Abstract
Transcription factors (TFs) are essential regulators of gene expression, and mutated TF genes have been shown to cause numerous human genetic diseases. Yet to date, no single, comprehensive database of human TFs exists. In this work, we describe the collection of an essentially complete set of TF genes from one depiction of the human ORFeome, and the design of a microarray to interrogate their expression. Taking 1468 known TFs from TRANSFAC, InterPro, and FlyBase, we used this seed set to search the ScriptSure human transcriptome database for additional genes. ScriptSure's genome-anchored transcript clusters allowed us to work with a nonredundant high-quality representation of the human transcriptome. We used a high-stringency similarity search by using BLASTN, and a protein motif search of the human ORFeome by using hidden Markov models of DNA-binding domains known to occur exclusively or primarily in TFs. Four hundred ninety-four additional TF genes were identified in the overlap between the two searches, bringing our estimate of the total number of human TFs to 1962. Zinc finger genes are by far the most abundant family (762 members), followed by homeobox (199 members) and basic helix-loop-helix genes (117 members). We designed a microarray of 50-mer oligonucleotide probes targeted to a unique region of the coding sequence of each gene. We have successfully used this microarray to interrogate TF gene expression in species as diverse as chickens and mice, as well as in humans.
Transcription factors (TFs) constitute a large and diverse group of regulatory proteins that, in the typical case, bind to DNA relatively close to a target gene to activate or repress its transcription. Gene regulation via TF binding is the primary mechanism by which complex processes of development and differentiation are controlled. In a recent review of 144 human developmental disorders in which the function of the causative gene had been identified, 49 (34%) were due to mutated TF genes, a number nearly double that of the next largest class (Boyadiev and Jabs 2000). The activity of TFs that are developmental regulators is commonly controlled at the level of mRNA synthesis (Semenza 1998). In contrast, many of the TFs that regulate physiological targets are constitutively present, and their activity is determined by posttranslational modifications such as phosphorylation (Semenza 1998; Brivanlou and Darnell 2002). Nevertheless, some of these latter classes of TFs have also been found to be developmentally regulated at the transcriptional level (Ranger et al 1998; Crabtree 1999).
There are two major types of TFs in eukaryotes. The first are those that participate in the ordered assembly of RNA polymerase II transcription-initiation complexes, which include the general TFs, coactivators, corepressors, and chromatin and histone modifiers. The second type are those that activate or repress the trans cription of particular genes directly by binding to characteristic regulatory sites (for reviews, see Lemon and Tjian 2000; Brivanlou and Darnell 2002). The groups that sequenced the human genome estimated that there are between two and three thousand TFs in the human genome (International Human Genome Sequencing Consortium 2001; Venter et al. 2001). However, this was only an estimate, and to date, no study has cataloged the entire TF gene content of man.
In the current report, we sought to identify the vast majority of human TFs with the intention of using this collection of sequences to design a more comprehensive microarray than is currently available for expression profiling this important set of regulatory genes. Furthermore, by designing oligonucleotide probes from the coding regions of these genes, we sought to use this same array to study TF expression in closely related organisms, such as mouse and chicken.
RESULTS
The initial informatics component of this study consisted of two steps: collecting a set of known human TFs (a seed set) and then using that seed set to search for additional uncharacterized TFs in the human transcriptome and ORFeome as defined by the ScriptSure collection of genome-nucleated expressed sequence tags (ESTs).
Known Human TFs
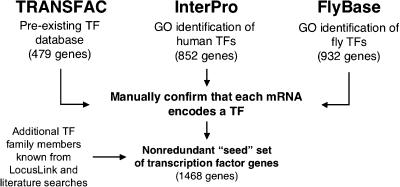
Figure 1 shows our strategy for building a set of known human TFs. We gathered known genes from three sources: TRANSFAC (http://www.gene-regulation.com; Wingender et al. 2000), InterPro (http://www.ebi.ac.uk/interpro/; Apweiler et al. 2001), and FlyBase (FlyBase Consortium 1999). TRANSFAC was chosen as a starting point because it is a pre-existing database devoted to the listing and binding characteristics of numerous TFs. The freely available version of TRANSFAC contained records for 479 human TF genes. InterPro was searched for human proteins that met the Gene Ontology (GO; http://www.geneontology.org) project definition of a TF (The Gene Ontology Consortium 2000). This resulted in the identification of 852 InterPro entries. We next turned to a well-studied organism with a fully sequenced genome that had been annotated with GO identifiers. At the time this work was performed, the human genome and the GO annotations of it were incomplete. The best available GO-annotated eukaryotic genome was Drosophila melanogaster, as represented in FlyBase (http:///flybase.bio.indiana.edu/). FlyBase contained 932 genes annotated as encoding TFs. For most of these, FlyBase listed human orthologs, and those it omitted were obtained via Homologene (Zhang et al. 2000).
Figure 1.
Creation of the seed set. Known TF genes were gathered from three databases: TRANSFAC, InterPro, and FlyBase. Each gene was manually confirmed to be described as a TF in the literature or annotated as a TF in LocusLink. After removing redundancies and adding some known TFs that were not present in our source databases, our seed set of human TFs contained 1468 members.
Genes gathered from the three sources were merged into one list, and redundancies were eliminated. In some cases, not every member of a known TF gene family was present in the nonredundant set. For example, LocusLink lists eight known human members of the chromobox gene family (CBX1-8), but we identified only three of these from TRANSFAC, InterPro, and FlyBase combined. In this case we manually added the five missing CBX genes to our initial list. At this stage we also eliminated core components of RNA and DNA polymerases from our seed set, because these are ubiquitously and highly expressed. The resulting nonredundant seed set of human TF genes contained 1468 members.
