Abstract
Understanding the molecular mechanisms of DNA double-strand break (DSB) repair processes, especially nonhomologous DNA-end joining (NHEJ), is critical for developing next-generation radiotherapies and chemotherapeutics for human and animal cancers. The localization, protein-protein interactions and post-translational modifications of core NHEJ factors, such as human Ku70 and Ku80, might play critical roles in controlling NHEJ activity. XRCC4-like factor (XLF) is a core NHEJ factor and plays a key role in the Ku-dependent NHEJ repair process in human cells. Recently, companion animals, such as canines, have been proposed to be a good model for many aspects of cancer research, including the development of chemotherapeutics. However, the localization and regulation of core NHEJ factors in canine cells have not been elucidated. Here, we show that the localization of canine XLF changes dynamically during the cell cycle. EYFP-canine XLF localizes in the nuclei of interphase cells and accumulates immediately at microirradiated DSB sites. The structure of a putative human XLF nuclear localization signal (NLS) and a putative 14-3-3 binding motif are evolutionarily conserved in canine, chimpanzee and mouse XLF. However, the putative β-TRCP-recognizable degron of human XLF is not conserved in canine and mouse. Additionally, some vital human XLF phosphorylation sites, including the ATM major phosphorylation site (S251), are not conserved in canine XLF. Our findings might be useful for the study of the molecular mechanisms of NHEJ in canine cells and for the development of new radiosensitizers that target XLF.
Keywords: canine, companion animal, DNA double-strand break, Ku, XLF
Resistance to radiation or chemotherapeutics in cancer is a common clinical problem in human and veterinary medicine. Despite evidence of drug efficacy in rodent models of cancer, many new chemotherapeutics are ineffective against cancers in other species or cannot be used, because of high levels of toxicity [9, 19, 23]. Naturally occurring cancers in human and companion animals, such as canine, share many features. These features include histological appearance, tumor genetics, molecular targets, biological behavior and response to conventional therapies [23]. Therefore, canines are proposed to be an excellent cancer model to provide valuable information for veterinary and human medical cancer research [9, 19, 23].
Cancer cells can become resistant to radiotherapy and chemotherapy treatments. Overcoming such resistance is essential for developing next-generation chemotherapeutics and radiotherapies. Accumulating evidence suggests that identifying chemotherapeutic compounds that target proteins involved in DNA double-strand break (DSB) repair pathways is a promising cancer therapy for patients, as a monotherapy or in combination with genotoxic drugs or radiation [10, 11]. Nonhomologous DNA-end joining (NHEJ) is a major pathway of DSB repair in human and rodent cells, and repairs most DSBs induced by radiation and chemotherapeutics in cancer cells [7, 21]. NHEJ starts with the binding of Ku, a heterodimer of Ku70 and Ku80, to the ends of the DSB. The DNA ends are then repaired by Ku and other core NHEJ proteins including Artemis, DNA-dependent protein kinase catalytic subunit (DNA-PKcs), DNA Ligase IV, X-ray repair cross-complementing protein 4 (XRCC4), XRCC4-like factor (XLF; also called Cernunnos or NHEJ1), and PAralog of XRCC4 and XLF (PAXX; also called C9orf142 or XLS) [5, 7, 12, 21, 22, 26]. Chemotherapeutics that target core NHEJ factors, such as DNA-PKcs and DNA Ligase IV, have been reported, but to date, there are no known inhibitors of XLF [10, 11].
XLF is encoded by the human NHEJ1 gene. XLF was discovered as an XRCC4-interacting protein in a yeast two-hybrid screen and as a molecule mutated in patients with growth retardation, microcephaly and immunodeficiency [1, 4]. XLF interacts with the XRCC4-DNA Ligase IV complex to promote the end-joining activity of DNA Ligase IV [1]. The Ku70 and Ku80 heterodimer, bound to XLF, might be essential for the recruitment of human XLF to DSB [17, 21]. Recently, it has been reported that XRCC4-XLF complexes form mobile sleeve-like structures around DNA that can rapidly reconnect the broken ends [3]. However, the function and regulation mechanisms of XLF in NHEJ remain unclear.
