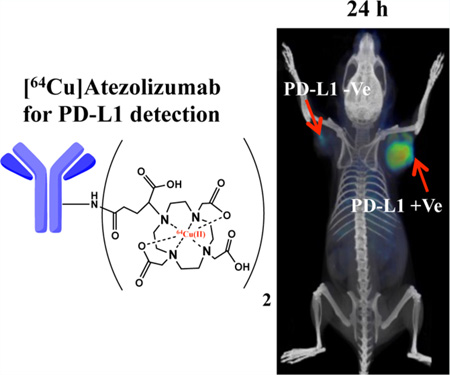
Abstract
The programmed death protein 1 (PD-1) and programmed deathligand 1 (PD-L1) pair is a major immune checkpoint pathway exploited by cancer cells to develop and maintain immune tolerance. With recent approvals of anti-PD-1 and anti-PD-L1 therapeutic antibodies, there is an urgent need for noninvasive detection methods to quantify dynamic PD-L1 expression in tumors and to evaluate the tumor response to immune modulation therapies. To address this need, we assessed [64Cu]atezolizumab for the detection of PD-L1 expression in tumors. Atezolizumab (MPDL3208A) is a humanized, human and mouse cross-reactive, therapeutic PD-L1 antibody that is being investigated in several cancers. Atezolizumab was conjugated with DOTAGA and radiolabeled with copper-64. The resulting [64Cu]atezolizumab was assessed for in vitro and in vivo specificity in multiple cell lines and tumors of variable PD-L1 expression. We performed PET-CT imaging, biodistribution, and blocking studies in NSG mice bearing tumors with constitutive PD-L1 expression (CHO-hPD-L1) and in controls (CHO). Specificity of [64Cu]atezolizumab was further confirmed in orthotopic tumor models of human breast cancer (MDAMB231 and SUM149) and in a syngeneic mouse mammary carcinoma model (4T1). We observed specific binding of [64Cu]atezolizumab to tumor cells in vitro, correlating with PD-L1 expression levels. Specific accumulation of [64Cu]atezolizumab was also observed in tumors with high PD-L1 expression (CHO-hPD-L1 and MDAMB231) compared to tumors with low PD-L1 expression (CHO, SUM149). Collectively, these studies demonstrate the feasibility of using [64Cu]atezolizumab for the detection of PD-L1 expression in different tumor types.
Graphical abstract
INTRODUCTION
Cancer cells exploit multiple mechanisms to evade immune recognition and establish immune tolerance.1 This tolerance is facilitated and maintained by regulatory immune cells, immunosuppressive cytokines and chemokines, and immune checkpoint pathways that downregulate immune cells’ antitumor activity. Over the past decade, immune modulation therapies that are focused on either antagonizing coinhibitory or activating costimulatory pathways have resulted in improved patient survival.2, 3 Therapeutic antibodies targeting immune checkpoint proteins are at the forefront of those advances.
A major therapeutic target in cancer immune modulation therapy is the axis between the two immune checkpoint proteins programmed death receptor 1 (PD-1, CD279) and its ligand PD-L1 (B7-H1, CD274).4 PD-1 is expressed on activated T, B, and NK cells, and PD-L1 is normally displayed as an autoimmune prevention mechanism on a subset of macrophages, endothelial cells, and other nonmalignant tissues. There is increased PD-L1 expression on a wide variety of tumors, including melanoma, nonsmall cell lung cancer, Merkel cell carcinoma, breast cancer, and squamous cell carcinomas.5–7 Promotion of PD-L1 expression in the tumor microenvironment (TME) is driven by both genetic and adaptive immune response mechanisms.8 Examples of the former include KRAS and PTEN mutations and translocation or amplification of the 9p24.1 gene in certain tumors.9–11 PD-L1 expression on tumor cells and immune cell infiltrates can also be induced as an adaptive mechanism in response to interferon-γ (INF-γ) secreted by antigen-bound tumor-infiltrating T cells.12 Increased expression of tumor and stromal cell PD-L1 leads to deactivation of PD-1-expressing tumor-infiltrating cytotoxic T cells, causing immune suppression in the TME.13
Clinical studies indicate that intratumoral PD-L1 expression, detected by immunohistochemistry (IHC), could be indicative of but not an effective biomarker of response to anti-PD-1 and anti-PD-L1 therapy.14, 15 Some of the contributing factors to the observed inconsistency are the unresolved issues with PD-L1 IHC, such as different antibodies for detection and variable criteria used for positive IHC in the evaluation of clinical trials14, 16 and the changes in PD-L1 expression in tumors induced by a dynamic TME. Obtaining repeated biopsies to evaluate PD-L1 expression in the TME is impractical, particularly in patients with metastatic disease, further limiting our ability to fully assess therapy-induced changes in PD-L1 expression in real time. Noninvasive imaging allows for repetitive measurements of target expression in all lesions in their entirety. Recent preclinical studies show that PD-L1 expression in tumors can be assessed by radionuclide imaging.17–21
We have previously shown in murine models that PD-L1 can be detected in the TME with 111In-radiolabeled atezolizumab using single photon emission computed tomography-computed tomography (SPECT-CT) imaging.19 Atezolizumab (MPDL3280a, PD-L1-mAb) is a humanized, human and mouse cross-reactive, PD-L1 antibody in clinical evaluation for treatment of advanced or metastatic bladder cancer,22 melanoma,23 nonsmall cell lung cancer,24 renal cell carcinoma, 25 and several other cancers. Atezolizumab binds both human and mouse PD-L1 with high affinity and provides an opportunity to evaluate PD-L1 expression in both human tumor xenografts and syngeneic mouse tumor models. Atezolizumab binds human and mouse PD-L1 with dissociation constants (Kd) of 0.43 and 0.13 nM, respectively.22, 26
The high sensitivity and quantitative features of positron emission tomography (PET) are increasingly used to image tumor target expression using radiolabeled antibodies.27, 28 For example, 64Cu-trastuzumab has been used to detect HER2 expression in tumors and metastases.29–31 Here, we report our evaluation of the ability of [64Cu]atezolizumab to detect PD-L1 expression in different tumor types using PET-CT.
RESULTS AND DISCUSSION
Fabrication of [64Cu]Atezolizumab
Atezolizumab was first conjugated with DOTAGA for stable chelation of 64Cu (Scheme S1). As indicated by MALDI-TOF spectrometry, the applied procedure resulted in conjugation of an average of two molecules of the chelator each antibody (Figure S1D). Subsequently, the atezolizumab−DOTAGA conjugate was radiolabeled with 64Cu, providing [64Cu]atezolizumab with radiochemical yields of 75 ± 5%, purity of >96% (as determined by ITLC), and specific activity of 8.6 ± 0.78 µCi/µg. Radiolabeling was further confirmed by SDS-PAGE, Coomassie blue staining, and autoradiography, under reducing and nonreducing conditions, indicating intact antibody upon chelation 64Cu (Figure S2).