Identifying Homologous Transcript Clusters
To identify more potential TF genes, we searched the ScriptSure transcript database (http://sapiens.wustl.edu/ScriptSure; J. Glasscock and W. Gish, in prep.). ScriptSure is a database of genome-anchored human transcript clusters (ESTs, mRNAs, and RefSeqs). ScriptSure clusters were built by using the genomic DNA sequence as a scaffold onto which transcripts were then aligned. Contaminants such as chimeric sequences and incorrect submissions were thus filtered out when they failed to correctly align to the genomic sequence. After passing strict criteria of minimum length and similarity to the genome, each EST was assigned to the locus with the highest scoring alignment in the genome, thereby reducing cases in which highly similar but distinct sequences merged into a single cluster. Overlapping EST-to-genome alignments created a genome-anchored transcript cluster. The underlying high-quality genomic sequence was used as the cluster consensus. These factors translate into a database that is less redundant, less contaminated, and composed of higher-quality sequence than are other available databases. We chose to search only the ScriptSure clusters that were annotated as spliced with multiple underlying transcripts to eliminate the spurious identification of processed pseudogenes (frequent for TFs in the human genome; Harrison et al. 2002; Zhang et al. 2003) and to eliminate rare and possibly spurious transcription events. Although this step ensures a high-quality data set, it will also inevitably result in a failure to identify any TFs that are encoded by single-exon genes, or TFs that are only rarely represented in the EST databases (for more on these points, see Discussion).
We were able to match 1361 of our 1468 TF sequences with ScriptSure clusters (92.7%). Thus, we failed to identify 107 genes out of our seed set within ScriptSure. This occurred because the version of ScriptSure we used was built on a draft genome assembly (International Human Genome Sequencing Consortium 2001) in which there were still gaps. However, this number is useful, because it provides us with an estimate of the rate of false negatives in our overall BLASTN analysis (7.3%).
Two methods of searching ScriptSure were used: a high-stringency BLASTN (http://blast.wustl.edu) of ScriptSure with each seed TF, and a query of each ScriptSure transcript, conceptually translated, against a collection of TF DNA-binding hidden Markov models (HMMs) extracted from Pfam (Fig. 2). We reasoned that the combination of these two approaches would provide a balance between sensitivity and specificity. The BLASTN search alone would yield false positives, and requiring each candidate to have a TF's DNA-binding domain would alleviate that problem. The conservative combination of these two search methods will, however, result in some false negatives (discussed below) and an overall slight underestimate of the total TF gene content.
Figure 2.
Search for paralogous TFs. By using the seed set of 1468 known human TF genes, we searched ScriptSure, a representation of the human transcriptome, using two methods: a high-stringency BLASTN search and an hmmpfam search for DNA-binding domains known to occur exclusively or primarily in TFs. The BLASTN search netted 3338 additional potential TFs, the domain search 2512. There were 494 genes that were found with both search methods; these 494 comprise the “found” set of human TF genes.
A repeat-masked version of the 1468 TF seed sequences was used as a BLASTN query against the complete ScriptSure database. The query identified 5130 potential new TF candidate clusters that met our criteria. The bit score cutoff used in the analysis was determined from our analysis of a bit score distribution of HOX gene family members (see Methods). We next filtered out clusters that contained no introns (possible spurious alignments to processed pseudogenes) and clusters that were represented by only one or two aligned ESTs. The number of newly identified clusters that passed through this filter was 3338.
Further Selection Through TF Pfam Signatures
We were concerned that our ScriptSure BLASTN searches would yield false positives from homologies within conserved domains that are not TF-specific, such as protein-protein interaction domains. Therefore, we sought to select the clusters that contained bona fide TF protein motifs. The database used in this analysis was a subset of Pfam, containing DNA-binding domains found in TF genes (see Methods). We searched the entire set of “Spliced| Multiple” ScriptSure clusters (i.e., >22,000 clusters and not just the 3338 clusters identified by BLASTN) for Pfam TF DNA-binding motifs and found 3748 clusters that contained at least one TF protein motif. These clusters included 1236 out of the 1369 seed set present in ScriptSure, indicating that this Pfam method has a false-negative rate of ∼10%. Therefore, the Pfam search identified 2512 new putative TFs. Because the HMM used in our Pfam search used a relatively low cutoff for motif similarities, we expected to also detect false positives by this route. To remove these and the BLASTN false positives, we tested for overlaps between the results from the two search methods. The overlap between the 3338 BLASTN clusters and the 2512 Pfam clusters comprised 494 clusters (Fig. 2). These 494 newfound clusters, plus the 1468 seed set constituted a total of 1962 potential TF gene sequences. Interestingly, when all of the ESTs comprising the 1962 potential TF sequences were compared with all spliced EST clusters from ScriptSure, we found that the medians of the two distributions were significantly different. The putative TF set had a threefold higher level of alternative splice forms when compared with the non-TF set, suggesting that extensive potential isoform diversity may be encoded by this set as a whole. These genes and their accession numbers are listed in the online Supplemental material.
