Abstract
The understanding of gene function increasingly requires the characterization of DNA segments containing promoters and their associated regulatory sequences. We describe a novel approach for linking multiple DNA segments, here applied to the generation of promoter::reporter fusions. Promoters from Caenorhabditis elegans genes were cloned using the MultiSite Gateway cloning technology. The capacity for using this system for efficient construction of chimeric genes was explored by constructing promoter::reporter gene fusions with a gfp reporter. The promoters were found to provide appropriate expression of GFP upon introduction into C. elegans, demonstrating that the short Gateway recombination site between the promoter and the reporter did not interfere with transcription or translation. The recombinational cloning involved in the Gateway system, which permits the highly efficient and precise transfer of DNA segments between plasmid vectors, makes this technology ideal for genomics research programs.
With genomes sequenced, biological investigations are increasingly organized on a genome-wide scale. The strategies followed depend on techniques that can be applied in parallel to a large number of samples, typically one for each gene in a genome. Microarrays are a major example used in the study of gene transcripts, but other aspects of the genome also need to be studied at this scale.
The Gateway cloning technology (Hartley et al. 2000) utilizes site-specific recombination to transfer DNA segments between vector backbones. This technology allows genome-wide sets of proteins, the functional products of most genes, as encoded by cloned open reading frames (ORFs), to be studied in multiple ways in a high-throughput mode. Once cloned, the ORFs can be efficiently recloned, in parallel, into numerous alternative plasmid vectors (Destination Vectors), each of which can be designed for expression and study of the protein products with a specific desired technique. The recombination reactions occur in small, λ-derived, recombination sites flanking the DNA segments of interest. As a result, the integrity of the transferred sequence remains intact and resulting fusions can be directly compared. A complete set of Gateway “ORF Entry Clones”, the starting point for recloning into any Gateway destination vector, thereby becomes a highly valuable genomic resource. Such a strategy has been demonstrated for the large scale determination of protein-protein interactions for Caenorhabditis elegans (Walhout et al. 2000).
Another major component of the genome that needs to be investigated on a genome-wide scale is the DNA containing the regulatory elements that direct the appropriate temporal and spatial expression of the genes as an organism develops or responds to external stimuli. The genome of C. elegans is relatively densely packed for a metazoan (The C. elegans Sequencing Consortium 1998) and transcriptional regulatory elements, including enhancers, tend to be close upstream of the translational initiation codon. These upstream regions could be the targets of Gateway cloning, in preparation for parallel study through recloning into alternative Destination Vectors designed for studying promoter function through different experimental approaches.
Further advancements in the Gateway Cloning technology have provided an expanded collection of recombination sites, each with unique specificities (Cheo et al. 2004). By developing strategies that employ three or more unique recombination site specificities, two or more DNA segments can be linked with high efficiency in an order and orientation-specific way. Using this approach, a collection of Promoter Entry Clones and a collection of ORF Entry Clones could be generated such that they could be efficiently fused in any combination. Although there are other recombination-based cloning technologies, in which the recombination reactions are carried out in vitro (Liu et al. 1998) or in vivo either in Escherichia coli (Court et al. 2002) or in Saccharomyces cerevisiae (Winzeler et al. 1999), compatibility with the established library of Gateway ORF clones for C. elegans (Walhout et al. 2000; Reboul et al. 2001) would be a significant advantage. The feasibility of using MultiSite Gateway technology for cloning C. elegans promoters is explored here.
RESULTS
Expression patterns have been determined for a few hundred of the 20,000 C. elegans genes, using reporter gene fusions generated by conventional cloning techniques (http://www.wormbase.org). Four promoters, for C. elegans genes B0464.4, F44B9.2, F54D5.1, and F56D5.8, that we had previously fused to a lacZ reporter (Lynch et al. 1995; Mounsey et al. 2002) were selected for recloning downstream of a gfp reporter to test the MultiSite Gateway system. These promoters were all contained on relatively small fragments, at less than 3 kb, and drove β-galactosidase expression in a range of tissues. The strategy followed is outlined in Figure 1 and the MultiSite Gateway system is described in detail in Cheo et al. (2004).