[64Cu]Atezolizumab Shows PD-L1-Specific Cellular Uptake in Vitro
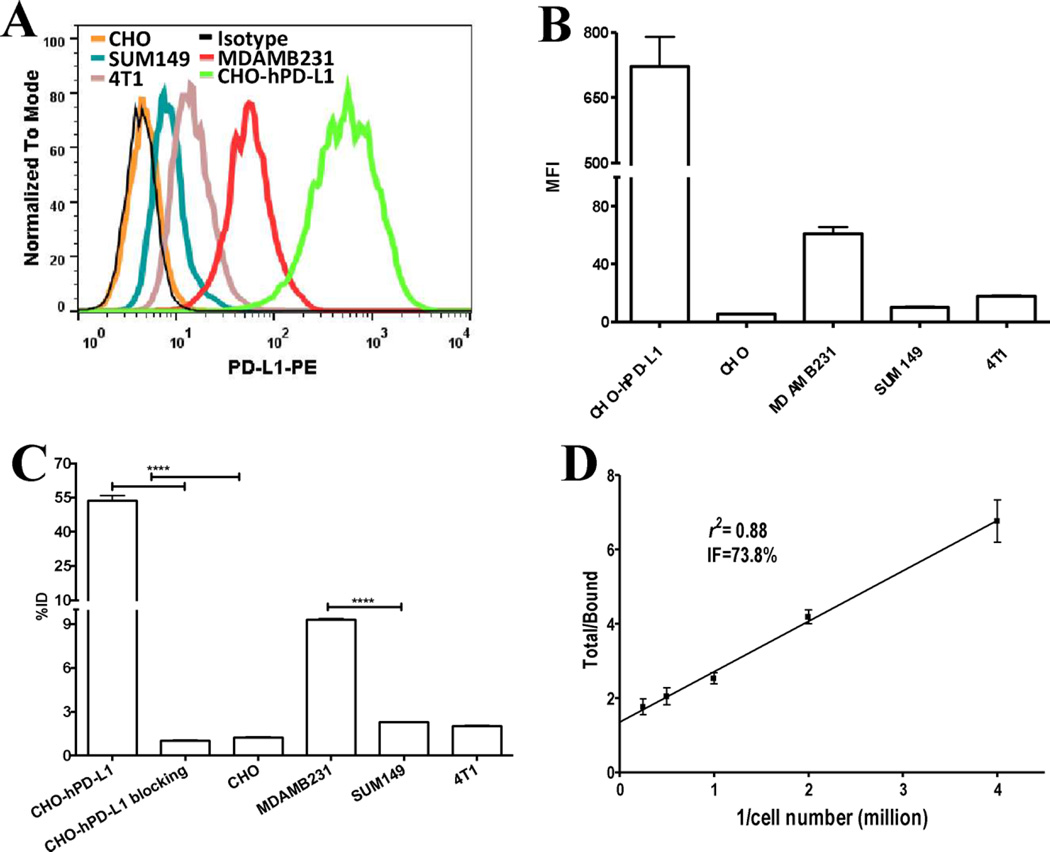
To evaluate the in vitro binding specificity of [64Cu]atezolizumab, we selected human and mouse cancer cell lines with varying PD-L1 surface expression. Flow cytometry analyses demonstrated the following order of PDL1 expression among cells tested: CHO-hPD-L1 > MDAMB231 > 4T1-Luc > SUM149 > CHO (Figure 1A and B). The in vitro binding assay detected significantly higher uptake of [64Cu]atezolizumab in the CHO-hPD-L1 and MDAMB231 cells with high PD-L1 expression than in the SUM149 and CHO cells with low PD-L1 expression (Figure 1C). The bound [64Cu]atezolizumab radioactivity correlated (correlation coefficient R = 0.997) with the flow cytometry mean fluorescence intensity (MFI) profile of the cells. The 4T1-Luc cells had low uptake, similar to that of SUM149 cells. A blocking study using unmodified nonradiolabeled atezolizumab in CHO-hPD-L1 cells showed significantly reduced uptake of [64Cu]atezolizumab, further confirming PD-L1-specific binding of [64Cu]atezolizumab. The immunoreactive fraction of [64Cu]atezolizumab assessed in CHO-hPD-L1 cells was 73.8% (Figure 1D).
Figure 1.
In vitro evaluation of [64Cu]atezolizumab. (A) Representative surface PD-L1 expression levels of studied cell lines analyzed by flow cytometry illustrated as a histogram and (B) mean fluorescence intensity (MFI). (C) In vitro binding specificity of [64Cu]atezolizumab for PD-L1 expression in various cell lines. (D) In vitro immunoreactive fraction of [64Cu]atezolizumab in CHO-hPD-L1 cells as determined by Lindmo analysis. ****, p < 0.0001.
Collectively, the in vitro results show that [64Cu]-atezolizumab can be used to detect varying levels of PD-L1 expression in cells of different cancer types.