Genomic Localizations of TF Genes
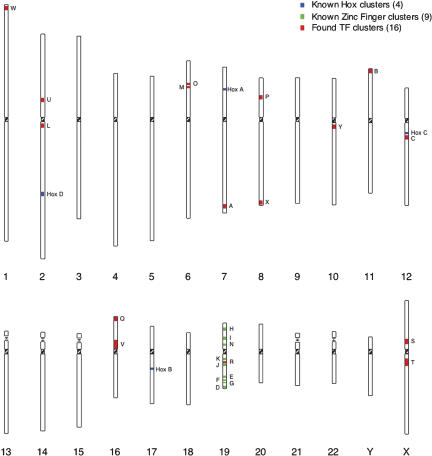
Tight clustering of genes is sometimes indicative of coregulated gene expression (Boutanaev et al. 2002; Lercher et al. 2002). For TFs in particular, there are precedents for biologically significant clusters of HOX genes on human chromosomes 7, 17, 12, and 2. We sought to determine if the newly identified set of putative TF genes were distributed randomly throughout the human genome or were found to cluster at discrete chromosomal locations.
Because heterochromatic and euchromatic regions of the genome are known to be relatively gene-poor and gene-rich, respectively, apparent clustering of genes was expected (International Human Genome Sequencing Consortium 2001). Taking this into consideration, we measured how often we observed three or more TF genes appearing in a window of eight clusters, translating to a probability of 0.37 under a binomial model (Fig. 3). This analysis identified 29 regions that passed the criteria. Four of these regions were Hox clusters, and another nine clusters were attributed to zinc finger clusters on chromosome 19 (19p12, 19q13.2, and 19q34; Eichler et al. 1998). The remaining 16 clusters had little underlying annotation of their transcript members. Interestingly, a comparison of the 29 clusters with the completed mouse genomic DNA sequence revealed that 18 out of the 29 were conserved in the mouse, supporting the notion that there may be functional reasons for some of this clustering. For a list of the genes comprising each putative cluster, see Supplemental materials.
Figure 3.
Genomic locations of TF clusters. Clusters of TF genes are shown on an ideogram representation of the 24 human chromosomes. As shown in the legend at the top right, the four canonical Hox gene clusters are shown in blue, the previously described chromosome 19 zinc finger gene clusters are shown in green, and putative TF clusters identified in this study are shown in red. The number in parentheses following each in the legend indicates the number of each type of cluster shown in this figure. Clusters containing known genes are labeled. Labels are not included for hypothetical and unnamed genes, and so clusters consisting entirely of these are unlabeled. For a list of the genes comprising each cluster, see Supplemental materials.
Microarray Design
From the set of 1962 TF genes, we designed a microarray of 50-mer oligonucleotide probes with which to interrogate their expression. One of the major issues in designing a microarray of TFs is that many of these genes fall into families that share significant regions of conserved sequence homology. For example, there are >500 TFs that contain zinc finger domains (Eichler et al. 1998; this study, see below). To design a probe that will measure the expression of only one gene, it was necessary to identify sequence regions in each gene that were unique to it. The obvious choice in this situation is to target the 3′ untranslated region (3′ UTR), which is usually the most evolutionarily divergent region in a transcript. However, our intention in building a TF microarray was to use it across species, at least for organisms that are evolutionarily close enough to retain a high degree of sequence similarity, such as the mouse and the chicken. Therefore, we chose to design 50-mer oligonucleotide probes from within each coding region (determined by conceptual translation of each putative TF) and as 3′ as possible within the coding sequence. It should be noted that designing probes far away from the 3′ ends of genes may result in a significant loss of sensitivity when used with 3′ biased amplification protocols. One way to circumvent this potential limitation is to use alternative amplification methods such as full-length amplification (Castle et al. 2003). We also selected these probes to be matched for Tm (Li and Stormo 2001). These probes are listed in Supplemental materials. This array has been successfully used to interrogate TF gene expression across species as distant as chicken, mouse, and man (Hawkins et al. 2003).
DISCUSSION
In this study we collected a set of known human TFs and used two complementary computational methods to search the human transcriptome for the entire set of human TF genes. Our analysis identified 1962 putative TF genes, a number that correlates well with previous estimates (International Human Genome Sequencing Consortium 2001; Venter et al. 2001).
This number is an estimate, but our seed set searches provide us with some idea of the error rates in that estimate. To count a gene as a TF, we required that it must either be previously annotated or described as such in the literature (the seed set), or be paralogous to a known TF gene and contain a DNA-binding domain known to occur exclusively or primarily in TFs (the found set). The term “transcription factor” encompasses many types of proteins. Those factors that do not bind directly to DNA are likely to be underrepresented in our analysis. However, our seed set did contain some non-DNA-binding TFs that had been experimentally verified and described in the literature.
There are four variables that influence our estimate of total TF genes: gaps in the human genome sequence, the incompleteness of Pfam, the degree of comprehensiveness of dbEST, and the exclusion of single-exon predicted genes from our analysis. Our finding that ∼7% of our 1468 input seed set failed to find matches in the genome-nucleated ScriptSure database indicates that we may be missing ∼35 genes in our “new” set of 494 at this filtering step. We also found that ∼10% of our seed set ScriptSure matches failed the Pfam criteria. Among these are oddities such as Myc-binding protein 1 (MBP-1), an alternate translation product of the glycolytic enzyme α-enolase (ENO1; Feo et al. 2000), and a bona fide TF that has been shown to bind to the c-myc P2 promoter and repress transcription of a reporter gene (Ray and Miller 1991; Subramanian and Miller 2000). Querying the MBP-1 mRNA sequence against the Pfam database, even at low stringency, yields no match to a known DNA-binding domain (data not shown). We would estimate that perhaps a further 50 genes (10% of the 494) may have failed this Pfam criteria. We can also make some estimate of the error rates inherent in confining our analysis to multi-exon genes and multiple ESTs. Multi-exon genes encode ∼92% of our original seed set genes. Choosing to work with the higher confidence spliced clusters in our analysis may have resulted in excluding ∼8% of the single exon TF genes in our found set. This 8% translates to 40 genes. EST databases appear to be quite comprehensive in terms of TF coverage. A survey of the rate of new TF discovery (using our seed set ScriptSure clusters as a benchmark) indicates that 92% of our clusters were represented as early as 1997 (data not shown). Rate of discovery after this year has declined drastically. Nevertheless, it remains possible that some transiently expressed or low abundance TF mRNAs remain to be discovered. Taken together, these variables indicate that our analysis may have missed ∼130 TF genes (6.6% of our total estimate).
