaminate drinking water systems and industrial equipment, and they form environmental reservoirs for pathogens such as Vibrio cholerae, Legionella pneumophila, and Mycobacterium species (4–7). The opportunistic pathogen Pseudomonas aeruginosa is one of the most formidable and best-studied biofilmforming organisms. P. aeruginosa biofilms cause airway infections that lead to respiratory failure in cystic fibrosis and other bronchiectasis patients (1, 8–10) and the endotracheal tube colonization that leads to ventilator-associated pneumonia (11). Biofilms also cause infections in medical devices such as urinary catheters (12) and contact lenses (13).
Physiological changes produced by biofilm growth can greatly enhance the survival of bacteria. The most notorious biofilm-mediated effect increases the resistance of organisms to antimicrobial agents; P. aeruginosa biofilms can be up to 1,000 times more resistant than the same bacteria in the planktonic (free-living) state (2, 14). Biofilm bacteria may also be less conspicuous to the immune system, because antigens may be hidden, and the expression of ligands used by phagocytic cells can be repressed (15–17). The biofilm matrix can provide protection from physical injury, and the close proximity of organisms may allow metabolic interactions (18), promote horizontal gene transfer of virulence traits (19), and enhance communication between cells, facilitating coordinated behavior (18, 20, 21). Importantly, all of these advantages spring from the organized group structure of biofilms. If the group is disrupted, resistance to killing and other benefits are lost, and the vulnerabilities of the individual bacterium return (14).
Because this group structure plays such a key role in their function, biofilms are often thought of as bacterial communities (2, 22). Ecologists have long recognized that the stability of many types of biological communities is enhanced by diversity. For example, simple communities, such as monospecies forests, are more susceptible to environmental perturbations (such as drought or insect attack) than diverse communities, such as mixed woodlands (23). This phenomenon has been explained by the “insurance hypothesis,” which posits that the presence of diverse subpopulations increases the range of conditions in which the community as a whole can thrive (23, 24). Insurance effects could be of great benefit to biofilms because, like other communities, their long-term success depends on their ability to withstand changing environmental conditions.
Here we report three main findings: First, we have found that short-term growth of P. aeruginosa in biofilms generates extensive genetic diversity in the resident bacteria. This diversity arises by means of a mechanism that requires the recA gene and likely involves recombination functions. Second, the genetic diversity produces bacterial subpopulations with specialized functions in biofilms. Third, as predicted by the insurance hypothesis, this functional diversity increases the biofilm community's ability to withstand an applied physiological stress.
Methods
Strains, Plasmids, and Growth Conditions. The P. aeruginosa strains used were derived from the wild-type strain PA01 (from B. Iglewski, University of Rochester, Rochester, NY). In addition, strains PA14, PA103, and five cystic fibrosis clinical isolates were tested. Strains visualized by using confocal microscopy carried the gfp-containing plasmid pMRP9–1 (20). Nonpolar recA mutants were constructed in the wild-type and wrinkly variants as follows: DNA fragments (1 kb) flanking recA were amplified by PCR (including the first and last nine nucleotides of recA). The PCR products were then sequentially ligated into pEX18TC to create a vector containing a recA deletion. Standard gene replacement methods were used to move this mutation onto the chromosome of PA01 (20). The recA mutants were complemented by recA (cloned into miniCTX1) inserted at the phage attachment site of the PA01 chromosome. The dinP mutant was obtained from the University of Washington library (PTL39660) (25). Quorum-sensing mutants PA0-JP3 and PA0-MW1 were obtained from E. P. Greenberg (University of Iowa). Trypticase soy broth (TSB) (Difco) was used as the growth medium unless otherwise specified.
Swimming motility plates consisted of 1% tryptone, 0.5% NaCl, and 0.3% Bacto agar (Difco). Pyomelanin production was detected on plates containing 0.7% K2HPO4, 0.3% KH2PO4, 0.05% trisodium citrate, 0.01% MgSO4, 0.1% (NH4)2SO4, 0.2% glucose, and 1% l-tyrosine (26). Auxotrophs were detected by stamping colonies from TSB plates onto minimal salts medium plates with 0.2% glucose (27). Colonies that failed to grow on minimal medium but grew on TSB were considered auxotrophs. To increase the cell density and the number of cell divisions (generations) that occurred in the planktonic cultures, PA01 was grown in 500 ml of 5× TSB with shaking. This produced cultures of 3 × 1010 colony-forming units/ml after ≈32 generations. For chemostat growth, the flow rate was 10 ml/h in a 100-ml vessel with 1% TSB as the growth medium.