[64Cu]Atezolizumab Specifically Accumulates in Tumors with High PD-L1 Expression
Validation of the Specificity of [64Cu]Atezolizumab in CHO-hPD-L1 and CHO Tumor Models
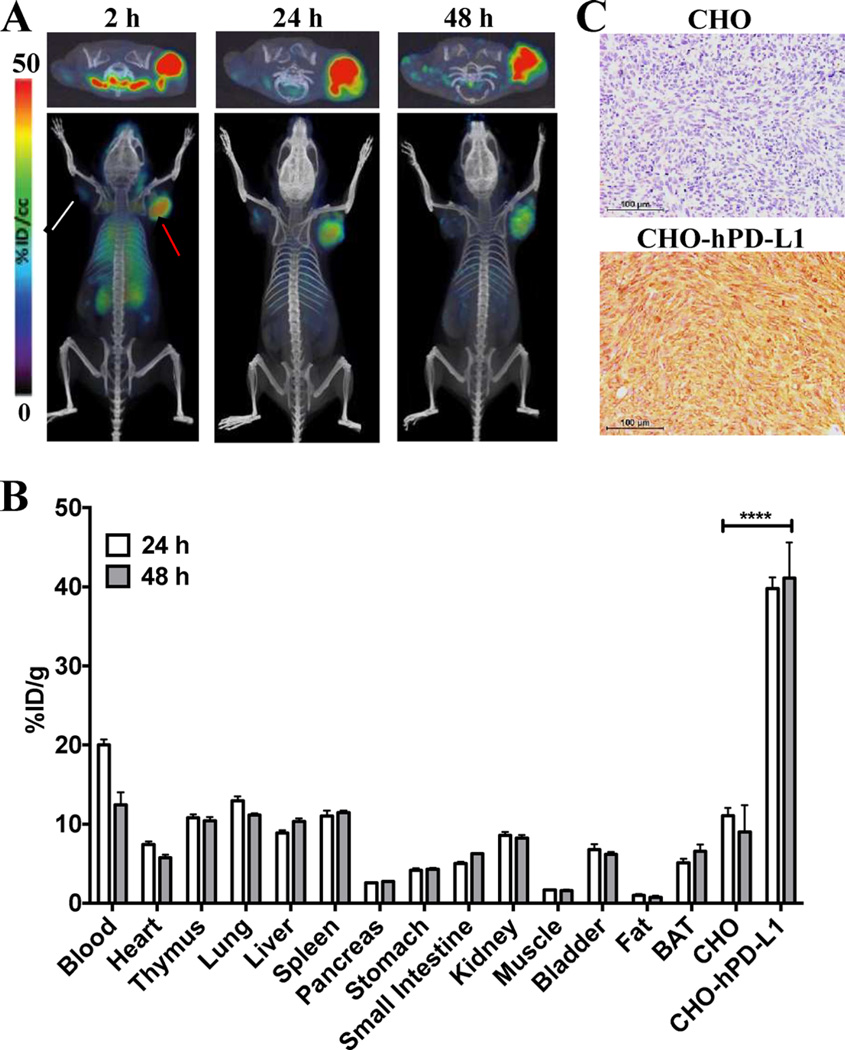
To validate the in vivo specificity of [64Cu]atezolizumab, we used NSG mice bearing CHO-hPDL1 tumors that stably expressed high levels of PD-L1. The PET-CT images of these mice indicated significantly higher accumulation of [64Cu]atezolizumab in CHO-hPD-L1 tumors compared to that in the control CHO tumors with low PD-L1 expression and other peripheral tissues at 24 and 48 h postinjection (pi) (Figure 2A). Biodistribution studies in the same model showed significantly elevated uptake of radioactivity in the CHO-hPD-L1 tumor with the percentage of injected dose per gram of tissue (%ID/g) of 39.8 ± 2.8 and 40.6 ± 6.9 at 24 and 48 h pi, respectively; in contrast, the %ID/ g values in the control CHO tumors were 11.1 ± 1.9 and 10.1 ± 3.3 at the same time points (Figure 2B). In peripheral organs (liver, spleen, heart, lungs, kidneys, and thymus) of the NSG mice bearing CHO-hPD-L1 tumors, uptake of [64Cu]-atezolizumab was similar to the level in CHO tumors. The imaging and biodistribution results were consistent with high PD-L1 immunoreactivity in CHO-hPD-L1 tumors compared to that in CHO tumors (Figure 2C).
Figure 2.
Evaluation of [64Cu]atezolizumab in CHO-hPD-L1 and CHO tumor models. (A) Transaxial (top panel) and volume rendered (bottom panel) PET-CT images of [64Cu]atezolizumab uptake in CHO-hPD-L1 (red arrow, high PD-L1 expression) and CHO (white arrow, low PD-L1 expression) tumors (n = 3) confirm PD-L1-mediated uptake of the radiotracer. (B) Ex vivo biodistribution analysis at 24 and 48 h after the injection of [64Cu]atezolizumab in the same tumor model (n = 4); ****, p < 0.0001. (C) Immunohistochemical analysis for PD-L1 expression demonstrating significantly higher immunoreactivity in CHO-hPD-L1 tumors (brown color) compared to that in CHO tumors.
Confirmation of the Specificity of [64Cu]Atezoluzumab in MDAMB321 and SUM149 TNBC Models
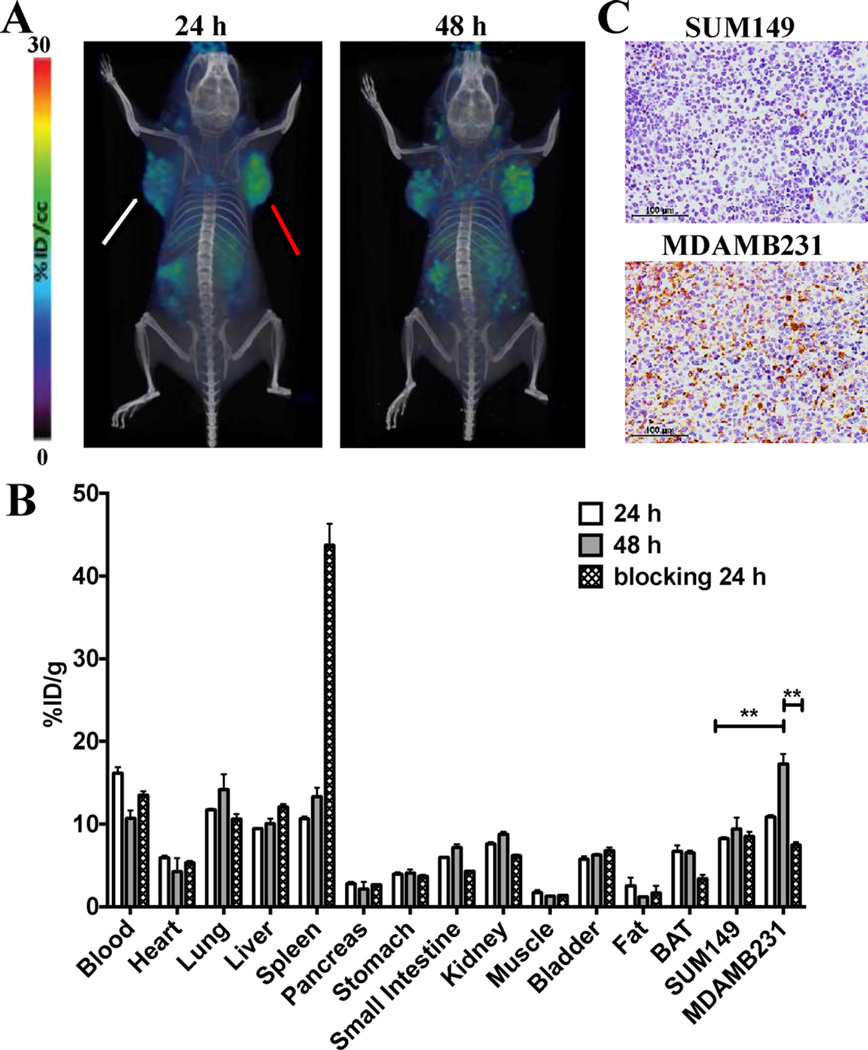
We further confirmed the specificity of [64Cu]atezolizumab to detect tumor PD-L1 expression in mice with MDAMB231 and SUM149 TNBC xenografts that have high and low endogenous cell surface PD-L1 expression, respectively. The PET-CT images of TNBC tumor-bearing mice showed a significantly increased uptake of [64Cu]atezolizumab in MDAMB231 tumors compared to that in SUM149 tumors (Figure 3A). These results were confirmed by ex vivo biodistribution, which showed high accumulation of the radioactivity in MDAMB231 tumors with %ID/g of 10.8 ± 0.4 and 17.2 ± 2.1 at 24 and 48 h pi, respectively; in contrast, the SUM149 tumors had values of 8.2 ± 0.26 and 9.4 ± 2.3 at 24 and 48 h pi, respectively (Figure 3B). The MDAMB231 tumor showed high tumor-to-muscle ratios of 13.2 ± 0.9 at 48 h pi. The intensity of PD-L1 staining in MDAMB231 tumors was significantly higher than in SUM149 tumors (Figure 3C) but lower than in CHO-hPDL1 tumors (Figure 2C), corroborating the lower uptake of [64Cu]atezolizumab in MDAMB231 tumors compared to that in CHO-hPD-L1 tumors. Unlike CHO-hPD-L1 tumors, [64Cu]atezolizumab uptake and retention was slow in MDAMB231 tumors and was prominent at 48 h, suggesting that PD-L1 expression levels and tumor physiology influence PD-L1 antibody accumulation and could potentially determine tumor response to therapy. The lowest uptake of radioactivity in the SUM149 xenografts agreed with the least intense PD-L1 immunoreactivity of the tumors (Figure 3C).
