Abstract
To find novel short coding sequences from accumulated full-length cDNA sequences, proteomic analysis of small proteins expressed in human leukemia K562 cells was performed using high-resolution nanoflow liquid chromatography coupled with electrospray ionization tandem mass spectrometry. Our analysis led to the identification of 54 proteins not more than 100 amino acids in length, including four novel ones. These novel short coding sequences were all located upstream of the longest open reading frame (ORF) of the corresponding cDNA. Our findings indicate that the translation of short ORFs occurs in vivo whether or not there exists a longer coding region in the downstream of the mRNA. This investigation provides the first direct evidence of translation of upstream ORFs in human cells, which could greatly change the current outline of the human proteome.
In parallel with human genome sequencing projects (Lander et al. 2001; Venter et al. 2001), the accumulation of sequence data of human full-length cDNAs has also been proceeding. The “RefSeq collection” (NCBI) provides us with representative resources of curated human full-length cDNAs, and the protein-coding sequence (CDS) of each cDNA is defined in the RefSeq database (Pruitt and Maglott 2001). Now a total of 19,995 proteins are stored in the RefSeq curated human protein database (as of January 27, 2004), and 19,271 (96.4%) of them are longer than 100 amino acids. This indicates that small proteins with ≦100 amino acids are only a limited fraction of all the proteins annotated in the RefSeq database.
According to the typical translation model, a 40S ribosomal subunit is first recruited to the cap structure of mRNA and linearly scans the 5′-UTR for the initiator ATG. When it recognizes the initiator ATG, it pauses until a large 60S subunit joins, and the complete ribosomal complex starts translation (Kozak 1989). Therefore, the most upstream ORF should be translated according to this model, much more with a good context around its ATG codon as previously analyzed (Kozak 1999). Some previous studies have reported that the short ORF in the 5′-untranslated region (UTR) functions as a regulator of the translation of its downstream CDS (Morris and Geballe 2000; Meijer and Thomas 2002). It has been considered that such translational control would be limited to some genes or conditions. However, the previous large-scale analyses focusing on the 5′-UTRs of human full-length cDNA sequences showed that 41%-49% of them had at least one ATG codon upstream of the CDS (Peri and Pandey 2001; Yamashita et al. 2003). This means that there are potential short coding regions in the 5′-UTRs of many genes if this classical model, indeed, represents a general mechanism of translation initiation. To our knowledge, few reports have presented evidence of the translation of upstream ORFs in vivo (Diba et al. 2001). Although there are also some mechanisms by which the ribosomal complex may evade the translation from the first ATG codon, such as leaky scanning (Kozak 1999) and IRES (internal ribosome entry site)-dependent translation (Meijer and Thomas 2002), we expect that the small proteins encoded by upstream ORFs in 5′-UTRs exist in vivo.
With a view to finding novel short upstream CDSs in accumulated cDNA sequences, we performed a proteomic analysis of small proteins expressed in vivo using direct nanoflow liquid chromatography (LC) coupled with the electrospray ionization (ESI)-tandem mass spectrometry (MS/MS) system (Natsume et al. 2002). This LC instrument can separate peptides and introduce them into a mass spectrometer with limited diffusion, leading to more sensitive detection than can be achieved with conventional LC systems. We aimed to identify novel short CDSs by searching not only against the RefSeq curated cDNA database but also against our in-house FLJ-unique cDNA data set, which contained as many as >10,000 full-length cDNA sequences that had no hit against the RefSeq cDNAs (Ota et al. 2004).
Here we report the proteomic analysis of small proteins (≦100 amino acids in length) expressed in human chronic myelogenous leukemia K562 cells. Our analysis led to the identification of 54 proteins in total, including four novel ones. Very intriguingly, these novel small proteins were all derived from the short ORFs in the presumed 5′-UTRs.
RESULTS
To carry out a proteomic analysis of small proteins expressed in K562 cells, we prepared the samples for mass spectrometric analysis by two different methods. Small proteins were isolated by either fractionation through SDS-PAGE or acid extraction. For the proteins resolved by SDS-PAGE, the part of the gel corresponding to the low molecular weight (<∼17 kDa) was excised, and the proteins trapped in the gel were digested with proteolytic enzymes (see “MS Sample Preparation 1” in Methods). On the other hand, the small proteins enriched by extraction in acid solution were digested directly without PAGE separation (see “MS Sample Preparation 2” in Methods). After concentrating the peptide mixtures prepared by each of the two methods, we applied them to the nanoflow LC-MS/MS system.