Biofilm Experiments. Drip flow reactors were used to grow biofilms at 37°C on stainless steel plates in aluminum chambers (28). Chambers were filled with 15 ml of 10% TSB and 250 μl of an overnight culture and incubated for 24 h without medium flow to allow attachment. Reactors were then set at a 10° angle, and 1% TSB was dripped over the plate at 50 ml/h. Biofilms were harvested into 10 ml of PBS and homogenized, and colony morphology was scored. 1,000 colonies were examined at each time point in Fig. 1 b–c and at 5 days for Fig. 2 b and d–e. The number of generations that occur during growth in drip flow reactors is difficult to exactly determine, because cells are continuously lost in the effluent. However, even if the number of lost cells is assumed to exceed the number in the biofilm by 100-fold, <17 generations would have occurred during biofilm growth. Variant colonies were produced in similar abundance in drip flow reactors (28), tube reactors (29), and after 5 days of biofilm growth in 96-well microtiter dishes with daily media changes. Variants appeared at low numbers in the rotating disk reactor (30) and in continuous culture flow cells (30). In some experiments, PA01 was tagged with a selectable marker (tetracycline resistance) on the chromosome (by using miniCTX1). Variants produced by the tagged strain contained this marker, confirming that variants were not contaminants. Flow cell experiments were performed as previously described (30).
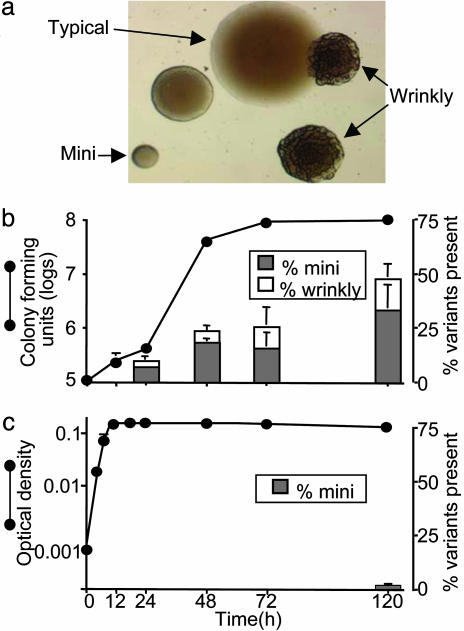
Fig. 1.
Variant colonies produced by biofilm growth. (a) Micrograph of variant colonies on agar produced by a 5-day-old P. aeruginosa biofilm. (b) Time course at which variants arise from biofilms. A simultaneous growth curve shows rate of cell accumulation. (c) Production of variants and growth curve in batch planktonic culture. Data in b and c are means of three experiments and representative of four others. Error bars show SEM.
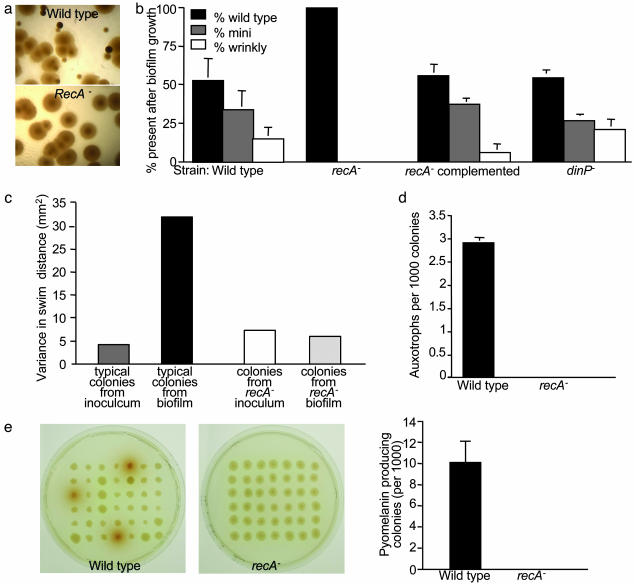
Fig. 2.