Understanding the detailed molecular mechanisms of NHEJ is critical for developing next-generation radiotherapies and chemotherapeutics for human and animal cancers. The localization and regulation of core NHEJ factors, such as human Ku70 and Ku80, might play pivotal roles in NHEJ [12, 13, 21]. However, there are no reports involving the localization, protein-protein interactions and post-translational modifications of canine XLF. In this study, we cloned XLF cDNA from a beagle dog testis library and performed comparative analysis to elucidate the regulatory mechanisms of XLF. In addition, we examined the localization of canine XLF in canine cells and whether canine XLF accumulates at DSB sites immediately after microirradiation.
MATERIALS AND METHODS
Cloning of canine XLF
Oligonucleotide primers used to amplify canine XLF cDNA from male beagle dog cDNA library (Biochain, Newark, CA, U.S.A.) were designed based on the predicted XLF genomic sequence of a female boxer dog, Canis lupus familiaris (XM_848099.2). EcoR1 and BamH1 restriction enzyme sites were incorporated on the 5’ end of the sense (F1) and antisense primers (R1), respectively. PCR amplification with sense and antisense primers was performed for 30 cycles in a Thermal Cycler PC-700 (ASTEC, Fukuoka, Japan) using LA Taq polymerase (Takara Bio Inc., Otsu, Japan). After pre-denaturation (94°C for 5 min), each cycle consisted of denaturation at 94°C for 1 min, annealing at 56°C for 1 min and extension at 72°C for 1 min, followed by a final extension (4 min). PCR products were subcloned into the pCR4-TOPO vector (Invitrogen, Carlsbad, CA, U.S.A.), and the nucleotide sequences were determined by sequencing. PCR primers used in this study were as follows: F1: 5’-CGAATTCGATGAAGGAACTGGAGCAAGGCC-3’, R1: 5’-CGGATCCTTAATTGAAGAGCCCCCTTAGCT-3’, F2: 5’-TTGAAGGGAGAAACAGGACGCGATGCAG-3’, R2: 5’-ATGACAGAGAAAAGCCGCAGGTGGAG-3’, F3: 5’-CCAAAGAGCTGATCTCTTCGGCAC-3’ and R3: 5’-TGCACATGAACTCCTGCTATTTCAC-3’.
Cell lines, cultures and transfections
A Madin-Darby canine kidney cell line (MDCK) (HSRRB, Osaka, Japan) was cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum. XLF cDNA from the pCR4-canine XLF plasmid was subcloned into the EcoRI and BamHI sites of pEYFP-C1 to produce the in-frame fusion gene, pEYFP-canine XLF. pEYFP-canine XLF or pEYFP-C1 was transiently transfected into cells using Lipofectamine 3000 (Invitrogen) or FuGene6 (Roche Diagnostics K.K., Indianapolis, IN, U.S.A.) according to the manufacturer’s protocol. Cells were cultured for two days and then monitored under an FV300 confocal laser scanning microscope (Olympus, Tokyo, Japan) as previously described [14, 15].
Immunofluorescence staining
Immunofluorescence staining was performed as previously described [15, 16] with the following modifications. Briefly, fixed cells were blocked using a blocking solution and then incubated for 30 min at room temperature with mouse anti-γH2AX monoclonal antibody (JBW301) (Upstate Biotechnology Inc., Charlottesville, VA, U.S.A.). After washing with PBS, detection was performed using an Alexa Fluor 568-conjugated secondary antibody (Molecular Probes, Eugene, OR, U.S.A.). DNA was stained with 0.025 μg/ml 4,6-diamino-2-phenylindole (DAPI) fluorescent dye (Boehringer Mannheim, Mannheim, Germany).
X-irradiation
Cells were exposed to X-rays at 10 Gy at a dose rate of 0.81–0.83 Gy/min. X-rays were generated at 200 kVp/20 mA and filtered through 0.5-mm Cu and Al filters using Pantak HF320S (Shimadzu, Kyoto, Japan).