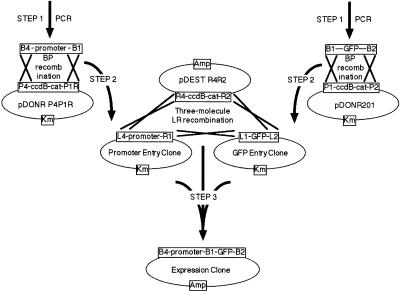
Figure 1.
Construction of reporter gene fusions through a three-molecule LR reaction. The Expression Clone, containing the promoter::gfp reporter gene fusion, is generated through a series of three steps (bold arrows) that involve specific recombination reactions (crosses). In Step 1, recombination sites of the appropriate specificity (determined by the sequence of the core 7 nucleotides within the att sites) are appended to the desired DNA segment by PCR. In Step 2, the Promoter Entry Clone is generated by BP recombination of attB4-promoter-attB1 PCR product with the attP4 and attP1R sites, respectively on pDONR P4P1R. The GFP Entry Clone is generated in a standard Gateway reaction by BP recombination of an attB1-ORF-attB2 PCR product with the attP1 and attP2 sites, respectively, on pDONR201. Selection for kanamycin resistance and against ccdB, upon DH5α transformation with the BP reactions, ensures the correct Entry Clone plasmids are obtained. In Step 3, these Entry Clones and the desired Destination Vector (pDEST R4R2) are combined in a three-molecule LR recombination. In this reaction, which depends on the specificity of the attL1 with attR1, attL2 with attR2, and attL4 with attR4 recombinations, the promoter and GFP DNAs are linked and transferred into the Destination Vector, pDEST R4R2. Selection for ampicillin resistance and against ccdB, upon transformation of DH5α, ensures only the desired Expression Clone is obtained.
The promoter regions were amplified by PCR from C. elegans total genomic DNA. Primers were designed with the appropriate Gateway recombination site, attB4 for the upstream primer and attB1.1 for the downstream primer. Downstream of the recombination sites, the primers included nucleotide sequences to hybridize to the ends of the promoter fragments, as cloned previously in the construction of the lacZ fusions. The PCR products were cloned into pDONR P4P1R through a BP recombination reaction to generate Promoter Entry Clones. In the BP reactions, recombination occurs specifically between the attB sites on the ends of the PCR products and the attP sites in the vector.
gfp was recloned in parallel with the Gateway cloning of the promoters. The template for the PCR was the plasmid pPD95.67 (Miller et al. 1999) which contains a gfp reporter designed specifically for studying expression in C. elegans. The upstream and downstream primers had attB1.1 and attB2.1 Gateway recombination sites, respectively. The PCR product was cloned into pDONR201 through a BP reaction to generate a gfp (“ORF”) Entry Clone (Fig. 1).
After transformation of E. coli with the products of recombination, plasmid was prepared from two independent clones for each of the promoter BP reactions and from four independent clones for the gfp BP reaction. Restriction enzyme analysis revealed all but one of the plasmids to contain the desired insert. Recombination with a small-sized PCR by-product, such as a primer dimer, despite the PEG precipitation step included to remove them, would explain the origin of the single clone that was not the desired product. Alternatively, background such as this could result from inactivation or loss of the ccdB gene in a small proportion of the vector plasmid preparation. This single example does not allow assessment of the frequency with which such background plasmids might be produced, but clearly the desired plasmids are the principal product.
One of the Promoter Entry Clones for each C. elegans gene was then used in a three-molecule LR reaction with a gfp Entry Clone and with the linearized vector plasmid, pDEST R4R2. In LR reactions, recombination occurs specifically between attL sites and the corresponding attR sites (Fig. 1). After transformation of E. coli with the products of recombination, plasmid was prepared from four independent clones for each promoter and restriction enzyme analysis revealed all 16 to be correct. Although the numbers are small, the success rate still testifies to the power of the MultiSite Gateway recombination system in ensuring only the desired molecules recombine in the desired manner in a three-molecule reaction.