Role of recA in biofilm-induced diversity. (a) Micrographs of colonies produced by 5-day-old wild-type and recA– biofilms. (b) Proportion of bacteria with variant colony morphology arising from biofilms after 5 days of growth. Biofilms were grown with isogenic wild-type, recA–, recA–-complemented, and dinP– strains. Data are means of three experiments; error bars show SEM. (c) Variance in swimming distance induced by biofilm growth. The swimming capability of bacteria from typical colonies from biofilms was compared with the capability of bacteria from the inoculum. The biofilm-induced variation required recA. Data are the variance of 50 randomly picked wild-type and recA– colonies. (d) Generation of auxotrophs by biofilms. Data are means of four experiments. Error bars show SEM. (e) Generation of strains overproducing pyomelanin by biofilms. Agar plates show pyomelanin-overproducing colonies from wild-type but not from recA– biofilms. Data in the graph are the mean of four experiments; error bars show SEM.
The rotating disk reactor (30) was used for generating biofilms for antimicrobial susceptibility testing and the quantitative detachment assay as previously described. The short growth period (24 h) and high-shear conditions in this system produced pure-culture wild-type and wrinkly-variant biofilms of comparable biomass. For antimicrobial susceptibility testing, chips were removed, washed in PBS, and treated for 4 h in 1 ml of H2O2 at indicated concentrations. For detachment assays, disks were washed and incubated in 1 ml of PBS for indicated times. The bacteria in the overlying fluid (detached) and bacteria that remained on the disk (attached) were enumerated by plate counting.
To test the ability of biofilms to withstand a stressful challenge, biofilms were grown for 3 days in the drip flow reactor, and 40 μM H2O2 was then added to the medium for 48 h, at which time the bacteria were enumerated. Adherence was measured by incubating 108 bacteria with glass slides for 1 h, washing, and counting the number of attached cells per ×40 field of view.
Results
In the course of other experiments, we observed that when wild-type P. aeruginosa was grown in biofilms for 2–7 days, and the bacteria were plated on standard agar, considerable variation in colony morphology was produced (Fig. 1a). Because such extensive variation occurred only after biofilm growth, we investigated the variants further, focusing on the two most abundant types. We termed one type “mini” (because of its small colonies) and the other “wrinkly” (because of its rough appearance). For clarity, we call the wild-type morphology “typical.”
The degree of variation in colony morphology increased with the duration of biofilm growth; by 5 days, an average of 48% of the population were mini or wrinkly variants in this system (Fig. 1b). To determine whether generation of the variants depended on particular conditions, we grew biofilms in five biofilm reactor types that used different growth conditions. Three of these reactors consistently produced large numbers of variants, whereas two other reactor types produced fewer variants (see Methods). We also tested different P. aeruginosa strains. Six of seven different wild-type strains (including four of five different clinical isolates) produced colony variants like the reference strain. Because cell-to-cell signaling (quorum-sensing) is involved in some biofilm processes (2), we tested a strain that lacked the two main quorum-sensing systems (las and rhl) and found that these mutations had no effect on the generation of variants by biofilms (data not shown).
Planktonic batch cultures grown to logarithmic, stationary, or late stationary phase in the same medium as the biofilm experiments produced no variants (Fig. 1c); however, if the culture period was extended for 5 days (4.5 days after the onset of stationary phase), a low number of small-colony variants did appear (0.6% of the population, Fig. 1c). To examine the role of cell density, we used concentrated medium to grow batch cultures. These cultures reached ≈1010 colony-forming units/ml after ≈32 bacterial generations [many more generations than likely occurs in the biofilm cultures (see Methods)]. Higher cell density did not increase production of the variants. One explanation for the much lower occurrence of variants after planktonic growth is that variant generation is induced by starvation; however, variants cannot be detected in batch cultures because their numbers cannot increase once nutrients are exhausted. To explore this possibility, chemostats were used to determine whether the slow infusion of medium would produce variants in starved planktonic cultures; however, no variants appeared in 5 days of growth. Thus, whereas high density and starvation in planktonic cultures failed to generate the observed variation, biofilm growth by numerous strains in different conditions generated large numbers of the same variant types.