Immunoblotting
The extraction of total cell lysates and Western blot analysis were performed as described previously [13, 18] with the following modifications. The membranes were blocked in Blocking One (Nacalai Tesque, Kyoto, Japan) or ECL Prime Blocking reagent (GE Healthcare Bio-Sci. Corp., Piscataway, NJ, U.S.A.) for 30 min at room temperature. The following antibodies were used: goat anti-XLF polyclonal antibody (SAB2501119) (Sigma, St. Louis, MO, U.S.A.), rabbit anti-GFP polyclonal antibody (FL) (Santa Cruz Biotechnology, Santa Cruz, CA, U.S.A.), mouse anti-γH2AX monoclonal antibody (JBW301) (Upstate Biotechnology Inc.) or mouse anti-β-actin monoclonal antibody (Sigma). The anti-XLF and anti-GFP antibodies were diluted in Signal Enhancer HIKARI (Nacalai Tesque). In accordance with the manufacturer’s instructions, the binding to each protein was detected using a Select Western blotting detection system (GE Healthcare Bio-Sci. Corp.) and visualized using the ChemiDoc XRS system (Bio-Rad, Hercules, CA, U.S.A.).
Local DNA damage induction using laser and cell imaging
Local DNA damage induction using laser and subsequent cell imaging was performed as described previously [15, 17]. Briefly, local DSBs were induced using a 3–10% power scan (for 1 s) from a 405 nm laser. Images of living or fixed cells expressing EYFP-canine XLF or EYFP alone were obtained using an FV300 confocal scanning laser microscopy system (Olympus).
RESULTS
Sequence analysis of canine XLF
Canine XLF cDNA was cloned and sequenced from a beagle dog testis library. We isolated a 900-nucleotide open reading frame encoding a protein of 299 amino acids (Fig. 1). The cDNA sequence obtained from the beagle dog was identical to that predicted sequence from a female boxer dog genomic sequence (XM_848099.2). The obtained canine sequence has been submitted to the DDBJ/ENA/NCBI database [accession number LC176889]. Comparative analysis of XLF sequences showed that canine XLF had 81.6%, 81.9% and 70.6% amino acid identity with human, chimpanzee and mouse, respectively. Protein-protein interaction and post-translational modifications of DNA repair proteins, including phosphorylation, might play a critical role in the regulation of various DNA repair pathways [2, 7, 21]. It is proposed that human XLF has a putative nuclear localization signal (NLS) sequence, KRKK, at amino acid position 290-293, a putative 14-3-3 binding motif at amino acid position 178-183 and a putative β-TRCP-recognizable degron at amino acid position 169-173 [1, 20]. Previously, we showed that the basic amino acids in the NLS of XLF are highly conserved in humans and domestic animal species, including cattle, goat, horse and avian [18]. We confirmed that the putative XLF NLS sequence is conserved in canine, chimpanzee and mouse (Fig. 1). However, the putative β-TRCP-recognizable degron of human XLF is not conserved in canine and mouse XLF (Fig. 1). Moreover, we found that the 14-3-3 binding site (P183) of human XLF is evolutionarily conserved in canine, chimpanzee and mouse. Next, we found that the phosphorylation sites of DNA-PK (S245), AKT-1 (T181) and CKI (S170) in human XLF [20, 27] are evolutionarily conserved in canine, chimpanzee and mouse. The ATM (S251) and CKI (S173) phosphorylation sites of human XLF [20, 27] are not conserved in canine and mouse (Fig. 1).
Fig. 1.
Amino acid sequences of XLF from canine (Canis lupus familiaris, GenBank accession number: LC176889), human (Homo sapiens, GenBank accession number: NM_024782.2), chimpanzee (Pan troglodytes, GenBank accession number: XM_001160321.5) and mouse (Mus musculus, GenBank accession number: NM_029342.4) species. The location of a putative nuclear localization signal (NLS) sequence (NLS: 290KRKK293), a putative 14-3-3 binding motif (14-3-3 consensus: 178RxxT/SxP183), and a putative β-TRCP-recognizable degron (β-TRCP-recognizable degron: 169ESGxT173) in human XLF are shown [1, 20]. Stars mark the location of the AKT phosphorylation site (T181), the ATM phosphorylation site (S251), the CKI phosphorylation sites (S170 and T173), the DNA-PK phosphorylation site (S245) and the putative 14-3-3 binding motif (P183) in the human sequence [20, 27].