We first tried identifying small proteins (≦100 amino acids in length) by searching against the RefSeq curated human “protein” database (NCBI). Accordingly, 36 proteins were identified from the gel-separated samples, and 23 proteins were identified from the acid-extracted samples. In total, 50 proteins (with nine overlaps) were identified out of 724 proteins (≦100 amino acids in length) stored in the RefSeq protein database (as of January 27, 2004; see Supplemental table). The range of amino acid length of the identified proteins was from 44 to 100. The list included various kinds of small proteins, such as ribosomal proteins, transporters, transcriptional regulators, cell cycle regulators, spliceo-some components, and proteins involved in energy metabolism. We also found several function-unknown proteins expressed in K562 cells.
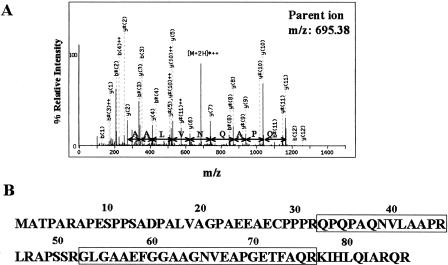
Next, to search for novel short CDSs that were not annotated in the RefSeq curated human protein database, all the MS/MS data that had no hit against RefSeq proteins were then searched against all the ORFs (in all three reading frames) of the RefSeq curated cDNA database and of our in-house “FLJ-unique” cDNA data set. As a result, four novel translated ORFs were identified (Table 1). Three of them were derived from RefSeq cDNAs, whereas the other was from an FLJ cDNA. As an example, the MS/MS spectrum matching the NM_015532 novel short CDS is shown in Figure 1A. An intense string of as many as 10 ions from the y ion series resulted in an excellent match for the corresponding peptide. The other peptides listed in Table 1 also yielded comparable search results, indicating the translation of these short ORFs. Moreover, the identification of the NM_015532 novel short CDS was also shown by matching of another peptide (Fig. 1B). This evidence gives us additional support for the presence of this novel CDS, which is also the case with the NM_005770 novel CDS (Table 1).
Table 1.
Novel Short CDSs Identified by Searching Against Human cDNAs
| GenBank Accession | Length (bp) | Novel CDS positiona | Novel CDS lengtha (amino acids) | Novel CDS initiator ATGa | Identified peptideb | Longest ORF positiona | Longest ORF lengtha (amino acids) |
|---|---|---|---|---|---|---|---|
| RefSeq cDNAs | |||||||
| NM_005770 | 1408 | 941...1120 | 59 | 6th | QRDSEIMQQKRDDGLSAAAR | 1023...1319 | 98 |
| NM_015532 | 4107 | 12...272 | 86 | 1st | QPQPAQNVLAAPRGLGAAEFGGAAGNVEAPGETFAQR | 127...1233 | 368 |
| NM_016215 | 1545 | 150...401 | 83 | 1st | ATPGLQQHQQPPGPGR (N-terminus acetylated) | 316...1137 | 273 |
| FLJ cDNAs | |||||||
| AK057257 | 1904 | 23...280 | 85 | 1st | LLPLGASPAGVVGGGLAPPR | 654...1013 | 119 |
Each data is based on the sequence information of the corresponding cDNA.
Peptides identified from MS samples (see Methods).
Figure 1.
Identification of a novel short coding sequence by mass spectrometry. (A) MS/MS spectrum corresponding to the peptide QPQPAQNVLAAPR at m/z 695.38 derived from the NM_015532 ORF. The corresponding amino acid differences based on a series of as many as 10 continuous y ions are represented. An asterisk (*) indicates an ion that has lost ammonia from its side chain. (B) Amino acid sequence of the NM_015532 upstream short ORF. The identified peptides are surrounded by rectangles.