The variant phenotypes we studied were heritable, suggesting that genetic changes produced them. None of 1,000 mini or wrinkly colonies switched morphotypes after overnight passaging. A prime candidate for mediating such variation is RecA, which can produce genetic changes by recombination (31) and by inducing error-prone DNA polymerases as part of the bacterial stress response (SOS response) (32). Inactivation of recA dramatically reduced biofilm-induced colony variation, and this defect was complemented by chromosomally inserted recA (Fig. 2 a and b). In contrast, recA mutation had no impact on the low number of variants produced by prolonged planktonic growth, suggesting that these variants arise by a different mechanism (data not shown). Mutation of dinP, the only error-prone polymerase gene so far identified in P. aeruginosa (33), did not decrease biofilm-associated variation, suggesting that recA acts by a recombination mechanism (Fig. 2b).
The involvement of RecA, which could mediate genetic change anywhere in the chromosome, led us to hypothesize that biofilm-generated diversity could extend to other functions. To test this hypothesis, we examined the effect of biofilm growth on three additional phenotypes: swimming motility, bacterial nutritional requirements, and the secretion of an extracellular product. In the swimming tests, we compared typical colonies from 5-day-old biofilms with those from the inoculum to detect diversity that was independent of colony morphology. The biofilm-grown bacteria exhibited much more variation in swimming capability (Fig. 2c) than did those from the inoculum. Notably, some motility variants showed increased swimming relative to the inoculum, whereas others had less swimming ability. This finding suggests that biofilm growth induces multiple genetic changes affecting motility, because such extensive variation is unlikely to be caused by a single mutation. Biofilm growth also produced auxotrophs and bacteria that overproduced pyomelanin, a pigment that can protect against oxidants and radiation (Fig. 2 d and e) (34). The differences in swimming, pyomelanin production, and auxotrophic phenotypes were heritable, were not produced by planktonic growth, and were dependent on recA function (Fig. 2 c–e).
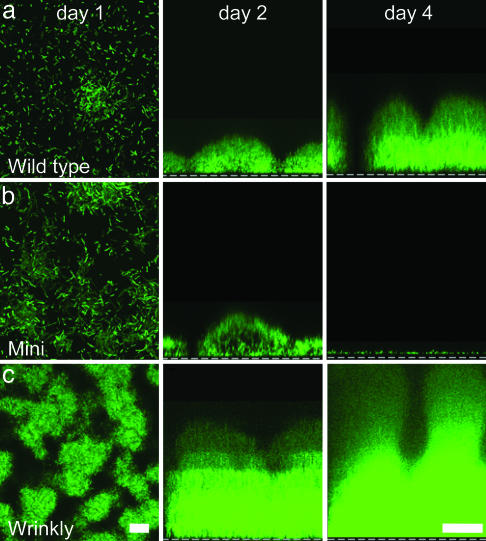
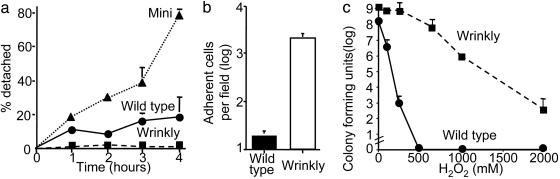
The fact that short-term biofilm growth generated variants in such high numbers led us to hypothesize that some of the variants may have specialized biofilm functions. To test this hypothesis, we picked two variants (one mini and one wrinkly) and grew them in pure-culture biofilms. In the growth conditions we used, the wild-type bacteria displayed the prototypical pattern of biofilm development (Fig. 3a). These bacteria attached to the growth surface, developed cell clusters, and eventually formed mature, tower-shaped biofilms. The mini variant we studied (Fig. 4a) exhibited similar attachment and cell cluster formation; however, after 2 days of growth, the mini-variant biofilm rapidly dispersed (Fig. 3b). We verified that their disappearance was not caused by cell death by using a different assay that measured the detachment of viable bacteria (Fig. 4a). These studies showed that the mini-variant biofilm detached at a 4-fold higher rate than did wild-type biofilms, and its detachment mechanism operates under different conditions. Interestingly, biofilms founded by the mini variant generated a degree of diversity comparable to that of biofilms formed by the wild-type parental strain (data not shown), suggesting that the hyperdetaching variants would have the capacity to reconstitute diverse biofilm populations at newly colonized sites.
Fig. 3.
Behavior of wild type and variants grown in biofilms. Confocal images of wild type (a) and mini (b) and wrinkly (c) variants expressing GFP; day 1 images are x–y views; scale, 10 μm. Day 2 and day 4 images, x–z views; dashed line represents biofilm attachment surface; scale, 50 μm. Results are representative of six experiments with each strain.