Figure 3.
Evaluation of [64Cu]atezolizumab in MDAMB231 and SUM149 tumor models. (A) PET-CT volume-rendered images of [64Cu]atezolizumab in MDAMB231 (red arrow, high PD-L1 expression) and SUM149 (white arrow, low PD-L1 expression) tumor-bearing mice (n = 3) confirm PD-L1 specific uptake of the radiotracer. (B) Ex vivo biodistribution analysis of [64Cu]atezolizumab at 24 and 48 h after injection in the same tumor models (n = 4); **, p < 0.01. (C) Immunohistochemical analysis for PD-L1 expression demonstrating high immunoreactivity in MDAMB231 tumors compared to that in SUM149 tumors.
The PD-L1-mediated uptake of [64Cu]atezolizumab in MDAMB231 tumors was further confirmed by reduced uptake of radioactivity in a blocking experiment in which mice were preinjected with unmodified nonradiolabeled antibody. The uptake of radioactivity in the MDAMB231 tumors was reduced by >50% at 24 h pi compared to that of mice that did not receive the blocking dose, suggesting PD-L1-specific blocking (Figure 3C). There was high accumulation of radioactivity in the spleen, possibly due to increased serum availability of the radiotracer after blocking. In contrast, there was low accumulation of the radioactivity in brown adipose tissue (BAT) and kidneys. BAT is an immunologically relevant tissue, and accumulated radioactivity is PD-L1 specific, as demonstrated previously by us and other groups in PD-L1 imaging studies.19, 32
TNBCs lack receptors for estrogen, progesterone, and human epidermal growth factor. Patients with TNBC have limited therapeutic options and have poorer outcomes than those with other subtypes of breast cancer.33 However, patients with TNBC treated with atezolizumab have a 30% increase in overall survival.3 Histological studies in untreated primary breast cancer specimens have indicated that tumor cell PD-L1 expression could serve as an indicator of active breast tumor immunity,7 suggesting that noninvasive PD-L1 detection could have predictive and prognostic implications for immune therapies targeting checkpoints in breast cancers. Results we observed in TNBC models (Figure 3) demonstrated that [64Cu]atezolizumab had the specificity to detect variable levels of PD-L1 with PET-CT and confirmed the feasibility of variable PD-L1 detection in TNBCs.
PD-L1 Detection in a Mammary Tumor Model in Immunocompetent Mice
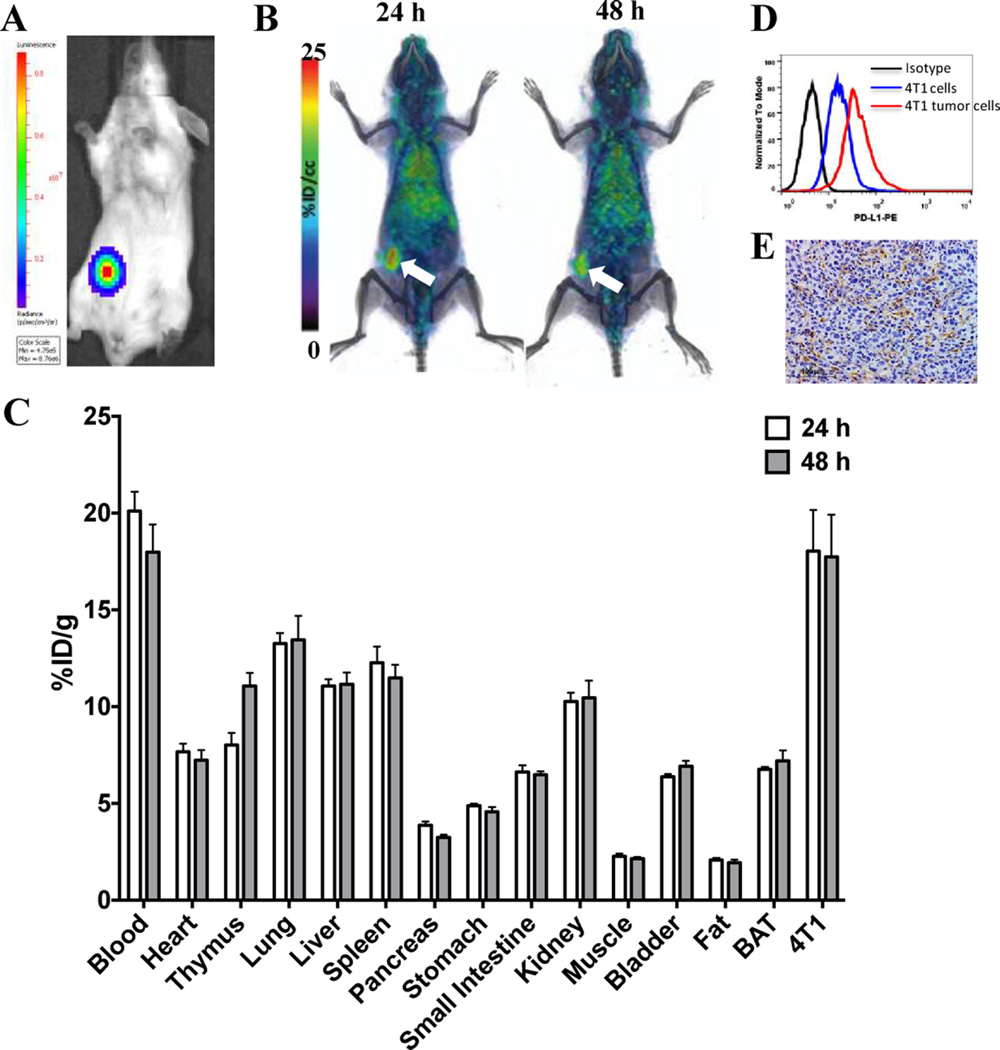
To evaluate the biodistribution of [64Cu]atezolizumab in immunocompetent mice, we performed its evaluation in Balb/C mice harboring syngenic 4T1 mouse mammary carcinoma, as PD-L1 is expressed on immune cells and atezolizumab is a human and mouse cross-reactive antibody. The 4T1-Luc cells were inoculated in the mammary fat pad, and formation of primary tumors was followed by bioluminescence imaging (Figure 4A). PET-CT imaging indicated high and persistent accumulation of [64Cu]-atezolizumab in the tumors (Figure 4B).
