Abstract
The first version of the Caenorhabditis elegans ORFeome cloning project, based on release WS9 of Wormbase (August 1999), provided experimental verifications for ∼55% of predicted protein-encoding open reading frames (ORFs). The remaining 45% of predicted ORFs could not be cloned, possibly as a result of mispredicted gene boundaries. Since the release of WS9, gene predictions have improved continuously. To test the accuracy of evolving predictions, we attempted to PCR-amplify from a highly representative worm cDNA library and Gateway-clone ∼4200 ORFs missed earlier and for which new predictions are available in WS100 (May 2003). In this set we successfully cloned 63% of ORFs with supporting experimental data (“touched” ORFs), and 42% of ORFs with no supporting experimental evidence (“untouched” ORFs). Approximately 2000 full-length ORFs were cloned in-frame, 13% of which were corrected in their exon/intron structure relative to WS100 predictions. In total, ∼12,500 C. elegans ORFs are now available as Gateway Entry clones for various reverse proteomics (ORFeome v3.1). This work illustrates why the cloning of a complete C. elegans ORFeome, and likely the ORFeomes of other multicellular organisms, needs to be an iterative process that requires multiple rounds of experimental validation together with gradually improving gene predictions.
The Caenorhabditis elegans genome sequence, released in December 1998, was nearly complete and highly accurate, with an error rate estimated at 1/30,000 (The C. elegans Sequencing Consortium 1998). The finished sequence was eventually released in November 2002, comprising 100,258,171 bp in six contiguous segments corresponding to the six C. elegans chromosomes (J. Sulston, pers com; http://elegans.swmed.edu/Announcements/genome_complete.html).
Although the technology required for rapid and accurate whole-genome sequencing is mature, the gene prediction tools currently available to identify protein-encoding open reading frames (ORFs) and to define their exon/intron structures still need improvements. For exon prediction in mammalian genomes, these tools have an overall sensitivity and specificity of only 60% (Burset and Guigo 1996), and ∼40% for the 5′ and 3′ gene boundaries specifically (Korf et al. 2001). Predicted genes can be truncated, extended, split, or merged (see Reboul et al. 2001), relative to their actual “observed” exon/intron structure.
Using GeneFinder, a gene prediction tool developed for C. elegans (http://ftp.genome.washington.edu/cgi-bin/genefinder_req.pl), a total of 19,477 ORFs were annotated in Wormbase release WS9 (August 1999; http://www.Wormbase.org; Stein et al. 2001). Approximately 50% of these ORFs were predicted ab initio, without experimental support.
The C. elegans ORFeome project was launched to test the accuracy of these gene predictions, while simultaneously creating a resource of cloned full-length predicted ORFs to be used in various functional genomics and reverse proteomics studies (Reboul et al. 2001, 2003). ORFs were PCR-amplified between their 5′- and 3′-ends, and cloned using the Gateway recombinational cloning system (Hartley et al. 2000; Walhout et al. 2000a,b). PCR amplification was performed on a highly representative cDNA library using gene-specific primer pairs for each of the 19,477 ORFs based on WS9 predictions. Gateway tails attached to all primers allowed the cloning of the ORFs into the pDONR201 vector, resulting in a total of 11,984 (61.5% of the ORFs) Entry clones in the first version of the ORFeome (version v1.1; Supplemental Table 1).
The C. elegans ORFeome version 1.1a (v1.1a) represents a consolidated set of 10,623 ORFs cloned in-frame, 11.4% (1361 out of 11,984) of all cloned ORFs in version 1 were cloned outof-frame because of mispredicted gene boundaries (v1.1b). This first version of the worm ORFeome contributed significantly to the reannotation of C. elegans gene structure. The alignment of OSTs (ORF Sequence Tags) to the corresponding predicted gene sequences allowed the improvement of C. elegans annotations by correcting the internal gene structure of 20% of v1.1a cloned ORFs. In addition, OSTs provided experimental verification for 45% of the set of “untouched” ORFs, that is, not detected yet by any mRNA or EST. For each gene, ORFeome v1.1a contains cloned pools that result from mixing ∼50 to ∼1000 Escherichia coli transformants for each Entry clone. Thus, such Entry pools might contain multiple splice variants and alleles corresponding to PCR misincorporations. We are in the process of generating a new resource, ORFeome v2 (Reboul et al. 2003), in which we isolate individual wild-type clones for all detected splice variants of ORFs cloned in v1.1a. We will shortly initiate similar attempts for the ORFs cloned in the ORFeome version 3 described below.
