Abstract
The sequence at the hemagglutinin (HA) cleavage site (CS) plays a key role in determining the pathogenicity of avian influenza viruses. Three types of HA CS sequences, QREKR/GL, QRKKR/GL and QRRKR/GL, were previously reported in Taiwanese H5N2 viruses that were isolated from chickens from 2003 to 2013. However, no HA CS sequence was reported for viruses isolated after 2013. This article presents the HA CS sequences and pathogenicity of H5N2 viruses that were isolated from chickens in Taiwan during 2013–2015. Two novel HA CS sequences, QKEKR/GL and KREKREKR/GL, were found in the viruses isolated in 2013 and 2014, and pathogenicity tests showed that the viruses with these novel HA CS sequences are low and high pathogenic viruses, respectively. In contrast, the HA CS sequence QREKR/GL was found in all viruses that were isolated in 2015, and all of these viruses were low pathogenic viruses. After 10 passages in embryonated chicken eggs, a virus strain that was isolated in 2003 evolved into a viral quasispecies that contained at least four distinct types of HA CS sequences. These results highlight the potential of Taiwanese H5N2 viruses to change their pathogenicity and HA CS sequences via mutations. Furthermore, viruses with the HA CS sequence QREKR/GL were more prevalent than others in 2015. These findings are useful for understanding the mechanism of sequence changes at the HA CS and for refining H5N2 virus control measures in Taiwan.
Keywords: avian influenza virus, H5N2, hemagglutinin cleavage site, Taiwan
Avian influenza (AI) is a highly contagious disease that is caused by type A influenza viruses, which have many subtypes that are determined on the basis of antigenic specificity of two surface glycoproteins: hemagglutinin (HA) and neuraminidase (NA). Sixteen HA (H1–H16) and 9 NA subtypes (N1–N9) have been identified [6]. Among the 16 HA subtypes, only the H5 and H7 subtypes are highly pathogenic to poultry [1]. The global dissemination of highly pathogenic H5N1 viruses has led to the deaths of hundreds of millions of birds and 450 human fatalities since 2003 [16].
HA is the major envelope glycoprotein of influenza A viruses. HA is produced as a precursor protein (HA0) that is cleaved by host proteases to generate two proteins, HA1 and HA2. This cleavage is essential for the function of HA and the infectivity of the virus [12]. Previous studies have demonstrated that the amino acid sequence at the HA cleavage site (CS) plays a critical role in viral virulence. Low-pathogenicity avian influenza (LPAI) viruses have a single arginine at the HA CS (the −1 position) and another basic amino acid at the −3 or −4 positions, whereas high-pathogenicity avian influenza (HPAI) viruses have multiple basic amino acids at the HA CS [10, 13]. LPAI viruses may evolve into HPAI viruses by acquiring multiple basic amino acids at the HA CS through mutations [2, 7].
The first outbreak of H5N2 virus in chickens occurred in Taiwan in 2003. The virus (strain 1209/03), which was isolated during this outbreak, has three basic amino acids at the HA CS (QREKR/GL) and an intravenous pathogenicity index (IVPI) of 0.00 [3, 4]. This H5N2 virus comprises HA and NA genes that were derived from H5N2 viruses circulating in Mexico in 1994 and six other genes from H6N1 viruses in Taiwan [3, 4]. In 2008 and 2012–2013, H5N2 viruses containing four basic amino acids at the HA CS (QRKKR/GL or QRRKR/GL) were isolated from chickens in Taiwan [4, 9]. Pathogenicity studies showed that the H5N2 viruses with four basic amino acids are more pathogenic than those with three [4, 9]. Soda et al. [14] reported that a Taiwanese LPAI H5N2 influenza virus evolved into HPAI viruses after consecutive passages in chickens. Similarly, Cheng et al. [4] reported that a Taiwanese LPAI H5N2 virus exhibited increased virulence after consecutive passages in embryonic chicken eggs, which was accompanied by sequence changes at the HA CS. However, the mechanism by which Taiwanese LPAI H5N2 viruses evolve into HPAI viruses is not well studied, and to date, there are no published reports regarding the HA CS sequence and viral pathogenicity of H5N2 viruses that were isolated after 2013 from chickens in Taiwan.
The goal of this study was to determine the pathogenicity and sequence at the HA CS of H5N2 viruses that were isolated from chickens in Taiwan during 2013–2015. A virus strain isolated in 2003 and its progeny viruses were also characterized in this study to elucidate the mechanism of sequence changes at the HA CS. The pathogenicity of the viruses was determined using IVPI or mortality rate tests, and the correlation between the HA CS sequences and viral pathogenicity was investigated. Finally, we put forth a proposal to refine control measures of H5N2 viruses in Taiwan.