Figure 4.
Evaluation of [64Cu]atezolizumab in 4T1 mammary carcinoma. (A) Representative bioluminescence image of a mouse with a 4T1-Luc tumor. (B) PET-CT volume rendered images of [64Cu]atezolizumab in 4T1-Luc tumor bearing mice (n = 3), demonstrating high accumulation of radioactivity in the tumors (white arrow). (C) Ex vivo biodistribution analysis of [64Cu]atezolizumab at 24 and 48 h after injection (n = 4). (D) Flow cytometry showing higher PD-L1 expression in tumor-derived cells compared to that in the cell line (black, isotype; blue, cell line; red-tumor-derived cells). (E) Immunohistochemical analysis for PD-L1 expression demonstrating positive immunoreactivity in 4T1-Luc tumors.
Ex vivo biodistribution showed 18.1 ± 4.2 and 17.0 ± 4.3% ID/g accumulated radioactivity in the 4T1-Luc tumors at 24 and 48 h pi, respectively (Figure 4C). Accumulation of radioactivity was also detected in the lymph nodes, heart, lungs, liver, and spleen. Interestingly, [64Cu]atezolizumab uptake in 4T1-Luc tumors was comparable to that in MDAMB231 xenografts at 48 h pi. Our in vitro flow cytometry analysis showed significantly lower expression of PD-L1 on 4T1-Luc cells than on MDAMB231 cells (Figure 1A and B), which is also consistent with the low [64Cu]atezolizumab binding observed in vitro (Figure 1C). PD-L1 expression in the tumor cells can be induced by increased IFN-γ that is secreted by inflammatory immune cell components recruited to the tumors.12 To characterize the [64Cu]atezolizumab uptake observed in the tumors, we performed flow cytometry of a single cell suspension of 4T1-Luc tumors. There was a significant increase in PD-L1 expression in tumor-derived 4T1-Luc cells compared to that in parental cultured cells (Figure 4D), which was further confirmed by IHC of tumors from the PET-CT imaging study (Figure 4E), indicating that observed radioactivity accumulation is PD-L1 specific. In immune competent mice, infiltrating leukocytes may partially contribute to accumulation of [64Cu]atezolizumab in the TME. Flow cytometry analysis of a single cell suspension from 4T1 tumors showed a small fraction of the total cells (~6%) to be CD45 and PD-L1 positive leukocytes, suggesting that accumulation of [64Cu]atezolizumab in the TME in untreated 4T1 tumor-bearing mice is mainly associated with expression of PD-L1 on tumor cells. Radioactivity uptake observed in tissues with endogenous PD-L1 expression, namely lungs, liver, spleen, and BAT, was significantly lower compared to that in 4T1-Luc tumors. Similar biodistribution results with a mouse PD-L1 cross-reactive antibody further support these observations.18, 32 Collectively, these results demonstrate PD-L1 detection in tumors in an immunocompetent model and demonstrate the use of human and mouse cross-reactive antibodies for the evaluation of targets for immune checkpoint therapy.
CONCLUSIONS
We have synthesized a positron-emitting version of atezolizumab and have demonstrated its capacity to image PD-L1 expression in the TME in a variety of cancers, including within an immunocompetent model. The development of such alternative methods for quantitative measurement of PD-L1 expression in the TME is necessary because of recent clinical developments and approvals of anti-PD1 and anti-PD-L1 therapeutic antibodies as well as unresolved issues in PD-L1 IHC analysis, dynamic changes in PD-L1 expression in the TME, and the impracticality of repetitive tissue sampling in advanced stage disease patients. Noninvasive PD-L1 measures could enhance immune checkpoint therapy management and reduce immune-related toxicities associated with immune checkpoint therapies.
Our studies in five tumor models of variable PD-L1 expression demonstrate the potential and specificity of this antibody for PD-L1 detection with PET-CT. We observed strong PD-L1 expression-dependent [64Cu]atezolizumab accumulation in the tumors, suggesting that radiolabeled atezolizumab PET-CT imaging could be further developed as a companion diagnostic test for PD-L1-targeted therapies.
MATERIALS AND METHODS
Reagents
The antihuman PD-L1 monoclonal antibody atezolizumab (PD-L1 mAb) was custom synthesized as described previously.22, 26 Chemicals were purchased from Sigma-Aldrich or Fisher Scientific with two exceptions: 2,2′,2″-(10-(2,6-dioxotetrahydro-2H-pyran-3-yl)-1,4,7,10-tetraazacyclododecane-1,4,7-triyl)triacetic acid (DOTAGA anhydrate) was purchased from CheMatech Macrocycle Design Technologies (catalog #C109; Dijon, France), and [64Cu]Cl2 was purchased from The University of Wisconsin. All cell culture reagents were purchased from Corning unless otherwise specified.
Preparation of Radiolabeled Antibody
The PD-L1 mAb was reacted with DOTAGA anhydrate in PBS at 1:60 mAb:DOTAGA molar ratio for 3 h at room temperature. Unconjugated DOTAGA was removed using an Amicon ultra-4 centrifugal filter with 10 kDa MWCO (#UFC801024, EMD Millipore, Billerica) and equilibriated with 0.1 M sodium acetate (pH 5). Conjugation of DOTAGA to antibody was confirmed by matrix-assisted laser desorption ionization mass spectrometry. Radiolabeling of mAb-DOTAGA with 64Cu was carried out in 0.1 M sodium acetate (pH 5) for 1 h at 37 °C under metal-free conditions. The resulting [64Cu]atezolizumab was incubated with ethylenediaminetetraacetic acid (EDTA) at a final concentration of 5 mM for 5 min to chelate unbound 64Cu2+ and then subsequently purified on a PBS preequilibrated Zeba spin desalting column (Thermo Scientific, catalog #89882). Radiochemical purity was tested by instant thin-layer chromatography (ITLC) (catalog #61885, Pall Life Science) using citrate-phosphate-dextrose solution as a mobile phase.
SDS-PAGE Analysis and Autoradiography
Antibodies were electrophoretically run in 1 mm NuPAGE Novex Gel, either in reducing conditions (sample with 0.7 M 2-mercaptoethanol heated at 95 °C for 2 min before loading) or in native nonreducing conditions (sample without 2- mercaptoethanol and not heated). After the completion of the run, the gel was stained with colloidal Coomassie G-250 as per manufacturer’s protocol (SimplyBlue, Cat. No. LC6060, Life Technologies). The stained gel was then exposed to X-ray film overnight for autoradiography.