The difficulties inherent in identifying ORFs within metazoan genomes and predicting their correct structure are not specific to C. elegans. Genome annotation initiatives in the model organisms Arabidopsis thaliana (Yamada et al. 2003) and Drosophila melanogaster (Hild et al. 2003) have also shown limited accuracy. The accuracy of current gene prediction algorithms is also a major issue for the human genome. High numbers of splice variants and lower signal-to-noise ratios caused by longer introns and intergenic regions render human genome annotations even more difficult than for the model systems experimentally validated so far. Hence, both in model organisms and in human, functional genomic and reverse proteomics studies, which require the use of large sets of full-length ORFs, are hampered by inaccuracies in gene prediction, limiting the usefulness of sequenced genomes.
Since the release of Wormbase WS9 in 1999, continuous efforts to reannotate the C. elegans genome have occurred. Reannotations are mainly based on new experimental data, such as mRNAs and ESTs (the EMBL nucleotide sequence database [http://www.ebi.ac.uk/embl/] and the Y. Kohara DNA databank [DDBJ, http://www.ddbj.nig.ac.jp/]), as well as splice-leader sequences (Blumenthal et al. 2002). Furthermore, more refined ab initio approaches have allowed the reprediction of genes for which no confirmatory experimental data are yet available. To experimentally validate these new predictions, improve gene annotation, and generate a more complete C. elegans ORFeome resource, we attempted to clone the 4232 ORFs originally missed in v1.1a and that have been either repredicted or newly predicted between the release of WS9 and that of WS100 (May 2003).
RESULTS
Design of Version 3 of the C. elegans ORFeome
Wormbase, the central repository for the C. elegans genome annotation, is updated biweekly, reflecting the continuous effort made both to correct the structure of previously predicted ORFs (referred to here as “repredicted ORFs”) and to predict new putative ORFs. To identify ORFs that could not be cloned or were cloned out-of-frame in ORFeome version 1, and have been repredicted in improved versions of the genome annotation, we chose to compare WS9 predictions to those of the recent Wormbase release WS100 (see Methods). WS100 is the first Wormbase release that has been archived in the public domain (“frozen”; http://ws100.Wormbase.org). For each of the 8854 ORFs that were not in v1.1a, we searched for repredictions that at least partially overlapped with the region between the previously predicted initiation and termination codons (“starts” and “stops”). We focused only on structure differences at the 5′- and 3′-boundaries, while ignoring internal structure differences.
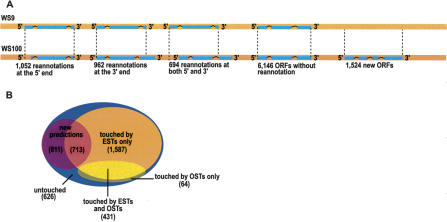
We found 2708 ORFs with repredicted starts or stops (Fig. 1A). These were classified into three categories: 1052 ORFs reannotated at the start, 962 at the stop, and 694 at both ends. Of these 2708 repredicted ORFs, 2213 correspond to uncloned ORFs, and 495 to ORFs found to be out-of-frame in the first version of the C. elegans ORFeome. The predicted structure of the remaining 6146 ORFs has not changed between WS9 and WS100. We also detected 1524 new WS100 genes that did not overlap with any predicted ORFs in WS9. In total, we attempted to clone and validate the structure of 4232 repredicted (2708) or new (1524) ORFs.
Figure 1.
The C. elegans genome annotation has evolved between WS9 and WS100. (A) For all ORFs that are missing in v1.1a, those with repredicted starts and/or stops in WS100 were identified. Between WS9 and WS100, 1052 ORFs have been repredicted at the start, 962 at the stop, and 694 at both ends. A total of 6146 ORFs had the exact same start and stop in the two Wormbase releases. We also identified 1524 newly predicted ORFs in WS100. (B) Venn diagram summarizing the classification of the 4232 repredicted and new ORFs based on the experimental data available. The blue oval represents all 4232 ORFs that we attempted to clone. The purple circle contains new predictions of which 713 are touched by ESTs and 811 are untouched. The large orange oval represents all ORFs touched by ESTs. The smaller oval in light yellow shows ORFs touched by OSTs. As no OST data are available for ORFs that we did not clone in ORFeome Version 1 or for newly predicted ORFs, only ORFs that we cloned out-of-frame earlier are touched by OSTs. A small portion (64) of the latter are not touched by any ESTs. Of all 4232 predicted ORFs that we attempted to clone, 34% (626 repredicted and 811 new ORFs) are not experimentally verified (untouched), whereas 66% are touched by ESTs, OSTs, or both.