MATERIALS AND METHODS
Viruses and isolation of viral RNA
The H5N2 viruses examined in this study were divided into two groups (Table 1). Group 1 contained eight field strains that were isolated from diseased or dead chickens submitted to the Animal Health Research Institute (AHRI) for diagnosis during 2013–2015. The two field strains, A2821/13 and A3190/14, were used in this study, because they were the only two H5N2 virus strains isolated by the AHRI during 2013–2014. The remaining six strains were used, because they were isolated from different areas in Taiwan in early 2015. All field strains were isolated using a previously described method [4].
Table 1. Summary of the Taiwanese H5N2 viruses used in this study.
| Virus strain | Abbreviation | Time of isolation | Place of isolation | Source of isolation |
|---|---|---|---|---|
| Group 1 | ||||
| A/chicken/PH/A2821/2013 (H5N2) | A2821/13 | 17/5/2013 | Penghu County | Lung |
| A/chicken/YL/A3190/2014 (H5N2) | A3190/14 | 17/4/2014 | Taipei City | Kidney |
| A/chicken/PT/a158/2015 (H5N2) | a158/15 | 15/1/2015 | Pinton County | Heart |
| A/chicken/GY/a451/2015 (H5N2) | a451/15 | 22/1/2015 | Chiayi County | Lung |
| /chicken/TC/a504/2015 (H5N2) | a504/15 | 23/1/2015 | Taichung City | Lung |
| A/chicken/CH/a556/2015 (H5N2) | a556/15 | 26/1/2015 | Changhua County | Lung |
| A/chicken/TY/a665/2015 (H5N2) | a665/15 | 29/1/2015 | Taoyuan City | Lung |
| A/chicken/SC/a688/2015 (H5N2) | a688/15 | 29/1/2015 | Hsinchu County | Lung |
| Group 2 | ||||
| A/chicken/Taiwan/1209/2003 (H5N2) | 1209/03 | 23/12/03 | Chunghua County | Lung |
| A/chicken/Taiwan/1209/2003-E5 (H5N2) | 1209/03-E5 | 1209/03 (five passages in eggs) | ||
| A/chicken/Taiwan/1209/2003-E10 (H5N2) | 1209/03-E10 | 1209/03 (10 passages in eggs) | ||
Group 2 contained a field strain that was isolated in 2003 (strain 1209/03) and its progeny viruses (1209/03-E5 and 1209/03-E10) that had undergone five and 10 passages in specific-pathogen-free (SPF) embryonated eggs [3]. Group 2 viruses were used to investigate the mechanism underlying the sequence changes at the HA CS.
Viral RNA was isolated using the MagNA Pure Compact Instrument (Roche Life Science, Branford, CT, U.S.A.) according to the manufacturer’s protocol. The virus isolation experiments were approved by the Institutional Animal Care and Use Committee (IACUC) at the AHRI (IACUC number A99010).
Reverse transcription-polymerase chain reaction (RT-PCR) amplification
Two primers, designated HA (+) and HA (−), were used to PCR-amplify a 194-bp DNA fragment containing the HA CS sequence of the virus. The sequences of the HA (+) and HA (−) primers were 5′–GAG TGC CCC AAG TAT GTC AAA–3′ and 5′–ATA TCC ACT ACC TTG CTC AT–3′, respectively. The RT-PCR mixture (25 µl) contained 16 µl of water, 2.5 µl of 10× buffer, 2.5 µl of a dNTP mixture (2.5 mM each of the four dNTPs), 2 µl of primers (10 pmol each), 1 µl of template RNA, 0.5 µl of SuperScript III reverse transcriptase (Thermo Fisher Scientific, Waltham, MA, U.S.A.) and 0.5 µl of SuperTaq XL DNA polymerase (Protech Technology, Taipei, Taiwan). The RT-PCR conditions were as follows: one cycle at 50°C for 15 min, followed by 35 cycles at 94°C for 15 sec (denaturation), 55°C for 30 sec (annealing) and 72°C for 30 sec (extension), followed by one cycle at 72°C for 5 min (final extension). The PCR products were separated by electrophoresis in 2% agarose gels, stained with ethidium bromide (0.5 µg/ml) and visualized with ultraviolet light.