Cell Lines
In this study, we used five cell lines for in vitro and in vivo studies: (i, ii) human triple negative breast cancer (TNBC) cell lines MDAMB231 and SUM149, (iii, iv) Chinese hamster ovary cell line CHO-K1 (henceforth referred to as CHO) and CHO cell line with stable human PD-L1 expression (CHO-hPD-L1), and (v) mouse mammary TNBC 4T1 cell line stably expressing firefly luciferase (4T1-Luc). The MDAMB231 and CHO lines were purchased from the American Type Culture Collection (ATCC) and passaged for no more than 3 months, after which new cultures were initiated from vials of frozen cells. SUM149 and 4T1-Luc cell lines were kindly provided by Dr. Stephen P. Ethier, Medical University of South Carolina, and Dr. Gary Luker of the University of Michigan, respectively. The CHO-hPD-L1 cell line was generated in our laboratory and maintained as described previously.19 The SUM149 cell line was authenticated by short tandem repeat (STR) profiling at the Johns Hopkins genetic resources facility.
The SUM149 cells were maintained in Ham’s F-12 medium with 5% FBS, 1% P/S, 5 µg/mL of insulin, and 0.5 µg/mL of hydrocortisone. All other cell lines were cultured in ATCC-recommended media in an incubator at 37 °C in an atmosphere containing 5% CO2.
Flow Cytometry
Adherent cells were detached using enzyme-free, PBS-based cell dissociation buffer (Gibco). The harvested cells were washed twice with flow cytometry buffer (1× PBS with 2 mM EDTA and 0.5% FBS) and stained with anti-human PD-L1 antibody conjugated with phycoerythrin (PE) (catalog #557924, BD) according to the manufacturer’s protocol.
For harvesting cells from the 4T1-Luc tumor, the tumor was dissected out, cut into small pieces, minced finely with scissors and scalpel, and then incubated in Hanks’ balanced salt solution (HBSS) containing 1 mg/mL of collagenase type I (Gibco) for 30 min at 37 °C. All of the digested tumor mass was passed through a 70 µM cell strainer to obtain a suspension of single cells. This suspension was then washed twice with flow cytometry buffer and then stained with anti-mouse PD-L1 antibody conjugated with phycoerythrin (PE) (catalog #558091, clone MIH5, BD) according to the manufacturer’s protocol.
Samples were aquired on a FACSCalibur flow cytometer (Becton Dickinson). At least 20,000 events were recorded and analyzed using FlowJo software (Tree Star).
In Vitro Binding Assay and Immunoreactive Fraction (IF) Determination
In vitro binding of [64Cu]atezolizumab to CHO-hPD-L1, CHO, MDAMB231, SUM149, and 4T1 cells was determined by incubating 1 µCi of [64Cu]atezolizumab with 1 × 106cells for 1 h at 37 °C. The PD-L1 blocking was performed by adding a 10-fold molar equivalent excess of the nonlabeled mAb. After incubation, cells were washed three times with cold PBS prior to counting on an automated gamma counter (1282 Compugamma CS, Pharmacia/LKBNuclear, Inc., Gaithersburg, MD). To determine the immunoreactive fraction (IF) using the Lindmo method,34 binding of [64Cu]atezolizumab to 4, 2, 1, 0.5, and 0.25 million CHOhPDL1 cells was carried out as described above. The ratios of total/bound activity vs 1/cell number were plotted, and IF was calculated as the inverse of the Y-intercept value when X = 0 (20). All in vitro studies were performed in triplicate and repeated three times.
Animal Models
Animal studies were performed according to the protocols approved by the Johns Hopkins University (JHU) Animal Care and Use Committee (ACUC). For the imaging and biodistribution experiments, six-to-eight week old, female, nonobese diabetic, severe-combined immunodeficient gamma (NSG) mice were obtained from the JHU Immune Compromised Animal Core. Mice were implanted subcutaneously in the upper flanks with CHO-hPD-L1 (10 × 106) and CHO (10 × 106) or orthotopically in the upper mammary fat pads with MDAMB231 (2 × 106) and SUM149 (2 × 106) cells in 100 µL of HBSS containing 50% matrigel (Corning). For the orthotopic mouse mammary carcinoma model in immunocompetent mice, six-to-eight week old female BALB/c mice were injected with 4T1-cells (5 × 105) in the lower mammary fat pad. Growth of tumors was monitored by bioluminescent imaging after injection of luciferin (Gold Biotechnology), and mice were used for the experiments when the tumor sizes reached volumes of 200−300 mm3.
PET-CT Imaging of Mouse Xenografts
Mice were injected intravenously with 450 µCi of [64Cu]atezolizumab (40 µg protein dose) in 200 µL of saline (n = 3−5) and anesthetized with 3% isofluorane prior to being placed in the scanner. During imaging, mice were maintained at 1% isofluorane levels. PET imaging (2 beds, 10 min per bed) was carried out using an ARGUS small-animal PET/CT scanner (Sedecal, Madrid, Spain). At the end of each PET scan, a CT scan (512 projections) was performed for anatomical coregistration.
The PET data were reconstructed using the two-dimensional, ordered-subsets-expectation maximization algorithm (2DOSEM) and corrected for dead time and radioactive decay. The values for percentage of injected dose per cc (%ID/cc) were calculated based on a calibration factor obtained from a known radioactive quantity. Final data visualization and image generation was accomplished using Amira (FEI, Hillsboro, OR).
Ex Vivo Biodistribution
Mice harboring subcutaneous or orthotopic tumors (n = 3−5/group) were injected intravenously with 40 µCi of [64Cu]atezolizumab (~10 µg of protein dose) combined with 30 µg of unmodified atezolizumab. The specific activity (40 µg of protein dose) used was selected based on our previous observations.19 For the blocking study, mice were injected with 1.5 mg of unmodified mAb 30 min before the injection of [64Cu]atezolizumab. In two groups of mice, blood, tumors, and selected tissues were harvested at 24 and 48 h after the [64Cu]atezolizumab injection, weighed, and counted in an automated gamma counter (1282 Compugamma CS, Pharmacia/LKBNuclear, Inc.). The values for percentage of injected dose per gram of tissue (%ID/g) were calculated based on signal decay correction and normalization to external [64Cu] standards measured in triplicate. Biodistribution data are shown as mean ± the standard error of the mean (SEM).