Table 1 shows a summary of the human TF genes we have found, classified by family. This table also shows the TF gene content of three other completed eukaryotic genomes: Saccharomyces cerevisiae, Caenorhabditis elegans, and Drosophila melanogaster. Zinc-binding TFs constitute the largest class, amounting to over one-third of all human TF genes. Homeobox-containing genes are the second largest family, with ∼200 members. The forkhead family has also expanded substantially in humans compared with the other species. This latter finding is of particular interest, as many of the metazoan-specific forkhead genes show tissue-specific expression and are involved in cell-type determination and differentiation (for review, see Carlsson and Mahlapuu 2002). For example, in the vertebrate embryo, forkhead genes are required for the development of the notochord (Hoodless et al. 2001; Yamamoto et al. 2001) and patterning of paraxial mesoderm and somites (Kume et al. 2001). Another forkhead gene (FOXP2) has been implicated in the ability to develop language (Enard et al. 2002). The “other” category in Table 1 contains a mixture of genes, mostly members of the core transcriptional machinery, and the “structural” category includes genes that are thought to regulate transcription by altering chromatin structure, such as the HMGA family (for review, see Reeves 2001).
Table 1.
A Comparison of Transcription Factors in Selected Eukaryotes
|
Homo sapiens
|
||||||
|---|---|---|---|---|---|---|
| Gene family | Seed | Found | All | S.c. | C.e. | D.m. |
| Zinc bindinga,b | 422 | 340 | 762 | 139 | 309 | 420 |
| Homeobox | 186 | 13 | 199 | 9 | 84 | 103 |
| BHLH | 92 | 25 | 117 | 8 | 25 | 46 |
| β-Scaffolda | 77 | 10 | 87 | 11 | 34 | 45 |
| BZip | 59 | 13 | 72 | 21 | 25 | 21 |
| NHR | 49 | 0 | 49 | 0 | 252 | 21 |
| Trp cluster | 38 | 8 | 46 | 10 | 13 | 14 |
| Forkhead | 36 | 4 | 40 | 4 | 15 | 18 |
| Bromodomaina | 14 | 3 | 17 | 10 | 13 | 16 |
| T-box | 16 | 1 | 17 | 0 | 21 | 8 |
| Jumonji | 6 | 7 | 13 | 1 | 1 | 2 |
| E2F | 9 | 1 | 10 | 0 | 4 | 3 |
| Dwarfin | 9 | 0 | 9 | 0 | 3 | 3 |
| Paired box | 9 | 0 | 9 | 0 | 7 | 5 |
| Heat shock | 6 | 2 | 8 | 5 | 1 | 1 |
| Tubbya | 5 | 2 | 7 | 0 | 1 | 1 |
| AF-4b | 7 | 0 | 7 | 0 | 0 | 2 |
| RFX | 6 | 0 | 6 | 1 | 1 | 1 |
| Methyl-CpG-bindinga | 4 | 1 | 5 | 0 | 2 | 4 |
| AP-2 | 4 | 0 | 4 | 0 | 4 | 1 |
| TEAa | 4 | 0 | 4 | 5 | 5 | 5 |
| Pocket domain (Rb)a | 3 | 0 | 3 | 0 | 1 | 2 |
| GCMc,d | 2 | 0 | 2 | 0 | 0 | 2 |
| Other | 214 | 14 | 228 | — | — | — |
| Coactivators and corepressors | 111 | 11 | 122 | — | — | — |
| Structural | 80 | 39 | 119 | — | — | — |
| Total | 1468 | 494 | 1962 | 224 | 821 | 744 |
The set of transcription factors is shown for four species, divided into families by the type of DNA-binding domain present and sorted by abundance in human. The data for human transcription factors show the seed and found set numbers separately, as well as the total number from the two sets added together. The Homo sapiens data are from this study. Unless otherwise specified, the data for S. cerevisiae, C. elegans, and D. melanogaster are from Riechmann et al. (2000). bHLH indicates basic helix-loop-helix; bZip, basic leucine zipper; C.e., Caenorhabditis elegans; D.m., Drosophila melanogaster; GCM, glial cell missing; NHR, nuclear hormone receptor; RFX, regulatory factor X; S.c., Saccharomyces cerevisiae; TEA, transcriptional enhancer activator (TEA/ATTS); and Trp cluster, tryptophan cluster.
S. cerevisiae, C. elegans, and D. melanogaster data on zinc binding subfamilies AN1, BTB/POZ-containing, MYND, and PHD, β-scaffold subfamily cold shock, bromodomain family, Tubby family, methyl-CpG-binding family, TEA family, and pocket domain (Rb) family from Rubin et al. (2000); supplemental information (http://www.sciencemag.org/feature/data/1049664.shl).