Direct sequencing of the RT-PCR products
The 194-bp PCR products were subjected to sequencing analysis using the BigDye Terminator Cycle Sequencing Kit (Thermo Fisher Scientific) in an automatic sequencer (ABI-3730XL DNA Analyzer, Thermo Fisher Scientific). Primer HA (+) was used to conduct the sequencing analysis. The sequences were compiled using the SeqMan program in the LASERGENE package (DNASTAR Inc., Madison, WI, U.S.A.) and then used to search for homologous in GenBank using the Basic Local Alignment Search Tool (BLAST) program at the National Center for Biotechnology Information website (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Sequencing of the cloned RT-PCR products
The 194-bp RT-PCR products were cloned into the pCR2.1-TOPO vector (Thermo Fisher Scientific) according to the manufacturer’s instructions. Recombinant plasmids harboring the 194-bp DNA fragment were purified using the QIAprep Spin Plasmid Purification Kit (Qiagen, Hilden, Germany) and individually sequenced with the pUC/M13 forward primer in an automatic sequencer (ABI-3730XL DNA analyzer). The sequences were compiled using the SeqMan program in the LASERGENE package.
Pathogenicity study
The pathogenicity of the H5N2 viruses was assessed by the IVPI or mortality rate tests in chickens using methods described previously by the Office Internationale des Epizooties [11]. In brief, the IVPI was determined by intravenous inoculation of 0.1 ml of allantoic fluid containing 106 mean egg infective doses of the H5N2 virus into 8-week-old SPF chickens. For each virus strain, 10 chickens were inoculated. The chickens were observed for clinical signs for 10 days, and the IVPI was calculated [11]. The mortality rate was determined by inoculating 0.2 ml of allantoic fluid containing the H5N2 virus (HA titer >1:16) into 8-week-old SPF chickens. For each virus, eight chickens were inoculated. The chickens were observed daily for 10 days, and the mortality rate was determined. Viruses that had an IVPI >1.2 or caused a mortality rate >75% were consider to be HPAI viruses. Note that the mortality test has been used by AHRI as the primary method to assess virus pathogenicity since 2015. This is because the IVPI test needs the differentiation between the “sick” and “severely sick” birds, which is a subjective assessment and sometimes problematic for AHRI. The pathogenicity tests were approved by the IACUC at the AHRI (IACUC number A04014).
RESULTS
RT-PCR amplification of the 194-bp DNA fragment containing the HA CS sequence
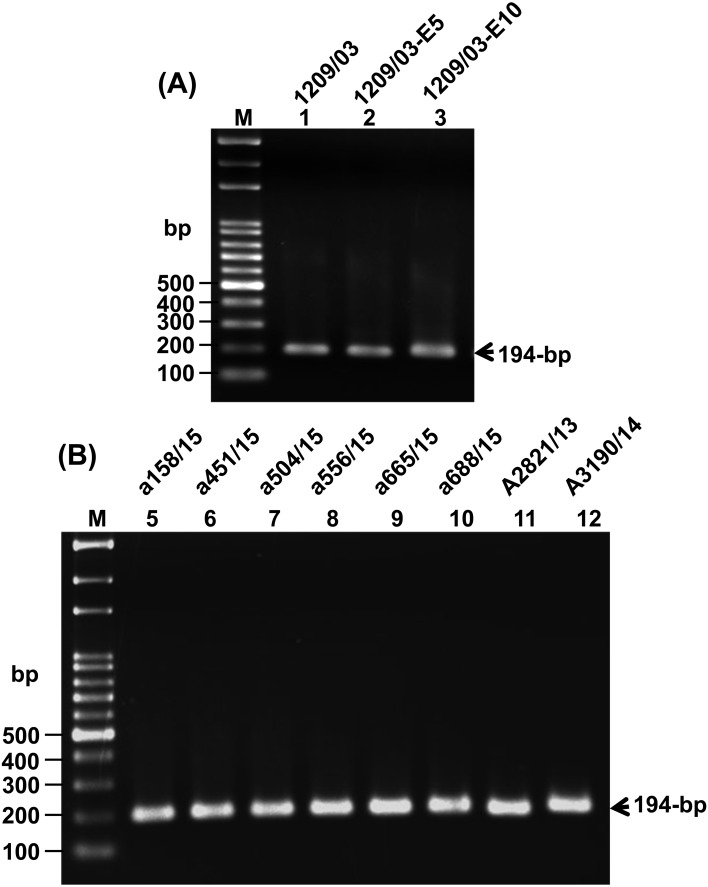
The H5N2 viruses used in this study are summarized in Table 1. All of the viruses were subjected to RNA isolation and RT-PCR amplification, and the results showed that RT-PCR products of approximately 194-bp, containing the HA CS sequences, were amplified successfully from all of the viruses (Fig. 1).
Fig. 1.