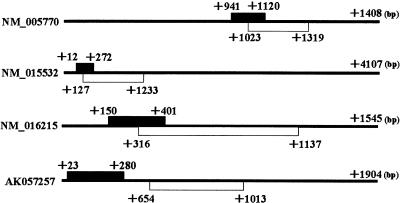
In Figure 2, we show the location of these novel CDSs within each corresponding cDNA. Interestingly, all the novel CDSs are located upstream of the longest ORF. Three of them overlap with each longest ORF, whereas the other is distant. A nucleotide deletion or insertion arising from a sequencing error can cause an erroneous short ORF to be produced from the longest ORF by a frameshift in the reading frame. Also, there might be splicing variants that can result in the fusion of the short ORF to the longest ORF. Therefore, careful confirmation of the corresponding nucleotide sequences was needed.
Figure 2.
Location of the identified novel CDS (black box) and the longest ORF (white box) of each full-length cDNA. The numbers indicate the positions from each full-length cDNA start site.
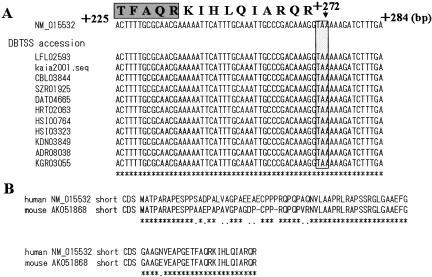
Here we tried aligning the EST data corresponding to the NM_015532 novel short CDS. As this novel short CDS is located near the 5′-end of the mRNA as shown in Figure 2, we aligned the 5′-end cDNA sequence data provided by the “oligo-capping” method, which was previously established by us for collecting accurate 5′-end nucleotide sequences from the mRNA start site (Suzuki et al. 1997, 2001). The multiple alignment of the sequence data of 11 corresponding cDNAs from 10 different resources showed no alternative splicing pattern over the entire region of this short CDS and a complete match for NM_015532 around the termination codon as shown in Figure 3A (the detailed sequence data can be seen at DataBase of Transcriptional Start Sites; DBTSS; Release 3.0; http://dbtss.hgc.jp/; Suzuki et al. 2002). As for the other three novel short CDSs, accumulated EST evidence showed that there were also no frameshift or alternative splicing variants that indicated the existence of a fused and longer ORF.
Figure 3.
Sequence analysis of the NM_015532 novel short CDS. (A) Multiple alignment of the 5′-end EST data around the termination codon. The dark box shows the C terminus of the secondarily identified peptide. An asterisk (*) indicates a complete sequence match between the RefSeq cDNA (NM_015532) and all the EST data. The shaded box indicates the termination codon of this short CDS. (B) Alignment of the NM_015532 short CDS with its mouse ortholog at the amino acid level. The amino acid sequence of the mouse ortholog was deduced from the sequence (from +13 to +267) of AK051868. An asterisk (*) and a dot (.) indicate identity and similarity in amino acid sequence, respectively. (DBTSS) DataBase of Transcriptional Start Sites (http://dbtss.hgc.jp/; Suzuki et al. 2002).
Furthermore, comparative sequence analysis of the NM_015532 novel CDS and its mouse ortholog counterpart showed 86% DNA identity with 16 conservative changes and 15 nonconservative ones over the aligned ORFs (261 nt in length), resulting in 85% identity and 95% similarity across the entire length of their deduced amino acid sequences (86 amino acids in length; Fig. 3B). Evidence of such a high degree of evolutionary sequence conservation indicates functional constraint on this novel short CDS. Table 2 shows that the NM_005770 upstream CDS is also functionally constrained, whereas that of NM_016215 is relatively loosened. As for that of NM_005770, the previous study has indicated that it shares homology with the protein encoded by a candidate modifying gene for spinal muscular atrophy (Scharf et al. 1998).
Table 2.
Sequence Conservation of the Upstream CDS and the Longest ORF of Each RefSeq cDNA
| RefSeq ID | Upstream CDSa (%) | Longest ORFa (%) | Longest ORF RefSeq definition |
|---|---|---|---|
| NM_005770 | 100 | 94 | Small EDRK-rich factor 2 (SERF2) |
| NM_015532 | 95 | 89 | Glutamate receptor, ionotropic, N-methyl D-aspartate-like 1A (GRINL1A) |
| NM_016215 | 71 | 92 | EGF-like domain, multiple 7 (EGFL7) |
Each amino acid sequence was deduced from the corresponding nucleotide sequence region of each cDNA. The mouse orthologous regions were extracted from the cDNA data below.
NM_005770 [upstream CDS: NM_011354 (+19−+198); longest ORF: NM_011354 (+101−+397)].