The Expression Clones, containing the promoter::gfp fusions, were used in transformation of C. elegans and GFP expression was examined in established transgenic lines (Fig. 2). B0464.4::gfp gave expression in a small number of nerve cells in the head, F44B9.2::gfp gave expression in all body wall and vulval muscle cells and F54D5.1::gfp gave expression in the pharyngeal and rectal valves, all from late embryogenesis until the adult stage, the same patterns as had been observed for the corresponding lacZ fusions. No expression was detected, however, for F56D5.8::gfp. This failure is unlikely to result from misincorporation during PCR amplification of the promoter as a second, independent Promoter Entry Clone was also assayed. Since the original lacZ fusion was constructed, F56D5.8 has been deemed a pseudogene in the C. elegans genome annotation (http://www.wormbase.org) and therefore the Gateway-generated result obtained using the gfp reporter may be the more reliable. These results demonstrate that the Gateway recombination site between the promoter and the gfp reporter does not appear to interfere with gene expression in C. elegans and MultiSite Gateway technology can be used in a large scale study of C. elegans promoters.
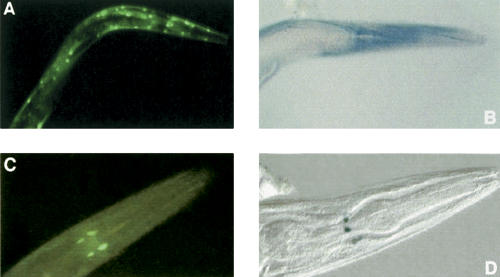
Figure 2.
GFP (A,C) and β-galactosidase (B,D) expression, in the head region of transgenic adult C. elegans, driven by F44B9.2 (A,B) and B0464.4 (C,D) promoter fragments. The gfp reporter gene fusions were generated using MultiSite Gateway while the lacZ reporter gene fusions had been generated previously by conventional cloning. Expression is in body wall muscle cells for F44B9.2 and in nerve cells for B0464.4. Micrographs of C. elegans strains UL1110 (A), UL484 (B), UL1111 (C), and UL42 (D) were captured at 400× magnification.
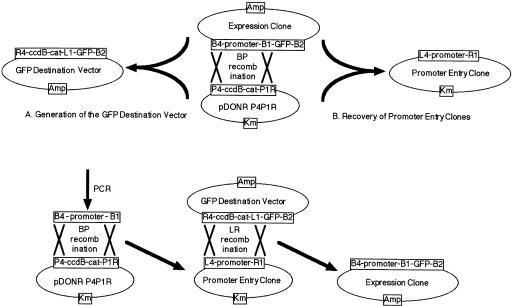
The flexibility of the system was demonstrated in experiments taking the Expression Clone containing the B0464.4::gfp fusion back through two different BP reactions. First, a GFP Destination Vector was generated (Fig. 3A) by performing a BP reaction between the B0464.4::gfp Expression Clone, linearized in the promoter fragment, and circular pDONR P4P1R, selecting for both ampicillin and chloramphenicol resistance, in DB3.1 cells, to allow replication of plasmids carrying ccdB. Plasmid preparations from five independent clones were all shown to be the desired product by restriction enzyme digestion. The GFP Destination Vector generated contains the ccdB cassette flanked by R4 and L1 recombination sites and so can be used in two-molecule LR reactions with Promoter Entry Clones to form promoter::gfp reporter gene fusions (Fig. 3C). It was anticipated that this would improve efficiency to a level allowing construction of promoter reporter gene fusions in a one-tube, combined, BP/LR protocol (see below). Second, if a one-tube protocol was utilized it would be important to be able to recover Promoter Entry Clones from Expression Clones so that selected promoters, such as those that had been demonstrated to drive GFP expression in C. elegans, could be used in LR reactions with other Destination Vectors for other experimental approaches. The B0464.4 promoter Entry Clone was indeed reliably derived from the Expression Clone in a BP reaction (Fig. 3B) with pDONR P4P1R linearized inside the ccdB cassette with selection for kanamycin resistance and against ccdB in DH5α cells.