RT-PCR amplification of the 194-bp DNA fragment from the H5N2 viruses that were investigated in this study. Lane M contains DNA size markers, and the names of the viruses are shown at the top of each lane.
Sequence diversity and pathogenicity of viruses isolated during 2013–2015
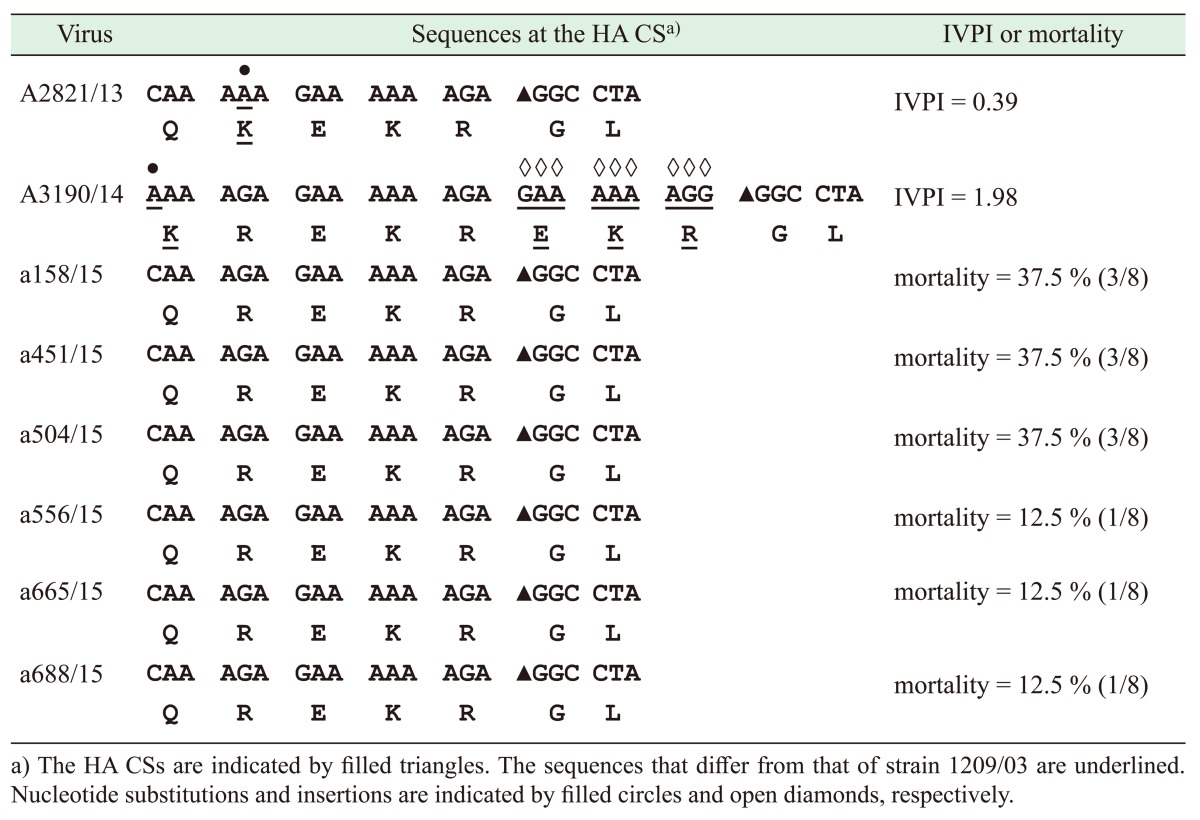
The HA CS sequences of the viruses were determined by directly sequencing the 194-bp RT-PCR products. A nucleotide BLAST search showed that the 194-bp sequences of these viruses were highly similar (96–98% identities) to H5N2 viruses isolated previously in Taiwan during 2003–2013 [4, 9]. However, two novel HA CS sequences, QKEKR/GL and KREKREKR/GL, were found in two virus strains (A2821/13 and A3190/14) investigated in this study (Table 2). These two strains were isolated in 2013 and 2014, respectively, and their novelty was due to nucleotide substitutions (G to A or C to A) and insertions (GAAAAAAGG) that occurred at positions close to the HA CS. Pathogenicity tests showed that strains A2821/13 and A3190/14 had IVPIs of 0.39 and 1.98; therefore, they were classified as LPAI and HPAI viruses, respectively (Table 2).
Table 2. HA CS sequences and pathogenicity of Taiwanese H5N2 viruses that were isolated during 2013–2015.