Immunohistochemistry
Tumor sections were evaluated for PD-L1 expression by immunohistochemistry (IHC). Harvested tumors were fixed in 10% neutral buffered formalin and embedded in paraffin, and 4 µm thick sections were obtained on slides. After deparaffinizing with xylene and alcohol gradients, antigen retrieval was carried out using 10 mM citrate buffer, pH 6.0 (#S1699, Dako target retrieval solution). Tumor sections were then treated with 3% H2O2 for 10 min, blocked with 5% goat serum for 1 h, and then incubated with a primary anti-PD-L1 antibody (clone SP142, Spring Bioscience) at 1:100 dilution at 4 °C overnight. Subsequently, using Dako CSAII Biotin-free Tyramide Signal Amplification System kit (#K1497), slides were incubated with secondary antibody, amplification reagent, and anti-fluorescein-HRP. Finally, staining was carried out by adding DAB chromogen. Sections were counterstained with hematoxylin, followed by dehydration with alcohol gradients, xylene washes, and mounting with a coverslip.
Data Analysis
Statistical analysis of in vitro receptor binding assay data and ex vivo biodistribution data were performed with Graphpad Prism 6 software using an unpaired two-tailed t test. When P < 0.05, the difference between the compared groups was considered to be statistically significant.
Supplementary Material
Acknowledgments
We thank Dr. Polina Sysa-Shah for help with PET image acquisition and University of Wisconsin team for Cu-64 production. Funding for this study was provided by Johns Hopkins University Career Catalyst Award (S.N.) and NIH R01CA16631 (S.N.). Flow cytometry, histology, and imaging resources were supported by NIH P30 CA006973 and NIH P50 CA103175.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.bioconjchem.6b00348.
[64Cu]atezolizumab radiolabeling schema, physiochemical characterization, and analysis (PDF)
REFERENCES
- 1.Chen DS, Mellman I. Oncology Meets Immunology: The Cancer-Immunity Cycle. Immunity. 2013;39:1–10. doi: 10.1016/j.immuni.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 2.Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27:450–461. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lipson EJ, Forde PM, Hammers HJ, Emens LA, Taube JM, Topalian SL. Antagonists of PD-1 and PD-L1 in Cancer Treatment. Semin. Oncol. 2015;42:587–600. doi: 10.1053/j.seminoncol.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swaika A, Hammond WA, Joseph RW. Current state of anti-PD-L1 and anti-PD-1 agents in cancer therapy. Mol. Immunol. 2015;67:4–17. doi: 10.1016/j.molimm.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 5.Lipson EJ, Sharfman WH, Drake CG, Wollner I, Taube JM, Anders RA, Xu H, Yao S, Pons A, Chen L, Pardoll DM, Brahmer JR, Topalian SL. Durable cancer regression off-treatment and effective reinduction therapy with an anti-PD-1 antibody. Clin. Cancer Res. 2013;19:462–468. doi: 10.1158/1078-0432.CCR-12-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Velcheti V, Schalper KA, Carvajal DE, Anagnostou VK, Syrigos KN, Sznol M, Herbst RS, Gettinger SN, Chen L, Rimm DL. Programmed death ligand-1 expression in non-small cell lung cancer. Lab. Invest. 2014;94:107–116. doi: 10.1038/labinvest.2013.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cimino-Mathews A, Thompson E, Taube JM, Ye X, Lu Y, Meeker A, Xu H, Sharma R, Lecksell K, Cornish TC, Cuka N, Argani P, Emens LA. PD-L1 (B7-H1) expression and the immune tumor microenvironment in primary and metastatic breast carcinomas. Hum. Pathol. 2016;47:52–63. doi: 10.1016/j.humpath.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lyford-Pike S, Peng SW, Young GD, Taube JM, Westra WH, Akpeng B, Bruno TC, Richmon JD, Wang H, Bishop JA, Chen LP, Drake CG, Topalian SL, Pardoll DM, Pai SI. Evidence for a Role of the PD-1:PD-L1 Pathway in Immune Resistance of HPV-Associated Head and Neck Squamous Cell Carcinoma. Cancer Res. 2013;73:1733–1741. doi: 10.1158/0008-5472.CAN-12-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parsa AT, Waldron JS, Panner A, Crane CA, Parney IF, Barry JJ, Cachola KE, Murray JC, Tihan T, Jensen MC, Mischel PS, Stokoe D, Pieper RO. Loss of tumor suppressor PTEN function increases B7-H1 expression and immunoresistance in glioma. Nat. Med. 2007;13:84–88. doi: 10.1038/nm1517. [DOI] [PubMed] [Google Scholar]
- 10.Green MR, Monti S, Rodig SJ, Juszczynski P, Currie T, O’Donnell E, Chapuy B, Takeyama K, Neuberg D, Golub TR, Kutok JL, Shipp MA. Integrative analysis reveals selective 9p24.1 amplification, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B-cell lymphoma. Blood. 2010;116:3268–3277. doi: 10.1182/blood-2010-05-282780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skoulidis F, Byers LA, Diao L, Papadimitrakopoulou VA, Tong P, Izzo J, Behrens C, Kadara H, Parra ER, Canales JR, Zhang J, Giri U, Gudikote J, Cortez MA, Yang C, Fan Y, Peyton M, Girard L, Coombes KR, Toniatti C, Heffernan TP, Choi M, Frampton GM, Miller V, Weinstein JN, Herbst RS, Wong KK, Zhang J, Sharma P, Mills GB, Hong WK, Minna JD, Allison JP, Futreal A, Wang J, Wistuba II, Heymach JV. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discovery. 2015;5:860–877. doi: 10.1158/2159-8290.CD-14-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taube JM, Anders RA, Young GD, Xu H, Sharma R, McMiller TL, Chen S, Klein AP, Pardoll DM, Topalian SL, Chen L. Colocalization of inflammatory response with B7-h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci. Transl. Med. 2012;4:127ra37. doi: 10.1126/scitranslmed.3003689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong H, Strome SE, Salomao DR, Tamura H, Hirano F, Flies DB, Roche PC, Lu J, Zhu G, Tamada K, Lennon VA, Celis E, Chen L. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat. Med. 2002;8:793–800. doi: 10.1038/nm730. [DOI] [PubMed] [Google Scholar]
- 14.Topalian SL, Taube JM, Anders RA, Pardoll DM. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat. Rev. Cancer. 2016;16:275–287. doi: 10.1038/nrc.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sunshine J, Taube JM. PD-1/PD-L1 inhibitors. Curr. Opin. Pharmacol. 2015;23:32–38. doi: 10.1016/j.coph.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel SP, Kurzrock R. PD-L1 Expression as a Predictive Biomarker in Cancer Immunotherapy. Mol. Cancer Ther. 2015;14:847–856. doi: 10.1158/1535-7163.MCT-14-0983. [DOI] [PubMed] [Google Scholar]