These 4232 ORFs can be divided into two classes depending on whether their predicted coding sequence has been verified, at least partially, by EST and/or OST data (“touched” ORFs) or not (“untouched” ORFs; Fig. 1B). Of the 4232 ORFs that we attempted to clone, 2795 (66%) are touched and 1437 (34%) are untouched. According to the information available in Wormbase, various approaches have been used to reannotate untouched genes. However, the criteria on which these repredictions are based are neither categorized into defined classes nor searchable in Wormbase. Repredictions or new predictions often seem based on sequence alignments between C. elegans predictions and coding sequences of other organisms. Also, the 5′- or 3′-ends of ORFs are often truncated to avoid an overlap with neighboring ORFs extended based on new ESTs. Other times the repredictions are based solely on the analysis of the genomic sequence. For instance, some gene repredictions are based on the presence of noncoding repetitive elements overlapping with the coding sequence in earlier predictions.
Overall Assessment of WS100
As the quality of WS100 repredictions and new predictions has not been experimentally validated yet, we first tested their overall accuracy using a subset of ORFs. We compared the ORF cloning success rate using new WS100 predictions to that of the WS9 predictions from ORFeome version 1 on ORFs that could not be cloned previously. ORFs were PCR-amplified from our highly representative C. elegans cDNA library (Walhout 2000b) and cloned into the Gateway Entry vector pDONR201. Following a second round of PCR amplification from the Gateway Entry clone to confirm that inserts were present and of the corrected size, ORF sequence tags (OSTs) were generated.
The OSTs were then aligned to the genome to confirm the identity of the clones. The cloning success rate was 59% (n = 111) using newly designed primers. In contrast, only 2.7% of attempted ORFs were successfully cloned using WS9-designed primers, used here as a negative control. These results clearly show that the C. elegans genome annotation has improved considerably between WS9 and WS100, and that primers designed based on these reannotations can amplify a substantial number of ORFs not originally cloned in ORFeome version 1.
C. elegans ORFeome Version 3
In Version 3 of the C. elegans ORFeome project, PCR amplifications were performed for 4232 repredicted or new ORFs, using ORF-specific primers (Supplemental Fig. 1). Alignment of the resulting OSTs to the C. elegans genome revealed that 56% (2315 ORFs corresponding to 1378 repredicted ORFs and 937 new ORFs) were successfully cloned. The cloning success for touched ORFs is much higher (63%) than for untouched ORFs (42%), and is slightly lower than the cloning success rate of touched ORFs in ORFeome Version 1 (71%; Supplemental Fig. 2).
We amplified 64% of ORFs that were cloned out-of-frame in ORFeome Version 1 (v1.1b). Among these, 87% are now cloned in-frame. Hence, reannotation efforts led to successful repredictions for 55.7% (64% × 0.87) of such ORFs, whereas wrong repredictions caused complete cloning failure in 36% of the cases. For the remaining 8.3% of originally out-of-frame ORFs, repredictions resulted again in out-of-frame PCR products.
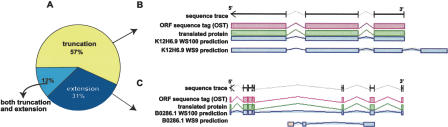
Of the ORFs cloned in ORFeome Version 3, 57% were shorter at one or both ends in WS100 relative to the gene annotation in WS9 (Fig. 2A). This explains why WS9-designed primers could not anneal to previously predicted ORF boundaries and did not amplify these ORFs in ORFeome Version 1 (Fig. 2B). Interestingly, a substantial number of ORFs (31%) extended at their 3′- and/or 5′-ends in WS100 were also successfully cloned in ORFeome Version 3, whereas the corresponding shorter ORFs, based on the WS9, failed to clone in ORFeome Version 1 (Fig. 2C). Given that these previously predicted ORFs are located completely within the repredicted genes, it seems surprising that previously designed primers failed to clone these truncated ORFs as internal primers. However, reannotation of ORF boundaries frequently alters the annotation of internal intron/exon structures such that primers initially designed to anneal to regions predicted to be exons in WS9 actually correspond to introns in the repredicted gene.
Figure 2.