All of the viruses that were isolated in 2015 had the same HA CS sequence QREKR/GL (containing three basic amino acids), which was found previously in the virus isolated in 2003 [4] (Table 2). Pathogenicity tests showed that all of the viruses that were isolated in 2015 caused mortality rates of 12.5% or 37.5% in chickens, and thus, they were classified as LPAI viruses (Table 2).
Sequence diversity in strain 1209/03 and its progeny viruses
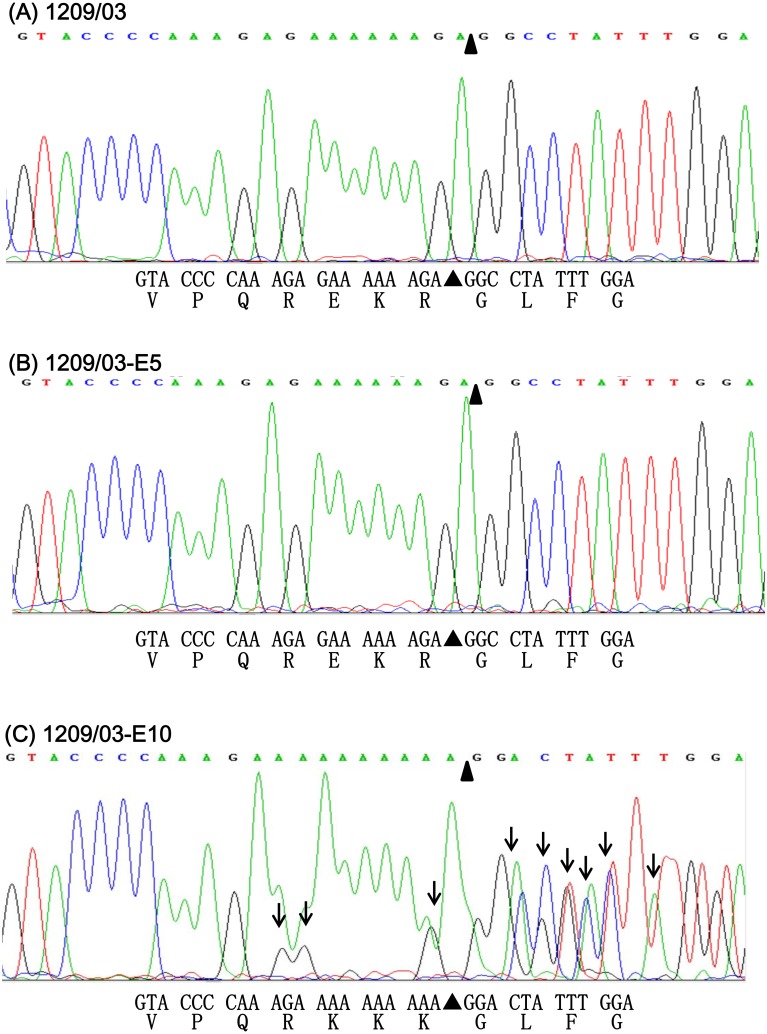
The HA CS sequences of strain 1209/03 and its progeny viruses (1209/03-E5 and 1209/03-E10) were determined by directly sequencing the 194-bp RT-PCR products. However, a problem occurred during the sequencing. In brief, a sequence analysis using an automatic DNA sequencer normally produces two files: one is a chromatogram that shows four-color peaks that represent the four DNA bases (A, G, C and T), and the other is a text file that shows the computer program’s best interpretation of the chromatogram data. Sequencing of strains 1209/03 and 1209/03-E5 produced chromatograms with distinct peaks, and the text files showed that the two strains had the same HA CS QREKR/GL (Fig. 2A and 2B). In contrast, the chromatogram of 1209/03-E10 presented many overlapping or ambiguous peaks (Fig. 2C). This result suggested that 1209/03-E10 might be a mixture of viruses or a viral quasispecies with different HA CS sequences. Although a text file was generated for 1209/03-E10, which enabled the prediction of the HA CS sequence (QRKKK/GL) (Fig. 2C), this HA CS might be erroneous because of the ambiguous peaks shown on the chromatogram.
Fig. 2.
Section of the chromatograms obtained by direct sequencing of the 194-bp RT-PCR products that were amplified from strains 1209/03 (A), 1209/03-E5 (B) and 1209/03-E10 (C). This section corresponds to nucleotide positions 1,006 to 1,038 of the HA gene. Positions containing mixed peaks are indicated by vertical arrows. Nucleotide and predicted amino acid sequences are shown under the chromatogram.