Fig. 4.
Biofilm phenotypes of variants. (a) Quantitative detachment rates of wild type and the mini- and wrinkly-variant biofilms. Data are means of three experiments; error bars show SEM. (b) Adherence of the wild type and wrinkly variant to the biofilm growth surface. Data are means of three experiments; error bars show SEM. (c) Susceptibility of pure-culture wild-type and wrinkly-variant biofilms to H2O2. Data are three replicates from one experiment and are representative of three others. Error bars show SEM.
The wrinkly variant we studied also functioned very differently from the wild type; however, in contrast to the mini variant, each step of biofilm development was accelerated: Initial attachment was increased, cell clusters formed earlier and were much bigger, and, by 5 days, the wrinkly-variant biofilm contained ≈100-fold more bacteria than the wild type (Figs. 3c and 4b). The wrinkly-variant biofilm also exhibited a 9-fold lower detachment rate than did the wild-type (Fig. 4a). In addition, antimicrobial susceptibility tests with comparably sized pure-culture biofilms (see Methods) showed that the wrinkly biofilm was more resistant to H2O2 (Fig. 4c), hypochlorite, and the antibiotic tobramycin (data not shown). In the planktonic state, no differences in susceptibility were seen. These experiments showed that the two variants we examined had specialized biofilm phenotypes, accelerated detachment (mini), and hyperbiofilm formation (wrinkly).
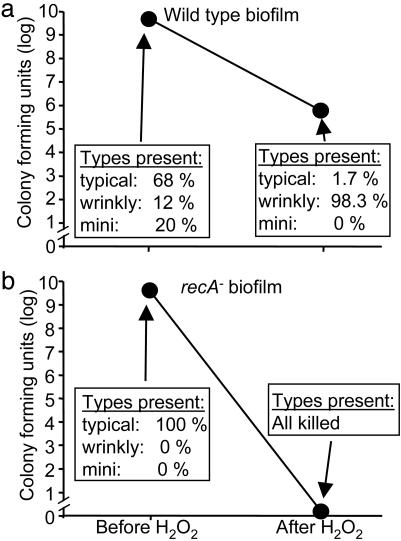
Studies in other biological systems indicate that functionally distinct subpopulations like these can produce insurance effects, particularly for communities under stress (23, 24, 35). To begin investigating whether the diversity arising in biofilms could produce insurance effects, we applied a stress to wild-type biofilms (that produced variant subpopulations) and biofilms formed by the recA mutant (that did not generate variants). An oxidative stress was chosen because bacteria frequently encounter this stress, and the experiments described above revealed that one subpopulation (wrinkly) was resistant. Treatment of wild-type biofilms with H2O2 reduced viable counts by ≈104, and, as expected, the vast majority of surviving bacteria were wrinkly variants (Fig. 5a). Biofilms formed by the recA mutant succumbed completely to the same treatment (Fig. 5b). Two points indicate that the increased resistance of wild-type biofilms is due to the presence of wrinkly variants rather than to some other action of recA. First, very few nonwrinkly bacteria within wild-type biofilms survived, even though these other subpopulations were more abundant, and all of them had functional recA genes (Fig. 5a). Second, the resistance phenotype of wrinkly-variant biofilms was independent of recA. This independence was demonstrated in susceptibility tests comparing pure-culture wild-type and wrinkly-variant biofilms (both of which were recA-competent, see Fig. 4c) and similar experiments with wild-type and wrinkly-variant biofilms in which recA had been inactivated (data not shown). Taken together, these studies suggest that biofilm communities can be strengthened by the presence of specialized subpopulations.
Fig. 5.
The presence of the wrinkly-variant subpopulation enhances biofilm resistance. (a) Number and types of bacteria in wild-type biofilms before and after exposure to H2O2.(b) Number and types of bacteria in recA– biofilms. No bacteria withstood H2O2 treatment. Data in a and b are means of four experiments; error bars indicating SEM are hidden by data points.