Figure 3.
Reactions concerned with the one-tube protocol for generation of promoter reporter gene fusions. (A) To generate the GFP Destination Vector, the B0464.4::gfp Expression Clone was linearized by digestion in the promoter segment with SalI before use in a BP recombination. Transformation of DB3.1 cells allowed recovery of plasmids containing the ccdB cassette. Selection for ampicillin resistance yielded the desired plasmids. (B) To recover the promoter Entry Clone, pDONR P4P1R was linearized by digestion in the ccdB cassette with BamHI before use in a BP reaction. Transformation of DH5α cells selected against plasmids containing the ccdB cassette and with selection for kanamycin resistance only the desired plasmid was obtained. (C) In the one-tube protocol, promoter::reporter gene fusions would be generated by carrying out the BP and LR recombination steps successively, in a single tube. Bold arrows indicate steps involved and crosses indicate recombination reactions.
If the primary aim was to prepare promoter::reporter gene fusions for as many C. elegans genes as possible, the one-tube, combined BP/LR reaction, protocol (Fig. 3C) could provide a significant gain in efficiency. The one-tube protocol would avoid a cloning step, halving the number of bacterial transformations and plasmid preparations required. In addition, it would be preferable to utilize larger promoter fragments, to increase the likelihood that an upstream region from an uncharacterized C. elegans gene would contain an intact promoter. Therefore the one tube protocol was attempted using one of the genes utilized above, F44B9.2, with a 1.8-kb promoter fragment, along with 5-kb promoter fragments for two more C. elegans genes, pes-1 and fkh-2 (Molin et al. 2000).
PCR products flanked by attB4 and attB1.1 recombination sites were generated as before, but this time for the promoters of the genes F44B9.2, pes-1, and fkh-2. The PCR products were used in BP reactions with pDONR P4P1R before adding linearized GFP Destination Vector and carrying out the LR reaction in the same tube. Transformation of DH5α cells with the products of the one-tube reaction, selecting for ampicillin resistance, should yield the desired Expression Clones containing the promoter/gfp reporter gene fusions (Fig. 3C). Plasmid DNA was prepared for three of the eight independent clones obtained from the F44B9.2 reaction and two of the three were the intended plasmid as determined by restriction enzyme analysis. This demonstrated the utility of the GFP Destination Vector and the feasibility of the one-tube protocol. However, reactions for pes-1 and fkh-2 failed to yield any bacterial transformants. Presumably, the longer promoter fragments for pes-1 and fkh-2, in comparison to that used for F44B9.2, reduced the efficiency of the procedure. Reliability of this one-tube protocol would need to be improved for utilization of this approach on a larger scale.
Promoter::gfp reporter gene fusions were generated using 5 kb promoter fragments, for both pes-1 and fkh-2, but using separate BP and LR reactions, with Promoter Entry Clones generated in the intervening step. The Expression Clones were used in C. elegans transformation. Both the pes-1::gfp and fkh-2::gfp fusion genes gave expression, and apparently in the same pattern, in specific cells of the early embryo, as had been observed previously for similar fusions made using conventional cloning procedures (Molin et al. 2000).
Finally, 16 genes from C. elegans chromosome I, for which expression patterns had not previously been determined, were selected for constructing reporter gene fusions using the MultiSite Gateway system. The primers used for PCR amplification of the promoter fragments (Supplemental Table 1) were designed such that fusion would be to the middle of the second exon of each C. elegans gene, a position that may maximize the likelihood of observing reporter gene expression (A. Mounsey and I.A. Hope, unpubl.). Separate BP and LR reactions, with intervening cloning of the Promoter Entry Clones, were used to join 5 kb, PCR generated, promoter fragments to gfp from the GFP Destination Vector. Reporter gene fusions were obtained for all 16 C. elegans genes, with 74% of the clones generated from the BP reactions and 86% of the clones generated from the LR reactions being that desired.