Detection of viral quasispecies with different HA CS sequences
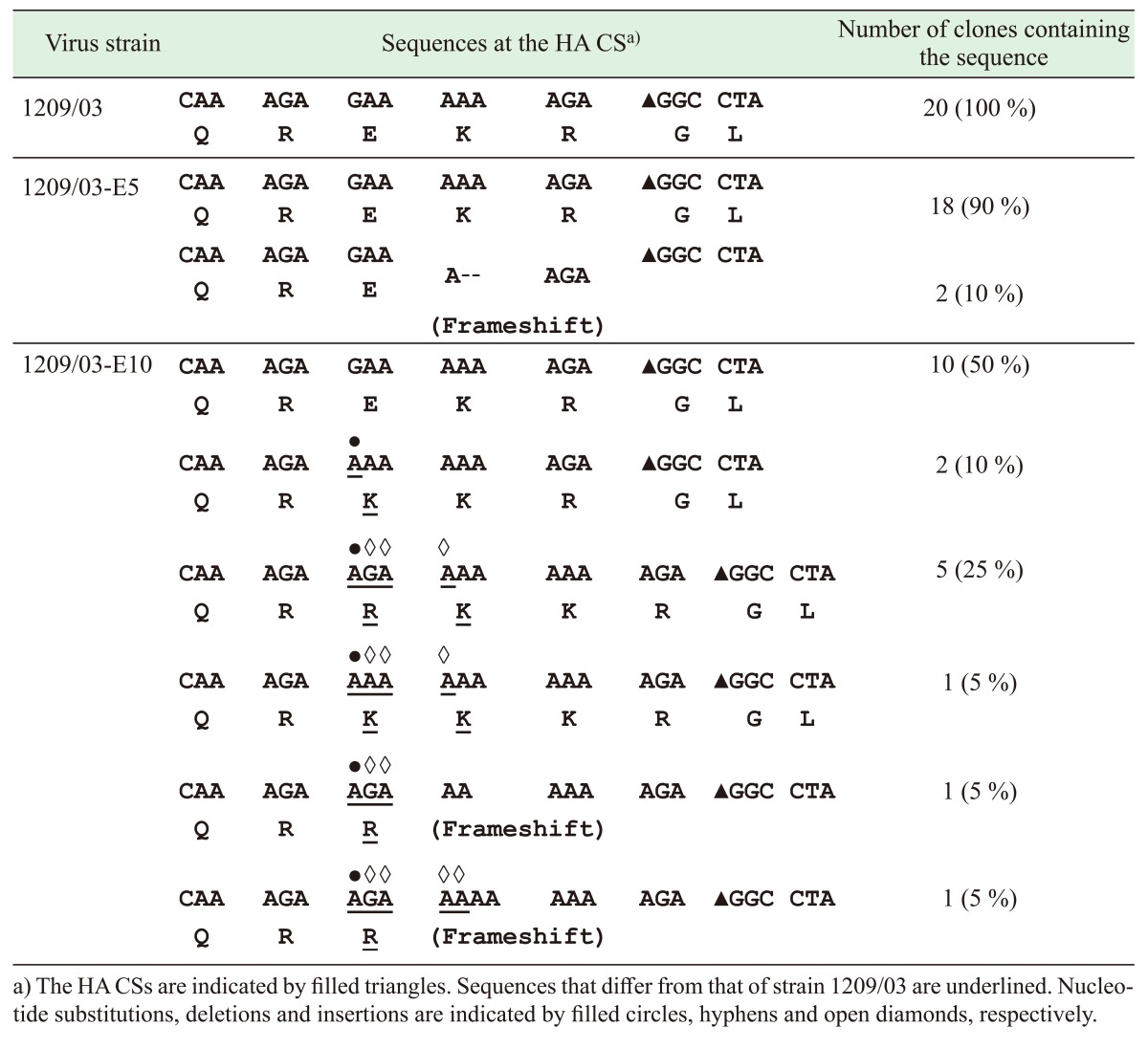
The possibility for the presence of a viral quasispecies in strain 1209/03 or its progeny viruses was explored by sequencing the cloned 194-bp RT-PCR products that were amplified from these viruses. Twenty clones were sequenced for each of the viruses, and the results are summarized in Table 3. All 20 clones of strain 1209/03 had the same HA CS sequence QREKR/GL. Similarly, 18 clones from strain 1209/03-E5 showed the same HA CS sequence QREKR/GL, whereas the two remaining clones contained two-nucleotide deletions that resulted in frameshift mutations. Interestingly, the 20 clones from strain 1209/03-E10 exhibited six distinct types of HA CS sequences. The most abundant type (10 clones) had a HA CS sequence that was identical to that of 1209/03 (QREKR/GL), while the other three types had HA CS sequences QRKKR/GL (two clones), QRRKKR/GL (five clones) and QRKKKR/GL (one clone), respectively. The remaining two types had nucleotide insertions (GA or GAAA) that caused frameshift mutations of the HA gene. Taken together, sequencing of the cloned RT-PCR product revealed that strain 1209/03-E10 is a viral quasispecies with at least four different HA CS sequences.