NM_015532 [upstream CDS: AK051868 (+13−+267); longest ORF: AK051868 (+122−+1222)].
NM_016215 [upstream CDS: NM_198724 (+125−+367); longest ORF: NM_198724 (+294−+1130)].
Each value represents the rate of similar amino acid residues over the entire region in the alignment of the human protein sequence with that of its mouse ortholog using CLUSTAL W (http://www.ddbj.nig.ac.jp/E-mail/clustalw-j.html).
DISCUSSION
Our proteomic analysis of small proteins expressed in K562 cells has enabled us to reveal the existence of the proteins encoded by upstream ORFs in 5′-UTRs. To our knowledge, this is the first direct evidence of translation of upstream ORFs in human cells. There were only four upstream short CDSs identified in our analysis, while leading to the identification of 50 RefSeq-annotated proteins. One of the reasons might be that some parts of upstream ORFs would not be efficiently translated in K562 cells because of the poor Kozak's context around their ATG codons. The previous studies indicated that 37%-57 % of the upstream ATGs in the 5′-UTRs had an unfavorable Kozak's context around the ATG codon (Kozak 1987; Suzuki et al. 2000). Secondly, our recent analysis also indicated that approximately three-fourths of the upstream ORFs analyzed were shorter than 40 amino acids (Yamashita et al. 2003). Considering that the smallest protein identified from the RefSeq curated database is 44 amino acids in length, it is very possible that such very small proteins were out of the detectable range in our analysis. They would be lost while preparing the samples for MS analysis.
However, these reasons from the statistical point of view cannot by themselves fully explain why there were no more than four proteins identified among thousands of upstream ORFs. One of the other possibilities is that many of the proteins derived from upstream ORFs might be selectively proteolyzed in the cells. Secondly, there might be some mechanisms that allow ribosomes to avoid the translation of upstream ORFs. Although IRES-dependent translation can permit ribosomes to directly enter a downstream site without encountering an upstream ATG, the previous studies have estimated that this mechanism would be applied to a limited fraction of genes (Meijer and Thomas 2002). There might exist another mechanism that enables ribosomes to escape the translation of an upstream ORF. Thirdly, the transcripts expressed in K562 cells might not reflect the corresponding cDNA sequences stored in the database. Our previous large-scale analysis on the 5′-UTRs has shown that the transcription start sites of many genes are more dispersed than was previously believed (Suzuki et al. 2001). Thus, it is likely that there are many genes whose transcripts in K562 cells have a shorter 5′-UTR that lacks an upstream ATG. Further analysis will be required to clarify this point.
Mapping of the novel short CDSs onto the corresponding full-length cDNAs indicates that three of these CDSs (but not the NM_005770 short CDS) use the most upstream ATG as an initiation codon (Table 1). As to the NM_005770 short CDS, the sixth ATG corresponds to its translation start site on the cDNA sequence of NM_005770. However, the accumulated oligo-capped 5′-end cDNA data of this gene obtained from various types of human tissues and cell lines uniformly showed the existence of the short transcript form whose first ATG corresponded to the initiation codon of this novel CDS (see the sequence data at DBTSS [Release 3.0]; http://dbtss.hgc.jp/). This indicates that this gene has two alternative transcript forms and the majority is the shorter one. Thus, it is very possible that translation of this short CDS initiates from the first ATG of the corresponding short transcript in K562 cells. In the conventional mechanism of translation initiation, the first ATG should be recognized as an initiation codon (the first ATG rule; Kozak 1989). Our finding of these four upstream CDSs is supportive evidence for this rule. In addition, the classification on the probable translation initiation sites of the small RefSeq proteins identified in our analysis shows that 42 (84%) out of the 50 listed proteins use the first ATG as an initiation codon (see Supplemental table). These results indicate that the small proteins relatively abundant in K562 cells were mainly produced according to the first ATG rule. Much more evidence of the translation of upstream ORFs could demonstrate that the translation from the first ATG generally occurs, indeed.