Cloning success based on the nature of repredictions. (A) Of ORFs cloned in ORFeome Version 3, 57% were repredicted to be shorter and 31% to be extended at one or both ends, whereas 12% of the cloned ORFs have been extended at one end and truncated at the other end. (B) Example of an ORF that was successfully cloned in ORFeome Version 3 after having been truncated at the 3′-end. The exon/intron structures in blue represent the old (K12H6.9WS9) and new (K12H6.9WS100) predictions of K12H6.9. Using primers based on WS100 and sequencing the resulting PCR product, we obtained a sequence trace (black arrow) that aligned to the WS100 prediction, showing a full-length OST (pink) of the exact structure predicted. The translated protein is shown in green, demonstrating that the cloned ORF is, indeed, in-frame. The primer designed for the 3′-end of the WS9 prediction cannot anneal to the coding sequence of the WS100 prediction explaining earlier cloning failure. (C) Example of an ORF that was successfully cloned in ORFeome Version 3 after having been extended at both ends. The 5′-primer based on WS9 is annealing in the middle of an intron in the new predicted gene model, explaining earlier cloning failure.
Corrections of Intron/Exon Organization
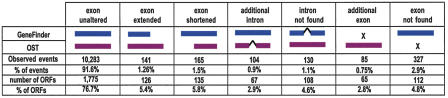
In ORFeome Version 3, we corrected internal exon/intron structures for 540 (23.3%) cloned ORFs. Compared with WS100 predictions, OSTs could be used to extend 141 exons, truncate 165 exons, add 85 unpredicted exons, and delete apparently wrongly predicted 327 exons. In addition, 104 and 130 introns were added or deleted, respectively (Fig. 3). These structural changes underestimate the number of actual structure differences, as we only analyzed OSTs from the 5′- and 3′-ends representing ∼1 kb of sequence in total. On the other hand, it is possible that WS100 predictions not observed here might correspond to genuine splice variants underrepresented in the worm cDNA library used here, and thus less likely to be represented in the Entry clone pools.
Figure 3.
Internal structure differences observed between WS100 predictions and their aligned OSTs. The structure of 540 ORFs has been corrected, each showing one or more differences compared with the corresponding OSTs. OSTs may have more, fewer, longer, or shorter exons than the prediction as well as additional or missing introns.
In comparison to ORFeome Version 1, the proportion of exons needing correction in ORFeome Version 3 decreased by 8%, which can be explained by a higher rate of EST coverage for the cloned ORFs. However, these additional EST data did not reduce the rate of ORFs cloned out-of-frame in ORFeome Version 3, because 11.7% (270) of all cloned ORFs display frame errors caused by mispredicted 3′- and 5′-boundaries. We have thus cloned 2045 (2315 - 270 out-of-frame) full-length ORFs in ORFeome Version 3.
Correction of Truncated Clones
As mispredictions of the 5′- or 3′-end of an ORF do not necessarily affect its internal gene structure, primers designed on mispredicted boundaries can give rise to truncated clones. Previously cloned ORFs that were subsequently merged, two or more at a time, into one single longer ORF in WS100 represent one class of such potentially truncated clones. Merges are typically based on additional EST data spanning the intergenic region between two individually predicted, neighboring ORFs.
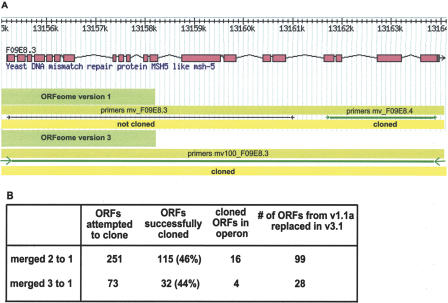
Our data set of 2708 repredicted ORFs contains 324 ORFs that resulted from a merge of two (251; Fig. 4A) or three ORFs (73), where at least one ORF of the pair or triplet was not cloned in ORFeome Version 1. Although C. elegans contains operons, it is unlikely that these merged genes are an artifact of polycistronic messages that are not transspliced (Blumenthal et al. 2002). Among the 147 merged ORFs that were successfully cloned in-frame in ORFeome Version 3, only 20 have been identified as being part of an operon (Fig. 4B).
Figure 4.
Merged genes account for a substantial number of repredictions in previously cloned ORFs. (A) Example of two ORFs that have been repredicted and merged into one longer ORF. In ORFeome Version 1 (upper lane), two pairs of primers were generated for the two predicted ORFs. The black arrows represent a primer pair (mv_F09E8.3) that did not amplify the previously predicted ORF F09E8.3. The green arrows represent primers (mv_F09E8.4) that successfully amplified a truncated version of the merged prediction. Using a new primer pair (mv100_F09E8.3), designed on the merged prediction in WS100 (green arrows, lower lane), this longer ORF was successfully cloned in-frame. (B) We have attempted to clone 324 merged ORFs in ORFeome Version 3 and confirmed former mispredictions of 99 pairs and 28 triplets of ORFs, each merged into one longer prediction in WS100.