Subcellular localization of canine XLF
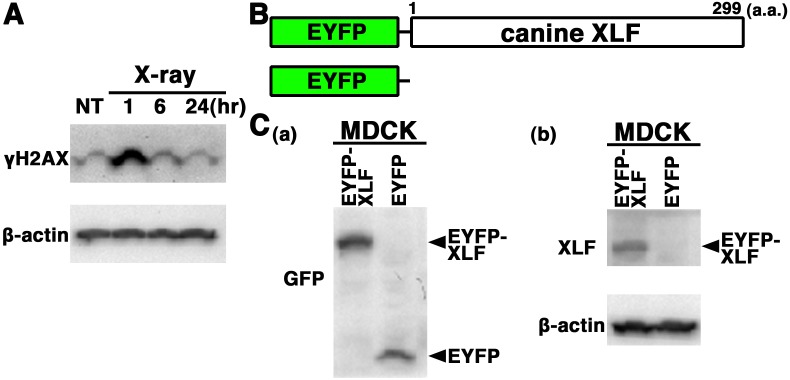
Histone H2AX is rapidly phosphorylated at serine 139 (γH2AX) in response to DSBs, and the elimination of γH2AX reflects DSB repair [25]. To test whether the DSB repair pathways are intact in MDCK cells, we examined X-irradiation-induced H2AX phosphorylation and γH2AX elimination in extracts from MDCK cells by Western blot analysis using the anti-γH2AX antibody. As shown in Fig. 2A, a high level of γH2AX was detected in extracts from MDCK cells at 1 hr postirradiation, and γH2AX elimination from 1 to 6 hr after X-irradiation was detected. This observation suggests that the fast DSB repair pathway, i.e., NHEJ, is intact in MDCK cells. We consider that this canine cell line is suitable for analyzing the localization and dynamics of DSB repair proteins, such as XLF. To examine subcellular localization of XLF in living canine cells, we generated cells transiently expressing EYFP-canine XLF in MDCK cells. The expression vector pEYFP-C1 containing canine XLF (pEYFP-canine XLF) was transfected into MDCK cells (Fig. 2B). Western blotting using anti-XLF and anti-GFP antibodies showed that EYFP-canine XLF was expressed in the transfectant cells (Fig. 2C). Confocal laser microscopy showed that EYFP-canine XLF was localized to the nuclei of living interphase cells in EYFP-canine XLF transfectants (Fig. 3A). During mitosis, fluorescence was detected throughout the cytoplasm of MDCK cells, but was not localized to mitotic chromosomes (Fig. 3B). EYFP, used as a control, was distributed throughout the cell excluding the nucleolus in EYFP transfectant cells (Fig. 3C). These data indicate that the localization of canine XLF changes dynamically during the cell cycle.
Fig. 2.
Expression of EYFP-canine XLF in canine cells. (A) Western blot analysis of γH2AX expression and γH2AX elimination in canine (MDCK) cells after X-irradiation. The MDCK cells were either nonirradiated (NT) or irradiated with 10 Gy X-rays. Each of the total protein samples from the cells was prepared 1, 6 or 24 hr after X-irradiation. γH2AX protein level was determined by Western blot analysis using a specific antibody against γH2AX. β-actin was used as the loading control. (B) Schematics of EYFP-canine XLF chimeric protein (EYFP-canine XLF) and control protein (EYFP). (C) Extracts from MDCK cells transiently expressing the EYFP-canine XLF or EYFP were prepared and subjected to Western blotting using anti-GFP (a), anti-XLF (b) or anti-β-actin (b) antibody.
Fig. 3.
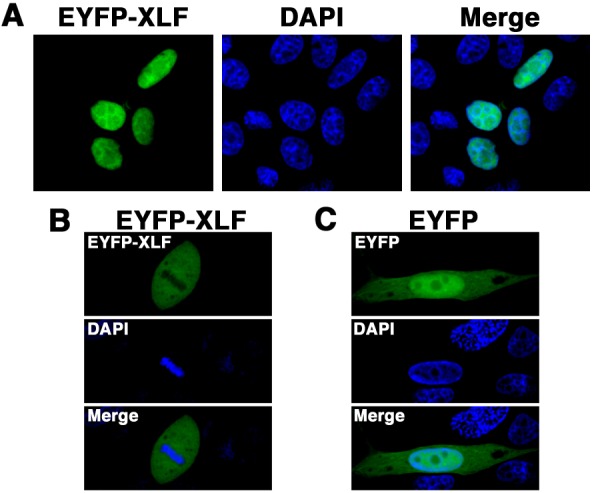
Subcellular localization of EYFP-canine XLF in canine cells. Imaging of living EYFP-canine XLF-transfected cells. MDCK cells transiently expressing EYFP-canine XLF (A, B) or EYFP (C) were fixed and stained with DAPI. The stained cells were analyzed by confocal laser microscopy. EYFP images for the same cells are shown alone (left or top panel) or merged (right or bottom panel) with DAPI images (center or middle panel). The images shown are a representative example for interphase cells (C) or mitotic phase cells (B).