Upon transformation of C. elegans with the Expression Clones generated, reporter gene expression was observed for 10 of the 16 genes (Fig. 4). This proportion compares favorably with success rates for reporter gene fusions made conventionally, for otherwise uncharacterized genes, in previous large-scale studies (Lynch et al. 1995; Mounsey et al. 2002); reporter gene expression had been observed for 178 of 364 genes assayed, or 49%. Different promoters directed expression in a wide range of cell types, including the hypodermis (C06G4.4), the spermatheca (C26E6.4), and pharyngeal nerve cells (C05D2.4/bas-1). The promoters for B0336.10 and C16A3.4 appeared to drive expression in all cell types. The fusion proteins encoded by the Expression Clones for C06E1.10 and C02F5.9, showed distinct and discreet subcellular localizations, with the weak, nuclear localization signal of the reporter gene being completely overridden by the portion of the fusion proteins encoded by the C. elegans genes (Fig. 4E,F). Reporter expression was also observed for B0393.1, C05D11.3, and C27D11.1 (but not B0280.9, B0361.5, B0393.5, C05D11.9, C23G10.8, or C45G9.2). The range of expression patterns observed is consistent with the general applicability of the MultiSite Gateway generation of reporter gene fusions.
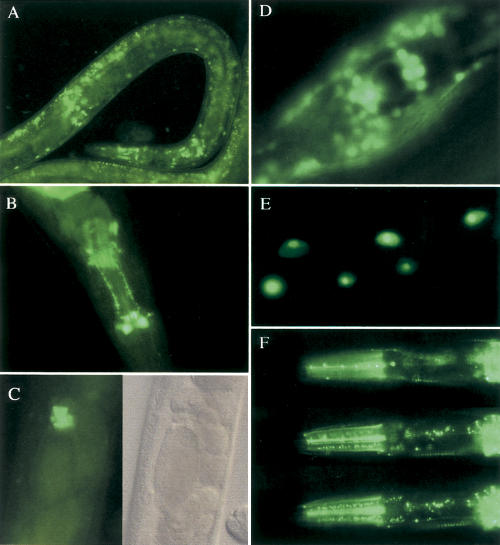
Figure 4.
Photomicrographs of GFP expression patterns for previously uncharacterized genes. (A) Hypodermal and pharyngeal expression for C06G4.4 in a late larva of strain UL1171. (B) Pharyngeal nerve cell expression for bas-1 / C05D2.4 in strain UL1164. (C) Expression in the spermatheca for C26E6.4 in a young adult of strain UL1173 presented with fluorescent and Nomarski images. (D) The general expression for C16A3.4 in UL1162 includes the developing vulva. (E) Nucleolar-localized expression in intestinal nuclei of strain UL1166 for C06E1.10. In WormBase (www.wormbase.org), C06E1.10 has been identified as an ATP-dependent RNA helicase, which would be consistent with the nucleolar localization. (F) Three different focal planes through the head of a young adult from strain UL1167 reveals discreet subcellular GFP localization, in subcellular foci and apparently at specific cellular surfaces, in different cell types, for C02F5.9. In WormBase (www.wormbase.org), C02F5.9 has been identified as a proteasome β-subunit.
DISCUSSION
Reporter gene fusions have been generated using the MultiSite Gateway technology and shown to be expressed in C. elegans. The attB1.1 Gateway recombination site between the promoter and reporter did not appear to interfere with expression and, with the ease of the steps involved, the Gateway system is thereby demonstrated to be suitable for genomic scale analysis of promoters. Use of the MultiSite system would allow promoters to be combined readily with any protein-coding region cloned as a Gateway ORF Entry Clone. This would allow efficient construction of chimeric genes for heterologous expression studies in C. elegans or fusion of promoters and their corresponding entire ORF for each C. elegans gene to a reporter gene to reveal subcellular gene product distributions (Dupuy et al. 2004). In addition, promoter fragments could be efficiently recloned into other Destination Vectors to allow promoter characterization using other experimental approaches.