Investigating the Existence of Clones Missing in Version 3.1
We next investigated whether the 44% of repredicted and newly predicted WS100 ORFs that could not be cloned here correspond to false-positive GeneFinder predictions, or genuine genes that need further exon/intron corrections. To obtain an estimate of the rate of repredicted ORFs not cloned in ORFeome Version 3 because of mispredicted ORF boundaries, we designed internal primers for a small subset of repredicted ORFs for which PCR amplification had failed (Reboul et al. 2001). These internal primers were designed to anneal to internally predicted exons, spanning at least one intron, and to amplify PCR products of 300 bp when the cDNA library is used as a template. As internal exons are easier to predict and hence more accurate than gene boundaries, many ORFs that are mispredicted at their 5′- and 3′-ends should be amplifiable using internal primers.
We amplified internal PCR products of the correct length for 52% of ORFs missed in Version 3. The most likely explanation why we could not clone these ORFs in ORFeome Version 3 is that their 5′- or 3′-ends are still mispredicted. There are two reasons why we were unable to amplify internal PCR products for the remaining 48% of ORFs: ORFs could be mispredicted at the level of their internal exon/intron structure, which consequently may render them undetectable in the cDNA library using internal primers. In addition, predicted ORFs that were not amplified might be absent from the cDNA library because they were wrongly predicted and do not actually exist.
We then investigated whether ORFs that we could not clone in ORFeome Version 3, were less supported by EST and Pfam data than ORFs that we successfully cloned. Of uncloned ORFs, 70% are either touched by EST data (16.5%), contain a Pfam motif (25.5%), or show evidence of both EST and Pfam data (28%). The number of cloned ORFs with EST and/or Pfam data is only slightly higher (74%). These results show that a substantial number of uncloned ORFs have experimental or bioinformatics evidence of their existence, supporting our conclusion that the main reason for cloning failure of C. elegans ORFs is the misprediction by Genefinder of their 3′- and 5′-boundaries.
DISCUSSION
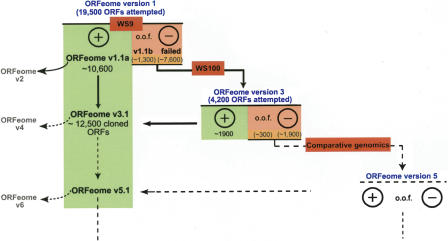
The examples presented in this paper illustrate that the goal of cloning a complete ORFeome should be organized in gradual steps (Fig. 5). In consecutive versions of the ORFeome, new, previously uncloned ORFs are added to the ORFeome resource, and previously cloned ORFs found to be a truncated version of a repredicted ORF are also replaced by the correct full-length equivalent. The updated version of the C. elegans ORFeome resource, ORFeome v3.1, represents all cloned ORFs from ORFeome Versions 1 and 3. Merged ORFs that were successfully cloned replace earlier truncated cloned versions if these are not detectably part of an operon. Version 3.1 of the C. elegans ORFeome contains 12,541 full-length, protein-coding clones (10,623 v1.1a + 2045 Version 3, -127 merged). For each predicted ORF, information about the cloning status, the cloned exon/intron structure, and the primers used for cloning can be found in WorfDB (Vaglio et al. 2003; http://worfdb.dfci.harvard.edu). Clones are available at MRC Geneservices (http://www.hgmp.mrc.ac.uk/geneservice/) and at Open Biosystems (http://www.openbiosystems.com/).
Figure 5.
The C. elegans ORFeome is an evolving resource. The cloning of a (nearly) complete ORFeome will be an iterative process. At each step, predicted ORFs that are successfully cloned in-frame (+) are added to the ORFeome resource. New attempts to clone ORFs that we cloned out-of-frame (o.o.f.) or that we did not clone (-) in earlier cloning steps are based on new or updated predictions (red box). The first two rounds of cloning, ORFeome Version 1 and Version 3, were based on two “snapshots” of the C. elegans genome annotation, WS9 and WS100, respectively. Further cloning steps will be based on different approaches to repredict ORFs, such as comparative genomics. Our current ORFeome resource, v3.1, contains ∼12,500 cloned ORFs. At this stage, our ORFeome resource contains pools of clones for each predicted gene. We are in the process of generating a new resource, ORFeome v2 (Reboul et al. 2003), in which we isolate individual wild-type clones for all detected splice variants of ORFs cloned in v1.1a.