S. cerevisiae, C. elegans, and D. melanogaster data on zinc binding subfamily MIZ and AF-4 family from “species distribution” feature of Pfam Web site (http://pfam.wustl.edu).
D. melanogaster data on GCM family from Akiyama et al. (1996).
S. cerevisiae and C. elegans data on GCM family from “species distribution” feature of Pfam Web site (http://pfam.wustl.edu).
Looking at the data listed in Table 1 in another way, we can estimate the proportion of the total transcriptome that encodes TFs in each of the four species (Table 2). We do not know the precise gene content of all of these species, but assuming that current numbers are approximately correct, it appears that TFs account for between 8% and 9% of all human genes. As one might expect, we see an upward trend in the proportion of TFs as the complexity of the organism increases. The development of more, and more finely tuned, regulatory mechanisms in higher eukaryotes has been hypothesized to explain their greater biological sophistication (Huang et al. 1999; Claverie 2001). This idea has been expounded upon in many recent studies in which “evolvability” (Kirschner and Gerhart 1998) and the evolution of development (Jacob 2001; Revilla-I-Domingo and Davidson 2003; Wray 2003) have been linked to increased complexity in regulatory networks. Our observation that humans devote at least 8% of their ORFeome to primary regulators of transcription (a number that is probably an underestimate given the levels of alternative splicing in TF genes) is consistent with the idea that developmental and body plan complexity are related to complexity in transcriptional regulatory networks.
Table 2.
Transcription Factors as a Proportion of Total Gene Content
| Organism | Approximate number of genes (TFs/total) | Percent |
|---|---|---|
| S. cerevisiae | 224/6569a | 3.4% |
| C. elegans | 824/19546b | 4.2% |
| D. melanogaster | 744/13525c | 5.5% |
| H. sapiens (NCBI) | 1962/24652d | 8.0% |
| H. sapiens (Ensembl) | 1962/21787e | 9.0% |
The number of transcription factor genes for S. cerevisiae, C. elegans, D. melanogaster, and H. sapiens is shown, divided by the approximate total number of genes for each organism, to estimate the percentage of the total gene content that transcription factor genes represent. Two estimates are given for H. sapiens, based on current gene predictions from Ensembl and NCBI. Transcription factor gene counts are the same as in Table 1. Total gene count sources are as follows: aSGD (July 2003), http://www.yeastgenome.org/VL-FAQ.html; bEnsembl C. elegans v19.102.1 (December 16, 2003), excluding 442 pseudogenes, http://www.ensembl.org/Caenorhabditis_elegans/stats/; cEnsembl D. melanogaster v19.3a.1 (January 7, 2003), http://www.ensembl.org/Drosophila_melanogaster/stats/; dNCBI human genome assembly build 34 (July 2003), http://www.ncbi.nlm.nih.gov/mapview/stats/BuildStats.cgi?taxid=9606&build=34&ver=1, and eEnsembl v19 build 34a (December 16, 2003), excluding 1744 pseudogenes, http://www.ensembl.org/Homo_sapiens/stats/.
The TF microarray we describe here is a versatile tool widely applicable to many areas of biological research. Two key features distinguish it from other microarrays. First, no other array described to date has probes for measuring the expression of as many TF genes. The Affymetrix U133 Genechip set, a large and commonly used human gene microarray, contains probes for ∼85% of the TF genes represented on this array (C. Helms, D. Messina, and M. Lovett, unpubl.). Second, although our probes were designed by using human DNA sequences, they represent coding sequences and not 3′ UTR. Thus, this microarray can (and has) been used to successfully measure TF gene expression in other species, including mouse and chicken (Hawkins et al. 2003). The evolutionary distance to the last common ancestor of human and chicken is ∼310 million years (Ureta-Vidal et al. 2003). Based on this distance, we would expect the TF array to be useful for studies in many vertebrate species, including chimpanzee, rhesus monkey, rat, dog, cat, horse, pig, cow, and sheep. A comparison of a random sampling of 50 TF genes from chicken and zebrafish to our collection of oligonucleotides revealed an average of 84% nucleotide identity for chicken and 79% nucleotide identity for zebrafish (diverged by 450 million years from human). Thus, this array may also prove useful for more divergent species such as zebrafish, pufferfish, and frog. However, we would urge caution in applying this tool to species more diverged than chicken. In these cases the rate of false negatives will increase (i.e., 50-mer oligonucleotides that fail to match their orthologous gene) and decreased hybridization stringencies will lead to an overall compression of dynamic range. Careful validation steps, and tuning of hybridization conditions, are required in all of these cross-species applications.
METHODS
Seed List
To build an initial set of TFs for the array, we gathered records from TRANSFAC (version 4.09-public; Wingender et al. 2000). The version of TRANSFAC we used did not have references to commonly used sequence identifiers, such as SWISS-PROT or GenBank sequence records. Therefore, we took gene names and descriptions from TRANSFAC records and correlated them by hand with GenBank records, from which we were able to obtain mRNA sequences. It should be noted that the latest version of TRANSFAC (TRANSFAC 6.0; Matys et al. 2003) contains more entries than the version we used, but when different splice forms are eliminated, these all appear to overlap with our final set of genes. Additional human TFs were extracted from InterPro (Apweiler et al. 2001) and FlyBase Consortium (1999). We searched InterPro for all records annotated as “human” and occurring at or below the “transcription factor” node of the GO hierarchy (GO ID 0003700). InterPro records contain protein, not mRNA sequence. We therefore used LocusLink (http://www.ncbi.nlm.nih.gov/LocusLink/) to identify the GenBank mRNA records that correspond to the EMBL protein identification in the InterPro record. If available, we chose mRNA sequences from the RefSeq database (Pruitt and Maglott 2001); RefSeqs comprise the majority of our set (1270/1468). Otherwise, we took the most complete GenBank mRNA sequence or EST representing that gene (198/1468). For four genes (GSH1, HMX3, DLX1, and CHX10), the best available sequences were RefSeq gene models (see http://www.ncbi.nlm.nih.gov/LocusLink/refseq.html for description). Thirteen genes were extracted from genomic sequence, and two (GSC and HMX2) were obtained from an Ensembl gene model (http://www.ensembl.org). For zinc finger genes, we manually removed subclasses of zinc finger proteins that are known not to bind DNA. However, it is possible that some of the zinc binding TFs in our final set will later be determined to be non-DNA-binding.