- 17.Heskamp S, Hobo W, Molkenboer-Kuenen JD, Olive D, Oyen WJ, Dolstra H, Boerman OC. Noninvasive Imaging of Tumor PD-L1 Expression Using Radiolabeled Anti-PD-L1 Antibodies. Cancer Res. 2015;75:2928–2936. doi: 10.1158/0008-5472.CAN-14-3477. [DOI] [PubMed] [Google Scholar]
- 18.Josefsson A, Nedrow JR, Park S, Banerjee SR, Rittenbach A, Jammes F, Tsui B, Sgouros G. Imaging, Biodistribution, and Dosimetry of Radionuclide-Labeled PD-L1 Antibody in an Immunocompetent Mouse Model of Breast Cancer. Cancer Res. 2016;76:472–479. doi: 10.1158/0008-5472.CAN-15-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chatterjee S, Lesniak WG, Gabrielson M, Lisok A, Wharram B, Sysa-Shah P, Azad BB, Pomper MG, Nimmagadda S. A humanized antibody for imaging immune checkpoint ligand PD-L1 expression in tumors. Oncotarget. 2016;7:10215–10227. doi: 10.18632/oncotarget.7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maute RL, Gordon SR, Mayer AT, McCracken MN, Natarajan A, Ring NG, Kimura R, Tsai JM, Manglik A, Kruse AC, Gambhir SS, Weissman IL, Ring AM. Engineering high-affinity PD-1 variants for optimized immunotherapy and immuno-PET imaging. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E6506–E6514. doi: 10.1073/pnas.1519623112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng R, Bumbaca D, Pastuskovas CV, Boswell CA, West D, Cowan KJ, Chiu H, McBride J, Johnson C, Xin Y, Koeppen H, Leabman M, Iyer S. Preclinical pharmacokinetics, pharmacodynamics, tissue distribution, and tumor penetration of anti-PD-L1 monoclonal antibody, an immune checkpoint inhibitor. MAbs. 2016;8:593–603. doi: 10.1080/19420862.2015.1136043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Powles T, Eder JP, Fine GD, Braiteh FS, Loriot Y, Cruz C, Bellmunt J, Burris HA, Petrylak DP, Teng SL, Shen X, Boyd Z, Hegde PS, Chen DS, Vogelzang NJ. MPDL3280A (anti-PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature. 2014;515:558–562. doi: 10.1038/nature13904. [DOI] [PubMed] [Google Scholar]
- 23.Hamid O, Sosman JA, Lawrence DP, Sullivan RJ, Ibrahim N, Kluger HM, Boasberg PD, Flaherty K, Hwu P, Ballinger M, Mokatrin A, Kowanetz M, Chen DS, Hodi FS. Clinical activity, safety, and biomarkers of MPDL3280A, an engineered PD-L1 antibody in patients with locally advanced or metastatic melanoma (mM) J. Clin. Oncol. 2013;31 (suppl; abstr 9010) [Google Scholar]
- 24.Spigel DR, Gettinger SN, Horn L, Herbst RS, Gandhi L, Gordon MS, Cruz C, Conkling P, Cassier PA, Antonia SJ, Burris HA, Fine GD, Mokatrin A, Kowanetz M, Shen XD, Chen DS, Soria JC. Clinical activity, safety, and biomarkers of MPDL3280A, an engineered PD-L1 antibody in patients with locally advanced or metastatic non-small cell lung cancer (NSCLC) J. Clin. Oncol. 2013;31 (suppl; abstr 8008) [Google Scholar]
- 25.Cho DC, Sosman JA, Sznol M, Gordon MS, Hollebecque A, Hamid O, McDermott DF, Delord JP, Rhee IP, Mokatrin A, Kowanetz M, Funke RP, Fine GD, Powles T. Clinical activity, safety, and biomarkers of MPDL3280A, an engineered PD-L1 antibody in patients with metastatic renal cell carcinoma (mRCC) J. Clin. Oncol. 2013;31 (suppl; abstr 4505) [Google Scholar]
- 26.Irving B, Chiu H, Maecker H, Mariathasan S, Lehar SM, Wu Y, Cheung J. In: Office USP, editor. USA: Genentech, Inc.; 2012. [Google Scholar]
- 27.Ulaner GA, Hyman D, Ross D, Corben A, Chandarlapaty S, Goldfarb S, McArthur H, Erinjeri J, Solomon S, Kolb H, Lyashchenko S, Lewis J, Carrasquillo J. Detection of HER2-positive metastases in patients with HER2-negative primary breast cancer using the 89Zr-DFO-trastuzumab PET/CT. J. Nucl. Med. 2016 doi: 10.2967/jnumed.115.172031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laforest R, Lapi SE, Oyama R, Bose R, Tabchy A, Marquez-Nostra BV, Burkemper J, Wright BD, Frye J, Frye S, Siegel BA, Dehdashti F. [Zr]Trastuzumab: Evaluation of Radiation Dosimetry, Safety, and Optimal Imaging Parameters in Women with HER2-Positive Breast Cancer. Mol. Imaging Biol. 2016 doi: 10.1007/s11307-016-0951-z. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mortimer JE, Bading JR, Colcher DM, Conti PS, Frankel PH, Carroll MI, Tong S, Poku E, Miles JK, Shively JE, Raubitschek AA. Functional imaging of human epidermal growth factor receptor 2-positive metastatic breast cancer using (64)Cu-DOTA-trastuzumab PET. J. Nucl. Med. 2014;55:23–29. doi: 10.2967/jnumed.113.122630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurihara H, Hamada A, Yoshida M, Shimma S, Hashimoto J, Yonemori K, Tani H, Miyakita Y, Kanayama Y, Wada Y, Kodaira M, Yunokawa M, Yamamoto H, Shimizu C, Takahashi K, Watanabe Y, Fujiwara Y, Tamura K. (64)Cu-DOTA-trastuzumab PET imaging and HER2 specificity of brain metastases in HER2-positive breast cancer patients. EJNMMI Res. 2015;5:8. doi: 10.1186/s13550-015-0082-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carrasquillo JA, Morris PG, Humm JL, Smith-Jones PM, Beylergil V, Akhurst T, O’Donoghue JA, Ruan S, Modi S, Hudis CA, Larson SM. Copper-64 trastuzumab PET imaging: a reproducibility study. Q J. Nucl. Med. Mol. Imaging. 2016 doi: 10.23736/S1824-4785.16.02867-3. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hettich M, Braun F, Bartholoma MD, Schirmbeck R, Niedermann G. High-Resolution PET Imaging with Therapeutic Antibody-based PD-1/PD-L1 Checkpoint Tracers. Theranostics. 2016;6:1629–1640. doi: 10.7150/thno.15253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bosch A, Eroles P, Zaragoza R, Vina JR, Lluch A. Triple-negative breast cancer: molecular features, pathogenesis, treatment and current lines of research. Cancer Treat. Rev. 2010;36:206–215. doi: 10.1016/j.ctrv.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 34.Lindmo T, Boven E, Cuttitta F, Fedorko J, Bunn PA., Jr Determination of the immunoreactive fraction of radiolabeled monoclonal antibodies by linear extrapolation to binding at infinite antigen excess. J. Immunol. Methods. 1984;72:77–89. doi: 10.1016/0022-1759(84)90435-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.