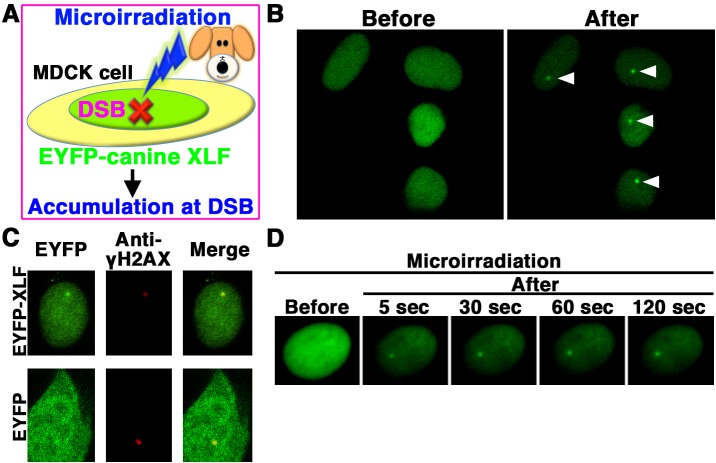
EYFP-canine XLF accumulates immediately at laser-microirradiation-induced DSBs
We next examined whether EYFP-canine XLF accumulates immediately at 405 nm laser-induced DSB sites in canine cells (Fig. 4A). EYFP-canine XLF accumulated at the microirradiated sites in living MDCK cells (Fig. 4B). To examine if EYFP-canine XLF accumulated at 405 nm laser-induced DSB sites, we immunostained cells with an antibody that detects γH2AX, a golden-standard marker of DSBs. EYFP-canine XLF, but not EYFP, accumulated and colocalized with γH2AX at microirradiated sites in MDCK cells (Fig. 4C). To investigate the temporal dynamics of XLF localization, we performed time-lapse imaging of EYFP-canine XLF transfected MDCK cells. We observed EYFP-canine XLF accumulation at the microirradiated sites 5 sec after irradiation (Fig. 4D). These results reveal that after irradiation, EYFP-canine XLF immediately accumulates and forms foci at laser-induced DSBs in living canine cells.
Fig. 4.
EYFP-canine XLF accumulated immediately at DSBs induced by laser microirradiation. (A) The localization and accumulation of EYFP-canine XLF at DSBs induced by 405 nm laser irradiation were examined in canine (MDCK) cells. (B) Imaging of living EYFP-canine XLF-transfected MDCK cells before (left panel) and after (right panel) microirradiation. Arrowheads show the microirradiated sites. (C) Immunostaining of microirradiated EYFP-canine XLF- or EYFP-transfected cells with anti-γH2AX antibody. Cells were fixed and stained with anti-γH2AX antibody 5 min postirradiation. Left panel, EYFP-canine XLF (upper panel) or EYFP (lower panel); Center panel, γH2AX image; Right panel, merged image. (D) Time-dependent EYFP-canine XLF accumulation in living cells (5-120 s) after irradiation.
DISCUSSION
To develop new chemoradiotherapies and targeted drugs for cancers, it is important to elucidate the molecular mechanisms of DNA repair processes, including NHEJ. Human XLF is a core NHEJ factor and appears to play essential roles in NHEJ [1]. XLF-deficient cells derived from human patients and XLF-deficient murine ES cells exhibit ionizing radiation sensitivity [4, 28]. Therefore, XLF might be a potential target molecule for the development of new radiosensitizers. The spatial and/or temporal regulation of XLF localization at DSBs, protein-protein interactions and post-translational modifications might play pivotal roles in regulating NHEJ. However, there have been no reports on the localization, protein-protein interactions and post-translational modifications of canine XLF. Here, we demonstrated that EYFP-canine XLF localizes to the nuclei and accumulates at micro-laser induced DSBs immediately after irradiation. Additionally, we showed that the localization of canine XLF changes dynamically during the cell cycle. Moreover, our XLF sequence alignment shows that the structure of protein-protein interaction motifs and sites of post-translational modification are not perfectly conserved between human and canine. Our findings might be useful for determining the molecular mechanisms of NHEJ and for the development of radiosensitizers that target human and canine XLF.