Table 3. HA CS sequences of the cloned RT-PCR products that were amplified from strain 1209/03 and its progeny viruses.
Sequencing of the cloned RT-PCR product in this study consistently generated a chromatogram that contained no overlapping peaks; this finding conformed to the notion that each of the cloned RT-PCR products was amplified from a single viral RNA molecule.
DISCUSSION
The current policy for the control of H5N2 viruses in Taiwan is to cull all poultry flocks infected with H5N2 viruses that contain four or more basic amino acids at their HA CS. However, for flocks infected with viruses that contain three basic amino acids, the pathogenicity of the infecting virus should be determined. If the virus has an IVPI greater than 1.2 or causes a mortality rate greater than 75% in chickens, then the entire flock should be culled; otherwise, the infected flock should only be monitored until they are free of the virus. This policy has been used in Taiwan for several years. However, the data presented here demonstrate that Taiwanese H5N2 viruses can change their HA CS sequences and viral pathogenicity via mutations. A total of five distinct types of HA CS sequences have now been identified in field viruses, and a single strain of the H5N2 virus might evolve into a quasispecies containing at least four different HA CS sequences after passages in eggs. These findings highlight the need to refine the policy for controlling H5N2 viruses in Taiwan. One option is to cull all poultry flocks that are infected with 1209/03-like H5N2 viruses, disregarding the number of basic amino acids that are present at the HA CS.
All six strains of the H5N2 virus that were isolated in 2015 had the same HA CS sequence (QREKR/GL, containing three basic amino acids), and no viruses with four or more basic amino acids were found in 2015. These six strains were considered to be representative strains of H5N2 viruses that were circulating in early 2015. Among these strains, the lack of an H5N2 virus with four or more basic amino acids at the HA CS suggests that H5N2 viruses with three basic amino acids at the HA CS are more prevalent. This result might be a consequence of our policy for controlling H5N2 viruses, that is, culling poultry flocks infected with viruses that contain four or more basic amino acids at the HA CS, but not those with three. In fact, all six strains of the H5N2 viruses isolated in 2015 caused a mortality rate lower than 75% in chickens, and thus, they were considered to be LPAI viruses; therefore, none of the flocks infected by these viruses were culled. This might allow these viruses to persist in poultry flocks and spread from one flock to another.
The viral sequences with two- or four-nucleotide deletions or insertions at the HA CS sequence were detected in the 1209/03-E5 and 1209/03-E10 strains. These mutations led to a frameshift in the viral HA gene, and thus, no functional HA protein could be produced from viruses with these sequences. It is unlikely that these mutations were due to RT-PCR errors, because a high-fidelity reverse transcriptase (SuperScript III) and Taq DNA polymerase (SuperTaq XL) were used in this study. Both polymerases are known to have higher fidelity than that of the avian influenza virus [5]. Moreover, viral RNA molecules with frameshift mutations in the HA and PB1 genes were previously found in other type A influenza viruses [8, 15]. Therefore, we speculate that viral RNAs with such mutations might represent by-products or intermediates during the evolution of the H5N2 viruses from low to high pathogenic viruses.
In summation, the results presented in this study highlight the potential of Taiwanese H5N2 viruses to change their pathogenicity and HA CS sequences via mutations. Moreover, viruses with the HA CS sequence QREKR/GL were more prevalent than others in 2015. These findings are useful for understanding the mechanism of sequence changes at the HA CS and for refining H5N2 virus control measures in Taiwan.
Acknowledgments
This work was supported by grant no. 102AS-10.5.1-BQ-B5 from the Council of Agriculture, and by the Aiming for the Top University Program from the Ministry of Education, Executive Yuan, R.O.C.