With the release of ORFeome v3.1, we have validated the existence of 2045 previously uncloned ORFs. Within this set, the internal structures of 540 ORFs were corrected. For most ORFs that were missed in ORFeome v1.1a, we relied on experimental data to obtain an accurate reprediction. Hence, a continuous supply of new experimental data is essential to reannotate the genome, correcting the gene structure of ORFs that were out-of-frame, mispredicted, or missed in previous versions. Sometimes, these data also reveal ORFs cloned in-frame that represent truncated versions of longer gene structures. ORFs cloned in-frame are thus also subject to change and consequently need to be replaced in the ORFeome resource, underlining the fluid character of an ORFeome resource. The reannotation of the genome and the experimental validation of these new predictions by cloning thus go hand in hand. Iteratively repeating these two steps increasingly generates a more complete representation of the C. elegans ORFeome.
The first two C. elegans ORFeome projects relied on different snapshots of the genome annotation. In both, ∼60% of the attempted ORFs were successfully cloned as Gateway Entry clones. The rate of wrongly predicted exons in ORFeome Version 3 decreased by 8% compared with ORFeome Version 1, probably as a consequence of additional EST coverage for the ORFs in WS100. However, the rate of cloned ORFs that displayed frame errors, indicating mispredicted ORF boundaries, remained the same (∼11%). Furthermore, the comparison of internal primer experiments performed in ORFeome Versions 1 and 3 showed a drop in the success rate of PCR amplification (73% vs. 52%) for ORFs missed in ORFeome v1.1. Although this lower success rate might be caused by a higher rate of false negatives and less optimal experimental conditions in ORFeome Version 3, it is more likely that, as the set of ORFs remaining to be cloned decreases, the proportion of ORFs that do not exist or that are difficult to predict increases. Ongoing cloning efforts based on continuously reannotated versions of the genome would thus have an increasing cost-to-benefit ratio.
The continuous efforts made to improve the C. elegans genome annotation during the last four years increased the size of the C. elegans ORFeome resource by ∼20%. A third iterative cloning step, based on a new snapshot of Wormbase predictions, might only marginally improve the resource. A coming leap in improved gene annotations will likely result from comparative genomics, which has proven useful for genome reannotations in yeast (Cliften et al. 2001, 2003; Kellis et al. 2003) and will soon be applied to the worm. The comparison of the C. elegans genome to the newly sequenced Caenorhabditis briggsae genome (Stein et al. 2003) should result in corrected annotations for many previously predicted genes, as well as the discovery of new genes. Genome sequencing is currently underway for three additional Caenorhabditis species, Caenorhabditis remaniae, Caenorhabditis japonica, and CB5161, and when available should enable accurate predictions of C. elegans ORF structures, upon which future iterations of the ORFeome project will be based.
The complete ORFeome for C. elegans is thus a long-term project relying on combined bioinformatics and experimental approaches. Besides providing a useful tool for functional genomics and reverse proteomics in the worm, these efforts might eventually define better models of metazoan genes, leading to improved gene prediction algorithms for numerous other genomes, including the human genome.
METHODS
Identification of Repredicted ORFs
To find ORFs that were repredicted between versions WS9 and WS100 of Wormbase, we compared the start and stop coordinates of each ORF to the genome sequence. The sequencing of the C. elegans genome was completed between those two versions, and, consequently, because of nucleotide additions, some of the nonrepredicted ORFs displayed had changed coordinates in WS100. Hence, it was necessary to update the start and stop positions of WS9 predictions by aligning their corresponding primers from ORFeome Version 1 to the current genome sequence. The set of ORFs that we attempted to clone consists of ORFs in WS100 that overlap with ORFs in WS9 while having a repredicted start, stop, or both, as well as ORFs that are newly predicted in WS100. We used the OSP program to design new primers (Hillier and Green 1991). For ORFs that were repredicted at only one end, we designed new primers at the repredicted ends and used the primers originally synthesized for ORFeome Version 1 (“v1 primers”) on the unchanged ends (mixed primer pairs). For ORFs that were repredicted at both ends, we designed new forward and reverse primers.
The overall quality of the v1 primers (synthesized a few years before this work) was tested by comparing PCR amplification of pairs of v1 primers, mixed new and v1 primers, and pairs of all new primers using worm genomic DNA as template. Given that the PCR success rate on genomic DNA is independent of the quality of the annotations, similar results are expected for all primer pairs. The comparison of v1 primer pairs to mixed and new primer pairs showed a PCR success rate of 74%, 76%, and 83%, respectively. These results indicate that only a small portion of old primers have decreased in quality since their synthesis, and can be used in mixed primer pairs without biasing the results.