Discussion
Whether they are living in natural environments or as pathogens within hosts, bacterial populations are continually faced with adverse conditions. The biofilm growth mode confers many advantages to bacteria that are facing stress, including antimicrobial resistance and physical protection by the matrix, among others (2). Our findings reveal another important advantage: the rapid development of diversity among members of the biofilm community. This diversity, which develops within days of biofilm formation, occurs under a wide range of biofilm growth conditions and with a number of different laboratory and clinical isolates. The diversity affects several different cellular functions, including motility, nutritional requirements, production of a secreted product, colony morphology, and at least three biofilm phenotypes: hyperbiofilm formation (Fig. 3c), accelerated detachment (Fig. 3b), and increased biofilm-mediated resistance (Fig. 4c). Furthermore, we have shown that the presence of this diversity enhanced the survival of biofilms that were subjected to a common environmental stress. Biofilm communities that were unable to diversify succumbed completely when subjected to the same challenge.
Whereas these experiments demonstrated increased resistance of diverse biofilms to a particular stress (mediated by a specific subpopulation), the extent of the diversity we observed suggests that other advantages may also be produced. For example, bacteria that secrete pyomelanin may manifest increased tolerance to UV radiation and certain host defenses (34). Because this pigment is secreted, other community members could share in this protection. The hyperdetachment phenotype of the mini variant could benefit the community by enhancing dissemination and the colonization of new locations, especially because the biofilms that the mini variant founds give rise to diverse populations of bacteria. This variant may also be better able to escape local stresses such as nutrient limitation and the accumulation of wastes. One could also imagine circumstances in which the presence of bacteria with differing motility, increased adherence, hyperbiofilm formation, or divergent nutritional requirements could have advantages.
An important point is that the insurance benefits of diversity do not require that any given subpopulation have increased fitness overall. Indeed, the fitness of each specialized population is likely to depend on prevailing conditions; a given phenotype may be advantageous in certain circumstances and detrimental or neutral in others. The insurance hypothesis relates to the effects of diversity on the community as a whole. It predicts that a community composed of functionally diverse populations is likely to perform better in general because of the likelihood that some subpopulation will thrive as prevailing conditions change. In addition to increased resistance (which our studies addressed), evidence from a number of systems indicates that diversity can also enhance the productivity and long-term sustainability of communities (23, 24, 35, 36). Enhanced productivity and sustainability result from positive interactions between subpopulations and because communities composed of members with complementary (rather than superimposed) niches use available resources and habitats more effectively. It remains to be determined whether the diversity we have observed impacts the productivity and sustainability of biofilm communities.
In environmental ecosystems, the functional differences that produce insurance effects usually derive from species diversity (37, 38). Our studies demonstrate that a clonal bacterial population grown in biofilms for short periods can generate sufficient functional diversity to produce such benefits, even in the absence of species differences. Previous work by other investigators also suggests a link between biofilms and the functional diversification of bacteria. In one study (39), growth of an environmental Pseudomonas strain in biofilms on hexadecane droplets produced variants with a number of different phenotypes. Investigators studying phenotypic variation of the bacterial capsule in Streptococcus pneumoniae (40) used in vitro biofilms to generate capsular variants because broth cultures failed to generate them. In another study (41) using Pseudomonas fluorescens, colony variants were generated by growth in a heterogeneous laboratory microcosm. Notably, many of the bacteria in this model grew in biofilm-like mats, suggesting that common mechanisms might mediate variation in the P. fluorescens studies and in our experiments. Interestingly, colony variants, auxotrophs, and strains that overproduce melanin are commonly isolated from patients with cystic fibrosis and bronchiectasis, diseases in which P. aeruginosa lives in biofilms (42–44). Such variants are not seen in infections associated with planktonic growth (44, 45). Thus, extensive genetic variation appears to be generated by biofilms both in vitro and in vivo.
Whereas our experiments show that diversity is rapidly produced in biofilms, we do not yet know how it is generated. One possibility is that the rate of genetic variation is similar in the biofilm and planktonic cultures, and the diversity we observed is caused by powerful selective pressures inherent to biofilms. Although further work will be required, we do not favor this as the sole explanation for our findings because it seems unlikely that strong selective pressures for auxotrophy would exist within biofilms, and recA gene function is not typically required for spontaneous growth-dependent mutation caused by replication errors (46). Another possibility is that the rate of genetic variation is somehow increased in biofilms. This increase could be triggered by conditions within biofilms (e.g., the accumulation of DNA-modifying agents), or as a programmed response to the biofilm state. It is also possible that both genetic variation and selective pressures are increased in biofilms. Together, these factors could have a powerful compound effect.