The one-tube protocol could expedite construction of promoter::reporter gene fusions, avoiding an entire cloning step and thereby improving efficiency for large scale studies. However, improvement in success rates for the one-tube protocol, through development of the procedure beyond that described here, would be needed, particularly if larger fragments are targeted. As Promoter Entry Clones may have more general value than the Expression Clones carrying the reporter gene fusions, the two-tube protocol may be preferable over the one-tube protocol anyway. The two-tube protocol is demonstrated here to have the efficiency and reliability to be applied to construction of promoter::reporter gene fusions on a genomic scale. Although clonase, the enzyme that catalyses the Gateway recombination reactions, is more expensive than the ligases used conventionally for joining DNA fragments and each primer used for PCR amplification requires an extra 28 bases, the ease and reliability of the technology, combined with the versatility of the Entry clones makes this approach more economical.
While Gateway technology facilitates the joining of reporter and promoters, the subsequent assay of the reporter gene fusions, on a genomic-scale, remains formidable. C. elegans needs to be transformed with each reporter gene fusion and the techniques currently available, microinjection (Mello et al. 1991) or micro-projectile bombardment (Praitis et al. 2001), are labor intensive. Furthermore, full interpretation of the expression patterns requires individual attention. Most transformed lines carry the transgenic DNA as extrachromosomal tandem arrays, which complicates expression pattern characterization due to mosaicism and silencing of the transgenes. Low-copy integrative transformation, as can be achieved with microprojectile bombardment (Praitis et al. 2001), reduces these difficulties but takes more time. While a few C. elegans laboratories dedicated to assaying of the reporter gene fusions would be able to assay a substantial fraction of the promoters in the genome, the more general value of the cloned promoterome (Dupuy et al. 2004) may lead to other C. elegans laboratories contributing to the task.
Data on developmental distributions of promoter activity are a crucial layer in the global understanding being generated by genomic analysis. The MultiSite Gateway cloning technology provides a robust approach for working with C. elegans promoters on a genomic-scale.
METHODS
PCRs
Promoter Entry Clones were generated from PCR products flanked by attB4 and attB1.1R sites. The 5′ end of the upstream attB4 primer was GGGGACAACTTTGTATAGAAAAGTTGTG. The 5′ end of the downstream attB1.1R primer was GGGGA CAACTTTTTTGTACAAAGTTGC. The GFP Entry Clone was generated from PCR products made using primers with flanking attB1.1 and attB2.1 recombination sites. The 5′ end of the upstream primer was GGGGACAACTTTGTACAAAAAAGTTGTG, thereby including the attB1.1 sequence. The 5′ end of the downstream primer was GGGGACAACTTTGTACAAGAAAGTTGC thereby including an attB2.1 site.
The gene-specific 3′ ends of the primers were designed to hybridize to the target DNA to be PCR amplified, with an annealing temperature of 60°C (http://alces.med.umn.edu/rawtm.html). For amplification of promoter fragments, the upstream and downstream promoter-specific sequences described as in Supplemental data Table 1 were added to the end of the attB4 and attB1.1 recombination sites, respectively. The downstream primer was designed to anneal in an exon to provide a translation fusion to the reporter gene, ensuring that the translational reading frame would be maintained. The reading frame used matches that used in the C. elegans ORFeome project (Reboul et al. 2001). For amplification of the gfp reporter, the gene-specific sequence added to the upstream attB1.1 recombination site was AATTGGCCAAAGGACCCAAA and the gene-specific sequence added to the downstream attB2.1 recombination site was GGCCGACTAGTAGGAAACAGTTATG.