Gateway Cloning of C. elegans ORFeome 3.1
Primer pairs were organized by the expected size of ORFs and aliquoted in 96-well format to optimize PCR conditions for individual plates and to facilitate size analysis of PCR products. PCR amplification for C. elegans ORFs was performed using Platinum Taq DNA polymerase (Invitrogen), and PCR cycling conditions were as previously described (Reboul et al. 2003). For one entire plate of 77 ORFs, we failed to obtain any PCR products, leaving 4155 PCR products to be further processed.
Entry clones were produced using the pDONR201 vector according to standard Gateway recombinant cloning technology protocols except that BP cloning reactions were done at one-fourth of the recommended volume (Invitrogen). Entry clones were subsequently transformed into DH5α cells rendered chemically competent with DMSO and cultured overnight in LB liquid media containing kanamycin (50 μg/mL). Cultures were then used to inoculate a second 1.0-mL liquid culture containing LB and kanamycin, which was grown overnight at 37°C. Recombinant products were archived for long-term storage as both bacterial glycerol stocks (15% glycerol in LB) and as plasmid DNA minipreps. A Qiagen 9600 robot was used to purify plasmid DNA. PCR was performed using recovered plasmid DNA and pDONR201 sequencing primers (Invitrogen), and the resulting PCR products were used as template for sequencing as described (Reboul et al. 2003).
C. elegans cDNA Library
The library used here as PCR template was described earlier (Walhout 2000b).
Sequencing and Bioinformatics Analysis
All cloned ORFs were sequenced at the 5′- and 3′-ends resulting in two OSTs (ORF Sequence Tags) for each ORF. ORFs that were not successfully cloned or sequenced (phred score below 20 over 200 bases) were not included in the analysis. All OSTs were aligned to the C. elegans genome stored in the ACeDB, using the acembly alignment software. The comparison between OSTs and corresponding predicted ORFs was done in two phases. First, all alignments were analyzed using a previously described protocol (Reboul et al. 2001), to detect OSTs that displayed a different internal exon/intron structure than their corresponding ORFs. In a second phase, these ORFs were analyzed manually to identify the type of structure difference and to detect frame problems in the OST. The information resulting from this analysis has been stored in a MySQL database.
We found 230 ORFs in which no splicing events could be identified. These ORFs could be categorized as having OSTs that arose from extremely short sequencing reads that did not span predicted introns, those that gave rise to average-length OSTs but for which splicing had not been predicted in that region and OSTs that were predicted to span an intron but for which no splicing event was identified. Of the latter category, only 62 ORFs could be interpreted in our analysis, as we require sequencing through a splice junction. Of these ORFs, 81% were found to be out-of-frame, suggesting that they were either mispredicted or represent pseudogenes.
Acknowledgments
We thank the C. elegans Sequencing Consortium for a complete and highly accurate genome sequence; L. Stein, D. Lawson, R. Durbin, K. Bradnam, N. Chen, and others from Wormbase for continuously improving genome annotations; the participants of the annual ORFeome meeting for their input and numerous suggestions; M. Cusick for critical reading of the manuscript; J.-F. Rual, N. Bertin, and T. Kishikawa for their input and help; T. Clingingsmith and C. McCowan for superb administrative assistance; the staff at Illumina and Agencourt for technical assistance; and C. Fraughton for laboratory support. This work was supported by grants 7 R33 CA81658-02 from the National Cancer Institute and 5R01HG01715-02 from the National Human Genome Research Institute and the National Institute of General Medical Sciences awarded to M.V.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2496804.