Additional known TF family members not identified by the above procedures were identified by using LocusLink or extensive literature searches and added to our database. Once data from these multiple sources have been collected, duplicates were eliminated, yielding a set of 1468 known human TFs (http://hg.wustl.edu/lovett/projects/nohr/Tfarray.html/).
Homologous Transcript Clusters
Each initial seed list member (Fig. 1) was matched to its best ScriptSure cluster, requiring a lower bound of 90% coverage of the seed sequence and 80% identity in the alignment between the seed sequence and the ScriptSure cluster. The 1369 seed sequences found to have a matching ScriptSure cluster were then masked for repetitive sequence. RepeatMasker with the parameters “-w -s -no_is -xsmall” was used for one round of masking, identifying interspersed repeats. RepeatMasker was used again in a separate round of masking by using the parameters “-s -noint -no_is -xsmall”; this works best for masking simple repeats. Lastly, low complexity sequence was masked by using nseg with the parameters “12 1.0 1.0 -z 1 -x”. A merge was performed on the results of these three masks, resulting in one tri-masked sequence for each of the 1369 seed sequences.
The tri-masked sequences were used in a BLASTN query against the complete ScriptSure database. WU-BLAST BLASTN 2.0 was used with the parameters “-S = 200 S2 = 100 gapS2 = 200 X = 26 gapX = 55 W = 11 gapW = 18 gapall Q = 11 R = 11 M = 5 N = -11 Z = 300000000 Y = 3000000000 V = 10000 B = 10000 gi novalidctxok nonnegok hspsepqmax = 200000 gapsepqmax = 1000000 lcmask topcomboN = 1 hspmax = 5000 -wordmask dust -mask-extra 15”. The seed set genes were subtracted out of the “Spliced|Multiple” results leaving 3338 potential additional TFs identified by the BLASTN search.
Bit Score Cutoff
The cutoff for determining positives in our BLASTN analysis was determined from a bit score distribution of known positives and known negatives. By using the well-characterized HOX gene clusters, ScriptSure clusters for each of the genes in the HOXA, HOXB, HOXC, and HOXD clusters were identified. Each of the HOX ScriptSure clusters was used as a query against the complete ScriptSure database. All clusters identified in the analysis (along with their bit scores) were put into one of two categories; HOX or non-HOX. An equivalency bit score, in which the number of false positives equaled the number of false negatives, was determined. This bit score was used as the cutoff in our BLASTN portion of the analysis.
Pfam TF Clusters
The starting point for this analysis was all the ScriptSure clusters that originated from spliced transcript clusters with multiple underlying transcripts (a total of 22,086 clusters). These clusters were translated in all six reading frames to amino acid sequences. The translated products were then searched by using hmmpfam from the HMMER 2.2 package (http://hmmer.wustl.edu). The database used in this query was a subset of the Pfam 7.1 database, the members (398) of which were annotated with the terms “DNA binding” or “transcription” (http://pfam.wustl.edu); 3748 clusters of the 22,086 clusters searched (17%) were found to match the transcription subset of the Pfam database with a P value of ≤0.0001; 2512 of these were clusters that did not match our seed list.
Mapping TFs to Genomic Contigs
The Human Genome Consortium's June 2002 draft of the human genome was used as the template for the version of ScriptSure we used in our analysis (June 2002b). Therefore, this same draft of the genome was used to place our identified clusters back onto the genome. The TF genes in our seed set as well as those identified in the overlap of the BLASTN and Pfam analysis were mapped back to the genome (1968 clusters total). Because ScriptSure reports its cluster coordinates relative to genomic contigs (rather than chromosome coordinates), UCSC's “lft” file was used to translate between contig and chromosome coordinates (http://genome.ucsc.edu). Loci were considered significant if three TF clusters were found in a colinear cluster of eight total clusters (P = 0.37 under binomial model).
Oligonucleotide Probe Design
For each TF we identified, we designed a 50-mer to represent that gene on our array. We designed probes with four criteria: (1) the probe must be from a unique region of the sequence of a gene to eliminate potential cross-hybridization to other genes; (2) to allow use of the array on nonhuman samples, the probe must be from protein coding sequence (CDS); (3) the design was targeted to a region of coding sequence as 3′ as possible; and (4) the probes were matched for melting temperatures (Tm). The vast majority of probes had a Tm of 72°C, with very occasionally a probe varying by as much as 3°C when severe design constraints existed. We were able to automate the selection of probes meeting criteria 1 and 4 with a microarray probe design program, Probes2 (Li and Stormo 2001); the other steps were semiautomated with custom Perl scripts. The Sanger Centre/Ensembl set of 27,395 verified human cDNA sequences (downloaded on July 14, 2001, current version available at ftp://ftp.ensembl.org/pub/current_human/data/fasta/cdna/) was used in conjunction with Probes2 to identify unique regions of each gene and design 50-mer probes. After candidate probes meeting these criteria were generated, we performed BLASTN similarity searches (default parameters) against the human genome sequence and inspected the results manually to confirm all criteria were met. Probes were synthesized (Sigma Genosys), resuspended at 60 μM in 1.5 M betaine and 6% DMSO, and spotted in duplicate on poly-L-lysine coated microscope slides with a GMS-417 arrayer (Affymetrix).