As for NM_015532, NM_016215, and AK057257, the splicing junctions are left downstream of the translation termination site of each upstream CDS. The nonsense-mediated decay (NMD) pathway triggers the degradation of the transcripts holding exon junction complexes (EJCs), which should be removed by a migrating ribosome during the process of translation (Maquat 2004). Therefore, these transcripts are considered to be susceptible to degradation by this pathway. Translation of the downstream longest ORF can protect the transcripts from degradation through removal of the remaining EJCs. As described in Table 2, the longest ORFs of the three RefSeq genes are highly conserved between human and mouse and the mouse ortholog corresponding to that of NM_016215 has been characterized as an endothelial repressor of smooth muscle cell migration (Soncin et al. 2003). As for the longest ORF of AK057257, it shares strong homology with α-tubulin, which also indicates its functionality (data not shown). The investigation on whether the translation of these downstream longest ORFs occurs in K562 cells will be needed to consider this point.
Further explorations based on mass spectrometric analysis will lead to the identification of more short CDSs through improvement of the method for the fractionation of small proteins or through sophistication of the LC system to acquire more MS/MS spectra. The analysis of small proteins expressed in other cultured cells or tissues will also reveal the existence of those expressed in a tissue-specific manner. Accumulating evidence of the translation of upstream short ORFs will make it possible for us to obtain a clearer outline of translatable regions of numerous mRNA species and help us to determine the real size and contents of the human proteome.
METHODS
Cell Culture
Human chronic myelogenous leukemia K562 cells were grown in RPMI/10% dialyzed FCS to a density of 1 × 106 cells/mL, harvested, and washed three times with PBS.
MS Sample Preparation 1
Harvested K562 cells (5 × 106) were lysed in lysis buffer (50 mM Tris-HCl at pH 7.6, 0.5 % [w/v] Triton X-100, 150 mM NaCl) supplemented with protease inhibitor cocktail Complete mini (Boerhinger Mannheim) and centrifuged for 10 min at 12,000 rpm at 4°C. The obtained supernatant was separated by SDS-PAGE with a 14% lower gel. The part of the lane corresponding to <∼17 kDa was cut into small pieces, and the proteins in the gel pieces were digested overnight at 37°C with 25 pmoles of trypsin, sequencing-grade (Roche Diagnostics) in 20 mM Tris-HCl (pH 8.8). These procedures were performed according to the previously described method (Shevchenko et al. 1996). The peptides were extracted from the gel pieces with a total of 300 μL of 50% (v/v) acetonitrile/5% formic acid by sonication and concentrated to ∼50 μL using a centrifugal vacuum concentrator. After the sample was desalted using a ZipTip (C18; Millipore), the peptides were eluted with 50 μL of 70% (v/v) acetonitrile/0.1% (v/v) formic acid and again concentrated to a final volume of 5 μL.
MS Sample Preparation 2
Harvested K562 cells (5 × 108) were boiled for 10 min at 95°C to inactivate proteases. The cells were then lysed in 1 M acetic acid using a Dounce homogenizer on ice and centrifuged for 10 min at 12,000 rpm at 4°C. After eliminating salts and other low-molecular-weight contaminants from the supernatant using a PD-10 Column (Amersham Biosciences) filled with Sephadex G-25, one-hundredth of the protein-enriched fraction was digested overnight at 37°C with 25 pmoles of trypsin in 20 mM Tris-HCl (pH 8.8). After the sample was desalted using a ZipTip (C18), it was processed in the same way as described above for MS Sample Preparation 1.
Automated Nanoflow LC-MS/MS Analysis and Protein Identification by Database Search
The peptide mixtures were analyzed using a high-resolution nanoflow reversed-phase capillary LC coupled with an electrospray quadrupole time-of-flight (Q-TOF) tandem mass spectrometer (Q-Tof-2; Micromass Ltd.). The acquired MS/MS spectra were converted to text files of peak lists and processed using the Mascot algorithm (Matrix Science Ltd.) with a maximum tolerance of 500 ppm in MS data and 0.5 Da in MS/MS data against each database. The RefSeq databases were downloaded periodically from the NCBI ftp site (ftp://ftp.ncbi.nih.gov/refseq/). The FLJ-unique cDNA data set was prepared and characterized as previously described (Ota et al. 2004). The results based on the RefSeq data were finally reviewed according to the RefSeq information as of March 5, 2004.
Acknowledgments
We thank T. Hasui for his technical support in the database construction. We are grateful to E. Nakajima for her critical reading of the manuscript. This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2384604.