Whatever mechanism is operative, diversity generated by biofilm growth may have implications beyond the insurance effects provided to biofilm communities. Many bacterial species have a strong proclivity for biofilm formation, particularly under stressful conditions. Our findings suggest that biofilms could serve as “hotbeds” of diversity and could promote adaptation to the harsh environments in which biofilms are commonly found. Such adaptation could be particularly important in chronic infections in which the ability to withstand severe and fluctuating conditions inside a host is essential for bacterial persistence.
Acknowledgments
We thank P. Bontu, E. Burtinel, T. O. Moninger, and M. Nevell for technical assistance and M. J. Welsh, M. R. Parsek, T. L. Yahr, and E. P. Greenberg for helpful discussions.
Author contributions: P.K.S. designed research; B.R.B., M.T., and P.K.S. performed research; and P.K.S. wrote the paper.
References
- 1.Hoiby, N., Krogh Johansen, H., Moser, C., Song, Z., Ciofu, O. & Kharazmi, A. (2001) Microbes Infect. 3, 23–35. [DOI] [PubMed] [Google Scholar]
- 2.Costerton, J. W., Stewart, P. S. & Greenberg, E. P. (1999) Science 284, 1318–1322. [DOI] [PubMed] [Google Scholar]
- 3.Lindsay, J. O., Ciesielski, C. J., Scheinin, T., Hodgson, H. J. & Brennan, F. M. (2001) J. Immunol. 166, 7625–7633. [DOI] [PubMed] [Google Scholar]
- 4.Parsek, M. R. & Singh, P. K. (2003) Annu. Rev. Microbiol. 677–701. [DOI] [PubMed]
- 5.Islam, M. S., Drasar, B. S. & Bradley, D. J. (1990) J. Trop. Med. Hyg. 93, 133–139. [PubMed] [Google Scholar]
- 6.Covert, T. C., Rodgers, M. R., Reyes, A. L. & Stelma, G. N., Jr. (1999) Appl. Environ. Microbiol. 65, 2492–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ridgway, H. F. & Olson, B. H. (1981) Appl. Environ. Microbiol. 41, 274–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lam, J., Chan, R., Lam, K. & Costerton, J. W. (1980) Infect. Immun. 28, 546–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh, P. K., Schaefer, A. L., Parsek, M. R., Moninger, T. O., Welsh, M. J. & Greenberg, E. P. (2000) Nature 407, 762–764. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi, H. (2001) Int. J. Antimicrob. Agents 17, 351–356. [DOI] [PubMed] [Google Scholar]
- 11.Levine, S. A. & Niederman, M. S. (1991) Clin. Chest Med. 12, 523–543. [PubMed] [Google Scholar]
- 12.Goto, T., Nakame, Y., Nishida, M. & Ohi, Y. (1999) Int. J. Antimicrob. Agents 11, 227–231, and discussion 237–239. [DOI] [PubMed] [Google Scholar]
- 13.Slusher, M. M., Myrvik, Q. N., Lewis, J. C. & Gristina, A. G. (1987) Arch. Ophthalmol. 105, 110–115. [DOI] [PubMed] [Google Scholar]
- 14.Stewart, P. S. (2002) Int. J. Med. Microbiol. 292, 107–113. [DOI] [PubMed] [Google Scholar]
- 15.Mahenthiralingam, E., Campbell, M. E. & Speert, D. P. (1994) Infect. Immun. 62, 596–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feltman, H., Schulert, G., Khan, S., Jain, M., Peterson, L. & Hauser, A. R. (2001) Microbiology 147, 2659–2669. [DOI] [PubMed] [Google Scholar]
- 17.Kelly, N. M., Kluftinger, J. L., Pasloske, B. L., Paranchych, W. & Hancock, R. E. (1989) Infect. Immun. 57, 3841–3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shapiro, J. A. (1998) Annu. Rev. Microbiol. 52, 81–104. [DOI] [PubMed] [Google Scholar]
- 19.Hausner, M. & Wuertz, S. (1999) Appl. Environ. Microbiol. 65, 3710–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davies, D. G., Parsek, M. R., Pearson, J. P., Iglewski, B. H., Costerton, J. W. & Greenberg, E. P. (1998) Science 280, 295–298. [DOI] [PubMed] [Google Scholar]