ATM, ATR and DNA-PK belong to a family of serine/threonine kinases, termed phosphatidylinositol 3-kinase related kinase (PIKKs), and play critical roles in the detection, signaling and repair of DSBs [2]. Accumulating evidence indicates that Akt can be activated upon DNA damage in a PIKK family member-dependent manner [8]. Recently, Liu et al. (2015) reported that Akt impairs NHEJ by phosphorylating XLF at T181 in human cells [20]. First, Akt phosphorylates XLF at T181 triggering its dissociation from the DNA Ligase IV/XRCC4 complex. This promotes the interaction of XLF with 14-3-3β and leads to XLF cytoplasmic retention. Next, the SCFβ-TRCP E3 ligase could target cytoplasmic T181-phosphorylated-XLF for ubiquitination and subsequent degradation in a CKI-dependent manner. Human XLF is a nuclear phosphoprotein that is phosphorylated by ATM and DNA-PK in response to radiation [27]. Expectedly, sequence alignment shows that the DNA-PK (S245), AKT-1 (T181) and CKI (S170) phosphorylation sites of human XLF [20] are evolutionarily conserved in chimpanzee, mouse and canine. Surprisingly, the human XLF CKI (S173) and ATM (S251) phosphorylation sites [20, 27] are not conserved in canine and mouse, although it remains unclear how the lacking of the sequences of the phosphorylation sites (S173 and S251) affects the regulatory mechanism of XLF in canine and mouse cells. Moreover, the putative β-TRCP-recognizable degron of human XLF [20] is not conserved in canine and mouse. Meanwhile, the structure of a putative 14-3-3 binding motif in human XLF is evolutionarily conserved in canine, chimpanzee and mouse. Further studies are necessary to elucidate whether Akt impairs NHEJ by phosphorylating XLF at T181 in canine and mouse cells.
In this study, we found that two structures putatively required for the regulation of human XLF localization, the NLS and 14-3-3 binding motif, are conserved among human, canine, mouse and chimpanzee. Previously, we showed that the putative NLS structure of human XLF is conserved among cattle and six other domestic animals including horse [18]. Additionally, we found that nuclear localization and accumulation of cattle XLF at DSB sites depend on 12 amino acids (288-299) in the XLF C-terminal region (XLF CTR). Furthermore, we reported that basic amino acids in the XLF CTR, containing the NLS, are highly conserved among all seven domestic animals examined, including goat and horse. This suggests that the CTR is essential for the function of XLF in domestic animals. We speculate that the CTR is critical for the function of XLF in canine cells, although further studies need to clarify this. Additionally, the 14-3-3 binding motif, leading to cytoplasmic retention of XLF, might be important for the regulation of XLF localization in canine and human cells.
Originally, inherited mutations in XLF have been reported in humans [4]. Loss of or variations in XLF cause inhibition of NHEJ activity and immune deficiency, and might contribute to cancer predisposition and other diseases, such as microcephaly. Parker et al. (2007) identified a disease-associated deletion within the XLF gene in dogs [24]. Collie eye anomaly (CEA) is an inherited eye disease similar to the human macular coloboma and affects the development of the choroids and sclera segregating in dogs [24]. The CEA-affected dogs share a 7.8 kb deletion in intron 4 of the XLF gene, although it is not clear whether and/or how the deletion causes CEA. We are interested in whether ectopic expression of canine XLF cDNA, which is cloned in this study, can rescue the defect in CEA-affected dogs. Furthermore, inherited mutations of another core NHEJ factor, DNA-PKcs, cause severe combined immune deficiency in humans and have also been identified in mice, horses and dogs [6]. Further comparative studies might provide useful information to identify novel diseases caused by mutations in XLF in humans and companion animals, including dogs.