Acknowledgments
This work was supported in part by a grant from the National Organization for Hearing Research Foundation (M.L.) and a training grant from the National Institutes of Health National Human Genome Research Institute (supporting D.M. and J.G.).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2584104.
References
- Akiyama, Y., Hosoya, T., Poole, A.M., and Hotta, Y. 1996. The gcm-motif: A novel DNA-binding motif conserved in Drosophila and mammals. Proc. Natl. Acad. Sci. 93: 14912-14916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler, R., Attwood, T.K., Bairoch, A., Bateman, A., Birney, E., Biswas, M., Bucher, P., Cerutti, L., Corpet, L., Croning, M.D.R., et al. 2001. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 29: 37-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutanaev, A., Kalmykova, A.I., Shevelyov, Y.Y., and Nurminsky, D. 2002. Large clusters of co-expressed genes in the Drosophila genome. Nature 420: 666-669. [DOI] [PubMed] [Google Scholar]
- Boyadiev, S.A. and Jabs, E.W. 2000. Developmental biology: Frontiers for clinical genetics. Clin. Genet. 57: 253-266. [DOI] [PubMed] [Google Scholar]
- Brivanlou, A.H. and Darnell Jr., J.E. 2002. Signal transduction and the control of gene expression. Science 295: 813-818. [DOI] [PubMed] [Google Scholar]
- Carlsson, P. and Mahlapuu, M. 2002. Forkhead transcription factors: Key players in development and metabolism. Dev. Biol. 250: 1-23. [DOI] [PubMed] [Google Scholar]
- Castle, J., Garrett-Engele, P., Armour, C.D., Duenwald, S.J., Loerch, P.M., Meyer, M.R., Schadt, E.E., Stoughton, R., Parrish, M.L., Shoemaker, D.D., et al. 2003. Optimization of oligonucleotide arrays and RNA amplification protocols for analysis of transcript structure and alternative splicing. Genome Biol. 4: R466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverie, J.M. 2001. Gene number: What if there are only 30,000 human genes? Science 291: 1255-1257. [DOI] [PubMed] [Google Scholar]
- Crabtree, G.R. 1999. Generic signals and specific outcomes: Signaling through Ca2+, calcineurin, and NF-AT. Cell 96: 611-614. [DOI] [PubMed] [Google Scholar]
- Eichler, E.E., Hoffman, S.M., Adamson, A.A., Gordon, L.A., McCready, P., Lamerdin, J.E., and Mohrenweiser, H.W. 1998. Complex β-satellite repeat structures and the expansion of the zinc finger gene cluster in 19p12. Genome Res. 8: 791-808. [DOI] [PubMed] [Google Scholar]
- Enard, W., Przeworski, M., Fisher, S.E., Lai, C.S.L., Wiebe, V., Kitano, T., Monaco, A.P., and Pääbo, S. 2002. Molecular evolution of FOXP2, a gene involved in speech and language. Nature 418: 869-872. [DOI] [PubMed] [Google Scholar]
- Feo, S., Arcuri, D., Piddini, E., Passantino, R., and Giallongo, A. 2000. ENO1 gene product binds to the c-myc promoter and acts as a transcriptional repressor: Relationship with Myc promoter-binding protein 1 (MBP-1). FEBS Lett. 473: 47-52. [DOI] [PubMed] [Google Scholar]
- FlyBase Consortium. 1999. The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res. 27: 85-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. 2000. Gene Ontology: Tool for the unification of biology. Nat. Genet. 25: 25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, P.M., Hegyi, H., Balasubramanian, S., Luscombe, N.M., Bertone, P., Echols, N., Johnson, T., and Gerstein, M. 2002. Molecular fossils in the human genome: Identification and analysis of the pseudogenes in chromosomes 21 and 22. Genome Res. 12: 272-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins, R.D., Bashiardes, S., Helms, C.A., Hu, L., Saccone, N.L., Warchol, M.E., and Lovett, M. 2003. Gene expression differences in quiescent versus regenerating hair cells of avian sensory epithelia: Implications for human hearing and balance disorders. Hum. Mol. Genet. 12: 1261-1272. [DOI] [PubMed] [Google Scholar]
- Hoodless, P.A., Pye, M., Chazaud, C., Labbe, E., Attisano, L., Rossant, J., and Wrana, J.L. 2001. FoxH1 (Fast) functions to specify the anterior primitive streak in the mouse. Genes & Dev. 15: 1257-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, L., Guan, R.J., and Pardee, A.B. 1999. Evolution of transcriptional control from prokaryotic beginnings to eukaryotic complexities. Crit. Rev. Gene Expr. 9: 175-182. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Jacob, F. 2001. Complexity and tinkering. Ann. N.Y. Acad. Sci. 929: 71-73. [DOI] [PubMed] [Google Scholar]
- Kirschner, M., and Gerhart, J. 1998. Evolvability. Proc. Natl. Acad. Sci. 95: 8420-8427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume, T., Jiang, H., Topczewska, J.M., and Hogen, B.L. 2001. The murine winged helix transcription factors Foxc1 and Foxc2, are both required for cardiovascular development and somitogenesis. Genes & Dev. 15: 2470-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon, B. and Tjian, R. 2000. Orchestrated response: A symphony of transcription factors for gene control. Genes & Dev. 14: 2551-2569. [DOI] [PubMed] [Google Scholar]