- 21.De Kievit, T. R., Gillis, R., Marx, S., Brown, C. & Iglewski, B. H. (2001) Appl. Environ. Microbiol. 67, 1865–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greenberg, E. P. (2003) Nature 424, 134. [DOI] [PubMed] [Google Scholar]
- 23.McCann, K. S. (2000) Nature 405, 228–233. [DOI] [PubMed] [Google Scholar]
- 24.Yachi, S. & Loreau, M. (1999) Proc. Natl. Acad. Sci. USA 96, 1463–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobs, M. A., Alwood, A., Thaipisuttikul, I., Spencer, D., Haugen, E., Ernst, S., Will, O., Kaul, R., Raymond, C., Levy, R., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 14339–14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogunnariwo, J. & Hamilton-Miller, J. M. (1975) J. Med. Microbiol. 8, 199–203. [DOI] [PubMed] [Google Scholar]
- 27.Barth, A. L. & Pitt, T. L. (1995) J. Clin. Microbiol. 33, 37–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang, C. T., Xu, K. D., McFeters, G. A. & Stewart, P. S. (1998) Appl. Environ. Microbiol. 64, 1526–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bagge, N., Schuster, M., Hentzer, M., Ciofu, O., Givskov, M., Greenberg, E. P. & Hoiby, N. (2004) Antimicrob. Agents Chemother. 48, 1175–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh, P. K., Parsek, M. R., Greenberg, E. P. & Welsh, M. J. (2002) Nature 417, 552–555. [DOI] [PubMed] [Google Scholar]
- 31.Kuzminov, A. (1996) BioEssays 18, 757–765. [DOI] [PubMed] [Google Scholar]
- 32.Shinagawa, H. (1996) EXS, 221–235. [DOI] [PubMed]
- 33.Ohmori, H., Friedberg, E. C., Fuchs, R. P., Goodman, M. F., Hanaoka, F., Hinkle, D., Kunkel, T. A., Lawrence, C. W., Livneh, Z., Nohmi, T., et al. (2001) Mol. Cell 8, 7–8. [DOI] [PubMed] [Google Scholar]
- 34.Nosanchuk, J. D. & Casadevall, A. (2003) Cell. Microbiol. 5, 203–223. [DOI] [PubMed] [Google Scholar]
- 35.Tilman, D., Wedin, D. & Knops, J. (1996) Nature 379, 718–720. [Google Scholar]
- 36.Tilman, D., Reich, P. B., Knops, J., Wedin, D., Mielke, T. & Lehman, C. (2001) Science 294, 843–845. [DOI] [PubMed] [Google Scholar]
- 37.Hooper, D. U. & Vitousek, P. M. (1997) Science 277, 1302–1305. [Google Scholar]
- 38.Tilman, D., Knops, J., Wedin, D., Reich, P., Ritchie, M. & Siemann, E. (1997) Science 277, 1300–1301. [Google Scholar]
- 39.Deziel, E., Comeau, Y. & Villemur, R. (2001) J. Bacteriol. 183, 1195–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waite, R. D., Struthers, J. K. & Dowson, C. G. (2001) Mol. Microbiol. 42, 1223–1232. [DOI] [PubMed] [Google Scholar]
- 41.Rainey, P. B. & Travisano, M. (1998) Nature 394, 69–72. [DOI] [PubMed] [Google Scholar]
- 42.Haussler, S., Tummler, B., Weissbrodt, H., Rohde, M. & Steinmetz, I. (1999) Clin. Infect. Dis. 29, 621–625. [DOI] [PubMed] [Google Scholar]
- 43.Haussler, S., Ziegler, I., Lottel, A., von Gotz, F., Rohde, M., Wehmhohner, D., Saravanamuthu, S., Tummler, B. & Steinmetz, I. (2003) J. Med. Microbiol. 52, 295–301. [DOI] [PubMed] [Google Scholar]
- 44.Sheehan, D. J., Janda, J. M. & Bottone, E. J. (1982) J. Clin. Microbiol. 15, 926–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oliver, A., Canton, R., Campo, P., Baquero, F. & Blazquez, J. (2000) Science 288, 1251–1254. [DOI] [PubMed] [Google Scholar]
- 46.Rosenberg, S. M. & Hastings, P. J. (2004) J. Bacteriol. 186, 4838–4843. [DOI] [PMC free article] [PubMed] [Google Scholar]