The present study showed that the localization of canine XLF changes dynamically during the cell cycle. Moreover, our findings showed that canine XLF is recruited to DSB sites at an early stage following irradiation. This is the first report examining the localization and accumulation of XLF at DSB sites in canine cells. Further comparative studies are necessary to elucidate the mechanisms regulating XLF at DSBs. Such studies could improve our understanding of the molecular mechanisms of NHEJ and contribute to the development of new XLF targeting radiosensitizers for humans and companion animals, including dogs.
Acknowledgments
This work was supported in part by JSPS KAKENHI Grant Number JP26450438.
REFERENCES
- 1.Ahnesorg P., Smith P., Jackson S. P.2006. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell 124: 301–313. doi: 10.1016/j.cell.2005.12.031 [DOI] [PubMed] [Google Scholar]
- 2.Bakkenist C. J., Kastan M. B.2004. Initiating cellular stress responses. Cell 118: 9–17. doi: 10.1016/j.cell.2004.06.023 [DOI] [PubMed] [Google Scholar]
- 3.Brouwer I., Sitters G., Candelli A., Heerema S. J., Heller I., de Melo A. J., Zhang H., Normanno D., Modesti M., Peterman E. J., Wuite G. J.2016. Sliding sleeves of XRCC4-XLF bridge DNA and connect fragments of broken DNA. Nature 535: 566–569. doi: 10.1038/nature18643 [DOI] [PubMed] [Google Scholar]
- 4.Buck D., Malivert L., de Chasseval R., Barraud A., Fondanèche M. C., Sanal O., Plebani A., Stéphan J. L., Hufnagel M., le Deist F., Fischer A., Durandy A., de Villartay J. P., Revy P.2006. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell 124: 287–299. doi: 10.1016/j.cell.2005.12.030 [DOI] [PubMed] [Google Scholar]
- 5.Craxton A., Somers J., Munnur D., Jukes-Jones R., Cain K., Malewicz M.2015. XLS (c9orf142) is a new component of mammalian DNA double-stranded break repair. Cell Death Differ. 22: 890–897. doi: 10.1038/cdd.2015.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding Q., Bramble L., Yuzbasiyan-Gurkan V., Bell T., Meek K.2002. DNA-PKcs mutations in dogs and horses: allele frequency and association with neoplasia. Gene 283: 263–269. doi: 10.1016/S0378-1119(01)00880-0 [DOI] [PubMed] [Google Scholar]
- 7.Downs J. A., Jackson S. P.2004. A means to a DNA end: the many roles of Ku. Nat. Rev. Mol. Cell Biol. 5: 367–378. doi: 10.1038/nrm1367 [DOI] [PubMed] [Google Scholar]
- 8.Gan W., Liu P., Wei W.2015. Akt promotes tumorigenesis in part through modulating genomic instability via phosphorylating XLF. Nucleus 6: 261–265. doi: 10.1080/19491034.2015.1074365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacob J. A.2016. Researchers turn to canine clinical trials to advance cancer therapies. JAMA 315: 1550–1552. doi: 10.1001/jama.2016.0082 [DOI] [PubMed] [Google Scholar]
- 10.Jekimovs C., Bolderson E., Suraweera A., Adams M., O’Byrne K. J., Richard D. J.2014. Chemotherapeutic compounds targeting the DNA double-strand break repair pathways: the good, the bad, and the promising. Front. Oncol. 4: 86. doi: 10.3389/fonc.2014.00086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelley M. R., Fishel M. L.2008. DNA repair proteins as molecular targets for cancer therapeutics. Anticancer. Agents Med. Chem. 8: 417–425. doi: 10.2174/187152008784220294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koike M.2002. Dimerization, translocation and localization of Ku70 and Ku80 proteins. J. Radiat. Res. (Tokyo) 43: 223–236. doi: 10.1269/jrr.43.223 [DOI] [PubMed] [Google Scholar]
- 13.Koike M., Awaji T., Kataoka M., Tsujimoto G., Kartasova T., Koike A., Shiomi T.1999. Differential subcellular localization of DNA-dependent protein kinase components Ku and DNA-PKcs during mitosis. J. Cell Sci. 112: 4031–4039. [DOI] [PubMed] [Google Scholar]