PCR reactions contained 0.5 pmol/μl of each primer, 2 mM MgCl2, 1 mM each dNTP, template DNA and 2.5 units of Platinum Taq DNA Polymerase High Fidelity (Invitrogen) in the provided buffer, appropriately diluted. C. elegans genomic DNA was prepared either in bulk (Sulston and Hodgkin 1988) for use as template at 0.2 ng/μl or as a single worm lysis (Epstein and Shakes 1995), one worm per PCR reaction. PCRs were incubated at 94°C for 1 min, followed by 30 cycles of 94°C for 30 sec, 55°C for 30 sec, and 68°C for 4 min (or 6 min for 5-kb fragments), with a final incubation at 68°C for 10 min PCR products were purified by diluting PCR reactions three-fold with TE (10 mM Tris, 1 mM EDTA, pH 8) before adding a half volume of 30% PEG8000/30mM MgCl2, microfuging for 15 min at room temperature and resuspending the DNA in TE.
Recombination Reactions
Gateway BP reactions contained PCR products or linearized plasmid DNA at 1 ng/μl and circular plasmid DNA at 7.5 ng/μl. The B0464.4::gfp Expression Clone was linearized in the promoter fragment, for generation of the GFP Destination Vector, by digestion with SalI. pDONR P4P1R was linearized in the ccdB cassette with BamHI, for recovery of promoter Entry Clones. The three-molecule Gateway LR reactions contained a promoter Entry Clone plasmid and the GFP Entry Clone plasmid, both circular, at 5 ng/μl and pDEST R4R2, linearized in the ccdB cassette by digestion with NcoI, at 15 ng/μl. The two-molecule LR reactions contained the circular promoter Entry Clone plasmids at 5 ng/μl and the GFP Destination Vector, linearized with SalI, at 15 ng/μl. Gateway BP or LR clonase (Invitrogen) and appropriate provided buffer were diluted five-fold in assembly of the reactions, which were then incubated at 25°C for 2 hr (or 6 hr for the three-molecule reaction). For the one tube protocol, after a BP reaction was performed as above, the following were added per 10 μl of BP reaction, with subsequent incubation at 25°C for 2 h; 0.5 μl of 0.75 M NaCl, 1.5 μl of 150 ng/μl GFP destination vector, linearized with SalI, and 3 μl LR clonase enzyme mix. A tenth volume of 2 mg/ml proteinase K was added at the end of recombination reactions, with incubation for 10 min at 37°C, before using the products in bacterial transformation.
Other Techniques
Bacterial transformation was carried out using library efficiency DH5α or DB3.1 frozen competent cells (Invitrogen). DH5α was used to select against plasmids containing the ccdB gene while DB3.1 was used to allow replication of plasmids containing the ccdB gene. Plasmids were prepared using Qiagen kits. Restriction enzyme analyses were performed using standard molecular procedures (Sambrook et al. 1989). Transformation of wild-type C. elegans (N2) with plasmids bearing reporter gene fusions were carried out by microinjection using the plasmid pRF4 as the coinjection marker to confer a rolling phenotype on transformed animals (Mello et al. 1991). GFP expression was observed and recorded, for established transgenic lines, using a Zeiss Axioplan microscope equipped for epifluorescence.
Acknowledgments
We thank the Medical Research Council-UK for support in the early stages of the project (G9810286) and the National Institute of Health/National Cancer Institute for support in the later stages of the project (R21 CA97516). The authors and the laboratories involved in this work do not have personal financial interests in the Invitrogen Corporation, the company responsible for the commercial development of the MultiSite Gateway cloning system. However, this work was facilitated by early access to the MultiSite Gateway technology, for testing purposes, prior to general release, for which the authors are grateful to Invitrogen.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2463804.