References
- Blumenthal, T., Evans, D., Link, C.D., Guffanti, A., Lawson, D., Thierry-Mieg, J., Thierry-Mieg, D., Chiu, W.L., Duke, K., Kiraly, M., et al. 2002. A global analysis of Caenorhabditis elegans operons. Nature 417: 797-798. [DOI] [PubMed] [Google Scholar]
- Burset, M. and Guigo, R. 1996. Evaluation of gene structure prediction programs. Genomics 34: 353-367. [DOI] [PubMed] [Google Scholar]
- The C. elegans Sequencing Consortium. 1998. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science 282: 2012-2018. [DOI] [PubMed] [Google Scholar]
- Cliften, P.F., Hillier, L.W., Fulton, L., Graves, T., Miner, T., Gish, W.R., Waterston, R.H., and Johnston, M. 2001. Surveying Saccharomyces genomes to identify functional elements by comparative DNA sequence analysis. Genome Res. 11: 1143-1144. [DOI] [PubMed] [Google Scholar]
- Cliften, P., Sudarsanam, P., Desikan, A., Fulton, L., Fulton, B., Majors, J., Waterston, R., Cohen, B.A., and Johnston, M. 2003. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301: 71-76. [DOI] [PubMed] [Google Scholar]
- Hartley, J.L., Temple, G.F., and Brasch, M.A. 2000. DNA cloning using in vitro site-specific recombination. Genome Res. 10: 1788-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hild, M., Beckmann, B., Haas, S.A., Koch, B., Solovyev, V., Busold, C., Fellenberg, K., Boutros, M., Vingron, M., Sauer, F., et al. 2003. An integrated annotation and transcriptional profiling approach towards the full gene content of the Drosophila genome. Genome Biol. 5: R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier, L. and Green, P. 1991. OSP: A computer program for choosing PCR and DNA sequencing primers. PCR Methods Appl. 1: 124-128. [DOI] [PubMed] [Google Scholar]
- Kellis, M., Patterson, N., Endrizzi, M., Birrent, B., and Lander, E.S. 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423: 233-234. [DOI] [PubMed] [Google Scholar]
- Korf, I., Flicek, P., Duan, D., and Brent, M.R. 2001. Integrating genomic homology into gene structure prediction. Bioinformatics 17: 140-148. [DOI] [PubMed] [Google Scholar]
- Reboul, J., Vaglio, P., Tzellas, N., Thierry-Mieg, N., Moore, T., Jackson, C., Shin-I, T., Kohara, Y., Thierry-Mieg, D., Thierry-Mieg, J., et al. 2001. Open-reading-frame sequence tags (OSTs) support the existence of at least 17,300 genes in C. elegans. Nat. Genet. 27: 332-336. [DOI] [PubMed] [Google Scholar]
- Reboul, J., Vaglio, P., Rual, J.F., Lamesch, P., Martinez, M., Armstrong, C.M., Li, S., Jacotot, L., Bertin, N., Janky, R., et al. 2003. C. elegans ORFeome version 1.1: Experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat. Genet. 34: 35-41. [DOI] [PubMed] [Google Scholar]
- Stein, L., Sternberg, P., Durbin, R., Thierry-Mieg, J., and Spieth, J. 2001. WormBase: Network access to the genome and biology of Caenorhabditis elegans. Nucleic Acids Res. 29: 82-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, L.D., Bao, Z., Blasiar, D., Blumenthal, T., Brent, M.R., Chen, N., Chinwalla, A., Clarke, L., Clee, C., Coghlan, A., et al. 2003. The genome sequence of Caenorhabditis briggsae: A platform for comparative genomics. PloS Biol. 1: E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaglio, P., Lamesch, P., Reboul, J., Rual, J.-F., Martinez, M., Hill, D., and Vidal, M. 2003. WorfDB: The Caenorhabitis elegans ORFeome Database. Nucleic Acids Res. 31: 237-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhout, A.J., Sordella, R., Lu, X., Hartley, J.L., Temple, G.F., Brasch, M.A., Thierry-Mieg, N., and Vidal, M. 2000a. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science 287: 116-122. [DOI] [PubMed] [Google Scholar]
- Walhout, A.J., Temple, G.F., Brasch, M.A., Hartley, J.L., Lorson, M.A., van den Heuvel, S., and Vidal, M. 2000b. Gateway recombinational cloning: Application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 328: 575-592. [DOI] [PubMed] [Google Scholar]
- Yamada, K., Lim, J., Dale, J.M., Chen, H., Shinn, P., Palm, C.J., Southwick, A.M., Wu, H.C., Kim, C., Nguyen, M., et al. 2003. Empirical analysis of the transcriptional activity in the Arabidopsis genome. Science 302: 842-846. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://elegans.swmed.edu/Announcements/genome_complete.html; The Caenorhabditis elegans WWW server.
- http://ftp.genome.washington.edu/cgi-bin/genefinder_req.pl; GeneFinder Web Server.
- http://worfdb.dfci.harvard.edu; WorfDB, the central repository of the C. elegans ORFeome.
- http://ws100.Wormbase.org; frozen release WS100 of Wormbase.
- http://www.ddbj.nig.ac.jp/; DNA Data Bank of Japan.
- http://www.ebi.ac.uk/embl/; EMBL nucleotide sequence database.
- http://www.hgmp.mrc.ac.uk/geneservice/; MRC geneservice.
- http://www.openbiosystems.com/; Open Biosystems.
- http://www.Wormbase.org; most updated version of Wormbase.