References
- 1.Alexander D. J.1995. The epidemiology and control of avian influenza and Newcastle disease. J. Comp. Pathol. 112: 105–126. doi: 10.1016/S0021-9975(05)80054-4 [DOI] [PubMed] [Google Scholar]
- 2.Banks J., Speidel E. S., Moore E., Plowright L., Piccirillo A., Capua I., Cordioli P., Fioretti A., Alexander D. J.2001. Changes in the haemagglutinin and the neuraminidase genes prior to the emergence of highly pathogenic H7N1 avian influenza viruses in Italy. Arch. Virol. 146: 963–973. doi: 10.1007/s007050170128 [DOI] [PubMed] [Google Scholar]
- 3.Cheng M. C., Lee M. S., Lee S. H., Wang C. H.2011. The Virulence Variation in a H5N2 Low Pathogenic Avian Influenza Virus after Passage in 14-day-old Chicken Embryonic Eggs. Taiwan V. J. 37: 221–226. (in Chinese) [Google Scholar]
- 4.Cheng M. C., Soda K., Lee M. S., Lee S. H., Sakoda Y., Kida H., Wang C. H.2010. Isolation and characterization of potentially pathogenic H5N2 influenza virus from a chicken in Taiwan in 2008. Avian Dis. 54: 885–893. doi: 10.1637/9208-120609-Reg.1 [DOI] [PubMed] [Google Scholar]
- 5.Duffy S., Shackelton L. A., Holmes E. C.2008. Rates of evolutionary change in viruses: patterns and determinants. Nat. Rev. Genet. 9: 267–276. doi: 10.1038/nrg2323 [DOI] [PubMed] [Google Scholar]
- 6.Fouchier R. A., Munster V., Wallensten A., Bestebroer T. M., Herfst S., Smith D., Rimmelzwaan G. F., Olsen B., Osterhaus A. D.2005. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J. Virol. 79: 2814–2822. doi: 10.1128/JVI.79.5.2814-2822.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horimoto T., Rivera E., Pearson J., Senne D., Krauss S., Kawaoka Y., Webster R. G.1995. Origin and molecular changes associated with emergence of a highly pathogenic H5N2 influenza virus in Mexico. Virology 213: 223–230. doi: 10.1006/viro.1995.1562 [DOI] [PubMed] [Google Scholar]
- 8.Iqbal M., Reddy K. B., Brookes S. M., Essen S. C., Brown I. H., McCauley J. W.2014. Virus pathotype and deep sequencing of the HA gene of a low pathogenicity H7N1 avian influenza virus causing mortality in Turkeys. PLOS ONE 9: e87076. doi: 10.1371/journal.pone.0087076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee C. C., Zhu H., Huang P. Y., Peng L., Chang Y. C., Yip C. H., Li Y. T., Cheung C. L., Compans R., Yang C., Smith D. K., Lam T. T., King C. C., Guan Y.2014. Emergence and evolution of avian H5N2 influenza viruses in chickens in Taiwan. J. Virol. 88: 5677–5686. doi: 10.1128/JVI.00139-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Londt B. Z., Banks J., Alexander D. J.2007. Highly pathogenic avian influenza viruses with low virulence for chickens in in vivo tests. Avian Pathol. 36: 347–350. doi: 10.1080/03079450701589134 [DOI] [PubMed] [Google Scholar]
- 11.OIE2008. Avian influenza, pp. 465–481. In: Manual of Diagnostic Tests and Vaccines For Terrestrial Animals (Mammals, Birds and Bees), 6th ed., vol. 1. Office Intl Des Epizooties, Paris. [Google Scholar]
- 12.Rott R.1992. The pathogenic determinant of influenza virus. Vet. Microbiol. 33: 303–310. doi: 10.1016/0378-1135(92)90058-2 [DOI] [PubMed] [Google Scholar]
- 13.Senne D. A., Panigrahy B., Kawaoka Y., Pearson J. E., Süss J., Lipkind M., Kida H., Webster R. G.1996. Survey of the hemagglutinin (HA) cleavage site sequence of H5 and H7 avian influenza viruses: amino acid sequence at the HA cleavage site as a marker of pathogenicity potential. Avian Dis. 40: 425–437. doi: 10.2307/1592241 [DOI] [PubMed] [Google Scholar]
- 14.Soda K., Cheng M. C., Yoshida H., Endo M., Lee S. H., Okamatsu M., Sakoda Y., Wang C. H., Kida H.2011. A low pathogenic H5N2 influenza virus isolated in Taiwan acquired high pathogenicity by consecutive passages in chickens. J. Vet. Med. Sci. 73: 767–772. doi: 10.1292/jvms.10-0532 [DOI] [PubMed] [Google Scholar]
- 15.Van den Hoecke S., Verhelst J., Vuylsteke M., Saelens X.2015. Analysis of the genetic diversity of influenza A viruses using next-generation DNA sequencing. BMC Genomics 16: 79. doi: 10.1186/s12864-015-1284-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.WHO. 2014. Cumulative number of confirmed human cases of avian influenza A(H5N1) reported to WHO. http://www.who.int/influenza/human_animal_interface/H5N1_cumulative_table_archives/en/. [accessed 16 August, 2016].