- Lercher, M.J., Urrutia, A.O., and Hurst, L.D. 2002. Clustering of housekeeping genes provides a unified model of gene order in the human genome. Nat. Genet. 31: 180-183. [DOI] [PubMed] [Google Scholar]
- Li, F. and Stormo, G.D. 2001. Selection of optimal DNA oligos for gene expression arrays. Bioinformatics 17: 1067-1076. [DOI] [PubMed] [Google Scholar]
- Matys, V., Fricke, E., Geffer, R., Gossling, E., Haubrock, M., Hehl, R., Hornischer, K., Karas, D., Kel, A.E., Kel-Margoulis, O.V., et al. 2003. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 31: 374-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt, K. and Maglott, D. 2001. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 29: 137-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranger, A.M., Grusby, M.J., Hodge, M.R., Gravallese, E.M, de a Brousse, F.C., Hoey, T., Mickanin, C., Baldwin, H.S., and Glimcher, L.H. 1998. The transcription factor NF-ATc is essential for cardiac valve formation. Nature 392: 186-190. [DOI] [PubMed] [Google Scholar]
- Ray, R. and Miller, D.M. 1991. Cloning and characterization of a human c-myc promoter-binding protein. Mol. Cell. Biol. 11: 2154-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves, R. 2001. Molecular biology of HMGA proteins: Hubs of nuclear function. Gene 277: 63-81. [DOI] [PubMed] [Google Scholar]
- Revilla-I-Domingo, R. and Davidson, E.H. 2003. Developmental gene network analysis Int. J. Dev. Biol. 47: 695-703. [PubMed] [Google Scholar]
- Riechmann, J.L., Heard, J., Martin, G., Reuber, L., Jiang, C., Keddie, J., Adam, L., Pineda, O., Ratcliffe, O.J., Samaha, R.R., et al. 2000. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 290: 2105-2110. [DOI] [PubMed] [Google Scholar]
- Rubin, G.M., Hong, L., Brokstein, P., Evans-Holm, M., Frise, E., Stapleton, M., and Harvey, D.A. 2000. A Drosophila complementary DNA resource. Science 287: 2222-2224. [DOI] [PubMed] [Google Scholar]
- Semenza, G.L. 1998. Transcription factors and human disease. Oxford University Press, Oxford, UK.
- Subramanian, A. and Miller, D.M. 2000. Structural analysis of α-enolase. J. Biol. Chem. 275: 5958-5965. [DOI] [PubMed] [Google Scholar]
- Ureta-Vidal, A., Ettwiller, L., and Birney, E. 2003. Comparative genomics: Genome-wide analysis in metazoan eukaryotes. Nat. Rev. Genet. 4: 251-262. [DOI] [PubMed] [Google Scholar]
- Venter, J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304-1351. [DOI] [PubMed] [Google Scholar]
- Wingender, E., Chen, X., Hehl, R., Karas, H., Liebich, I., Matys, V., Meinhardt, T., Prüß, M., Reuter, I., and Schacherer, F. 2000. TRANSFAC: An integrated system for gene expression regulation. Nucleic Acids Res. 28: 316-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray, G.A. 2003. Transcriptional regulation and the evolution of development. Int. J. Dev. Biol. 47: 675-684. [PubMed] [Google Scholar]
- Yamamoto, M., Meno, C., Sakai, Y., Shiratori, H., Mochida, K., Ikawa, Y., Saijoh, Y., and Hamada, H. 2001. The transcription factor FoxH1 (FAST) mediates Nodal signaling during anterior-posterior patterning and node formation in the mouse. Genes & Dev. 15: 1242-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Schwartz, S., Wagner, L., and Miller, W. 2000. A greedy algorithm for aligning DNA sequences. J. Comp. Biol. 7: 203-214. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., Harrison, P.M., Liu, Y., and Gerstein, M. 2003. Millions of years of evolution preserved: A comprehensive catalog of the processed pseudogenes in the human genome. Genome Res. 13: 2541-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.ensembl.org; Ensembl genome browser.
- http://flybase.bio.indiana.edu/; FlyBase, a database of the Drosophila genome.
- http://www.geneontology.org/; Gene Ontology Consortium.
- http://www.ebi.ac.uk/interpro/; InterPro.
- http://www.ncbi.nlm.nih.gov/LocusLink/; LocusLink.
- http://pfam.wustl.edu; The Pfam database of protein families and HMMs.
- http://sapiens.wustl.edu/ScriptSure/; ScriptSure homepage.
- http://www.gene-regulation.com; TRANSFAC, the transcription factor database.
- http://genome.ucsc.edu/; UCSC genome Web site.
- http://hg.wustl.edu/lovett/projects/nohr/Tfarray.html/; Washington Univ. Human Transcription Factor Microarray.
- http://blast.wustl.edu; WU-BLAST Web site.
- http://www.sciencemag.org/feature/data/1049664.shl; Eukaryote comparative genomics.
- http://www.yeastgenome.org/VL-FAQ.html; yeast genome database.
- ftp://ftp.ensembl.org/pub/current_human/data/fasta/cdna/; the Ensembl database.
- http://hmmer.wustl.edu; sequence analysis using profile Hidden Markov Models.