- 14.Koike M., Ikuta T., Miyasaka T., Shiomi T.1999. Ku80 can translocate to the nucleus independent of the translocation of Ku70 using its own nuclear localization signal. Oncogene 18: 7495–7505. doi: 10.1038/sj.onc.1203247 [DOI] [PubMed] [Google Scholar]
- 15.Koike M., Koike A.2008. Accumulation of Ku80 proteins at DNA double-strand breaks in living cells. Exp. Cell Res. 314: 1061–1070. doi: 10.1016/j.yexcr.2007.11.014 [DOI] [PubMed] [Google Scholar]
- 16.Koike M., Shiomi T., Koike A.2001. Dimerization and nuclear localization of Ku proteins. J. Biol. Chem. 276: 11167–11173. doi: 10.1074/jbc.M010902200 [DOI] [PubMed] [Google Scholar]
- 17.Koike M., Yutoku Y., Koike A.2011. Accumulation of Ku70 at DNA double-strand breaks in living epithelial cells. Exp. Cell Res. 317: 2429–2437. doi: 10.1016/j.yexcr.2011.07.018 [DOI] [PubMed] [Google Scholar]
- 18.Koike M., Yutoku Y., Koike A.2015. Dynamic changes in subcellular localization of cattle XLF during cell cycle, and focus formation of cattle XLF at DNA damage sites immediately after irradiation. J. Vet. Med. Sci. 77: 1109–1114. doi: 10.1292/jvms.14-0516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawrence J., Cameron D., Argyle D.2015. Species differences in tumour responses to cancer chemotherapy. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370: 20140233. doi: 10.1098/rstb.2014.0233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu P., Gan W., Guo C., Xie A., Gao D., Guo J., Zhang J., Willis N., Su A., Asara J. M., Scully R., Wei W.2015. Akt-mediated phosphorylation of XLF impairs non-homologous end-joining DNA repair. Mol. Cell 57: 648–661. doi: 10.1016/j.molcel.2015.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahaney B. L., Meek K., Lees-Miller S. P.2009. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem. J. 417: 639–650. doi: 10.1042/BJ20080413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ochi T., Blackford A. N., Coates J., Jhujh S., Mehmood S., Tamura N., Travers J., Wu Q., Draviam V. M., Robinson C. V., Blundell T. L., Jackson S. P.2015. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair. Science 347: 185–188. doi: 10.1126/science.1261971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paoloni M., Khanna C.2008. Translation of new cancer treatments from pet dogs to humans. Nat. Rev. Cancer 8: 147–156. doi: 10.1038/nrc2273 [DOI] [PubMed] [Google Scholar]
- 24.Parker H. G., Kukekova A. V., Akey D. T., Goldstein O., Kirkness E. F., Baysac K. C., Mosher D. S., Aguirre G. D., Acland G. M., Ostrander E. A.2007. Breed relationships facilitate fine-mapping studies: a 7.8-kb deletion cosegregates with Collie eye anomaly across multiple dog breeds. Genome Res. 17: 1562–1571. doi: 10.1101/gr.6772807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rogakou E. P., Pilch D. R., Orr A. H., Ivanova V. S., Bonner W. M.1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273: 5858–5868. doi: 10.1074/jbc.273.10.5858 [DOI] [PubMed] [Google Scholar]
- 26.Xing M., Yang M., Huo W., Feng F., Wei L., Jiang W., Ning S., Yan Z., Li W., Wang Q., Hou M., Dong C., Guo R., Gao G., Ji J., Zha S., Lan L., Liang H., Xu D.2015. Interactome analysis identifies a new paralogue of XRCC4 in non-homologous end joining DNA repair pathway. Nat. Commun. 6: 6233. doi: 10.1038/ncomms7233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu Y., Mahaney B. L., Yano K., Ye R., Fang S., Douglas P., Chen D. J., Lees-Miller S. P.2008. DNA-PK and ATM phosphorylation sites in XLF/Cernunnos are not required for repair of DNA double strand breaks. DNA Repair (Amst.) 7: 1680–1692. doi: 10.1016/j.dnarep.2008.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zha S., Alt F. W., Cheng H. L., Brush J. W., Li G.2007. Defective DNA repair and increased genomic instability in Cernunnos-XLF-deficient murine ES cells. Proc. Natl. Acad. Sci. U.S.A. 104: 4518–4523. doi: 10.1073/pnas.0611734104 [DOI] [PMC free article] [PubMed] [Google Scholar]