References
- The C. elegans Sequencing Consortium. 1998. Genome sequence of the nematode C. elegans A platform for investigating biology. Science 282: 2012-2018. [DOI] [PubMed] [Google Scholar]
- Cheo, D.L., Titus, S.A., Byrd, D.R.N., Hartley, J.L., Temple, G.F., and Brasch, M.A. 2004. Concerted assembly and cloning of multiple DNA segments using in vitro site-specific recombination: Functional analysis of multi-segment expression clones. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Court, D.L., Sawitzke, J.A., and Thomason, L.C. 2002. Genetic engineering using homologous recombination. Annu. Rev. Genet. 36: 361-388. [DOI] [PubMed] [Google Scholar]
- Dupuy, D., Li, Q.-R., Deplancke, B., Boxem, M., Hao, T., Lamesch, P., Sequerra, R., Bozak, S., Doucette-Stamm, L., Hope, I.A., et al. 2004. A first version of the Caenorhabditis elegans promoterome. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Epstein, H.F. and Shakes, D.C. 1995. Caenorhabditis elegans: Modern biological analysis of an organism. Academic Press, San Diego, CA.
- Hartley, J.L., Temple, G.F., and Brasch, M.A. 2000. DNA cloning using in vitro site-specific recombination. Genome Res. 10: 1788-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q., Li, M.Z., Leibham, D., Cortez, D., and Elledge, S.J. 1998. The univector plasmid-fusion system, a method for rapid construction of recombinant DNA without restriction enzymes. Curr. Biol. 8: 1300-1309. [DOI] [PubMed] [Google Scholar]
- Lynch, A.S., Briggs, D., and Hope, I.A. 1995. Developmental expression pattern screen for genes predicted in the C. elegans genome sequencing project. Nat. Genet. 11: 309-313. [DOI] [PubMed] [Google Scholar]
- Mello, C.C., Kramer, J.M., Stinchcomb, D., and Ambros, V. 1991. Efficient gene transfer in C. elegans: Extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10: 3959-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, D.M., Desai, N.S., Hardin, D.C., Piston, D.W., Patterson, G.H., Fleenor, J., Xu, S., and Fire, A. 1999. Two-color GFP expression system for C. elegans. BioTechniques 26: 914-921. [DOI] [PubMed] [Google Scholar]
- Molin, L., Mounsey, A., Aslam, S., Bauer, P., Young, J., James, M., Sharma-Oates, A., and Hope, I.A. 2000. Evolutionary conservation of redundancy between a diverged pair of forkhead transcription factor homologues. Development 127: 4825-4835. [DOI] [PubMed] [Google Scholar]
- Mounsey, A., Bauer, P., and Hope, I.A. 2002. Evidence suggesting that a fifth of annotated Caenorhabditis elegans genes may be pseudogenes. Genome Res. 12: 770-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis, V., Casey, E., Collar, D., and Austin, J. 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157: 1217-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboul, J., Vaglio, P., Tzellas, N., Thierry-Mieg, N., Moore, T., Jackson, C., Shin-i, T., Kohara, Y., Thierry-Mieg, D., Thierry-Mieg, J., et al. 2001. Open-reading-frame sequence tags (OSTs) support the existence of at least 17,300 genes in C. elegans. Nat. Genet. 27: 332-336. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. 1989. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sulston, J. and Hodgkin, J. 1988. Methods. In The nematode Caenorhabditis elegans (ed. W.B. Wood), pp. 587-606. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Walhout, A.J.M., Sordella, R., Lu, X., Hartley, J.L., Temple, G.F., Brasch, M.A., Thierry-Mieg, N., and Vidal, M. 2000. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science 287: 116-122. [DOI] [PubMed] [Google Scholar]
- Winzeler, E.A., Shoemaker, D.D., Astromoff, A., Liang, H., Anderson, K., Andre, B., Bangham, R., Benito, R., Boeke, J.D., Bussey, H., et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901-906. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.wormbase.org; the primary database for Caenorhabditis elegans.
- http://alces.med.umn.edu/rawtm.html; used for prediction of oligonucleotide primer melting temperatures.