References
- Diba, F., Watson, C.S., and Gametchu, B. 2001. 5′UTR sequences of the glucocorticoid receptor 1A transcript encode a peptide associated with translational regulation of the glucocorticoid receptor. J. Cell Biochem. 81: 149-161. [PubMed] [Google Scholar]
- Kozak, M. 1987. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 15: 8125-8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1989. The scanning model for translation: An update. J. Cell Biol. 108: 229-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1999. Initiation of translation in prokaryotes and eukaryotes. Gene 234: 187-208. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Maquat, L.E. 2004. Nonsense-mediated mRNA decay: Splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell Biol. 5: 89-99. [DOI] [PubMed] [Google Scholar]
- Meijer, H.A. and Thomas, A.A. 2002. Control of eukaryotic protein synthesis by upstream open reading frames in the 5′-untranslated region of an mRNA. Biochem. J. 367: 1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, D.R. and Geballe, A.P. 2000. Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol. 20: 8635-8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsume, T., Yamauchi, Y., Nakayama, H., Shinkawa, T., Yanagida, M., Takahashi, N., and Isobe, T. 2002. A direct nanoflow liquid chromatography-tandem mass spectrometry system for interaction proteomics. Anal. Chem. 74: 4725-4733. [DOI] [PubMed] [Google Scholar]
- Ota, T., Suzuki, Y., Nishikawa, T., Otsuki, T., Sugiyama, T., Irie, R., Wakamatsu, A., Hayashi, K., Sato, H., Nagai, K., et al. 2004. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 36: 40-45. [DOI] [PubMed] [Google Scholar]
- Peri, S. and Pandey, A. 2001. A reassessment of the translation initiation codon in vertebrates. Trends Genet. 17: 685-687. [DOI] [PubMed] [Google Scholar]
- Pruitt, K.D. and Maglott, D.R. 2001. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 29: 137-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf, J.M., Endrizzi, M.G., Wetter, A., Huang, S., Thompson, T.G., Zerres, K., Dietrich, W.F., Wirth, B., and Kunkel, L.M. 1998. Identification of a candidate modifying gene for spinal muscular atrophy by comparative genomics. Nat. Genet. 20: 83-86. [DOI] [PubMed] [Google Scholar]
- Shevchenko, A., Wilm, M., Vorm, O., and Mann, M. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68: 850-858. [DOI] [PubMed] [Google Scholar]
- Soncin, F., Mattot, V., Lionneton, F., Spruyt, N., Lepretre, F., Begue, A., and Stehelin, D. 2003. VE-statin, an endothelial repressor of smooth muscle cell migration. EMBO J. 22: 5700-5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, Y., Yoshitomo-Nakagawa, K., Maruyama, K., Suyama, A., and Sugano, S. 1997. Construction and characterization of a full length-enriched and a 5′-end-enriched cDNA library. Gene 200: 149-156. [DOI] [PubMed] [Google Scholar]
- Suzuki, Y., Ishihara, D., Sasaki, M., Nakagawa, H., Hata, H., Tsunoda, T., Watanabe, M., Komatsu, T., Ota, T., Isogai, T., et al. 2000. Statistical analysis of the 5′ untranslated region of human mRNA using “Oligo-Capped” cDNA libraries. Genomics 64: 286-297. [DOI] [PubMed] [Google Scholar]
- Suzuki, Y., Taira, H., Tsunoda, T., Mizushima-Sugano, J., Sese, J., Hata, H., Ota, T., Isogai, T., Tanaka, T., Morishita, S., et al. 2001. Diverse transcriptional initiation revealed by fine, large-scale mapping of mRNA start sites. EMBO Rep. 2: 388-393.x1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, Y., Yamashita, R., Nakai, K., and Sugano, S. 2002. DBTSS: DataBase of human transcriptional start sites and full-length cDNAs. Nucleic Acids Res. 30: 328-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter, J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304-1351. [DOI] [PubMed] [Google Scholar]
- Yamashita, R., Suzuki, Y., Nakai, K., and Sugano, S. 2003. Small open reading frames in 5′ untranslated regions of mRNAs. C.R. Biol. 326: 987-991. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- ftp://ftp.ncbi.nih.gov/refseq/; NCBI RefSeq ftp site.
- http://dbtss.hgc.jp/; DBTSS: DataBase of human Transcriptional Start Sites. [DOI] [PMC free article] [PubMed]
- http://www.ddbj.nig.ac.jp/E-mail/clustalw-j.html; CLUSTAL W.