Abstract
Canine distemper virus (CDV) is one of the most serious pathogens found in many species of carnivores, including domestic dogs. In this study, hemagglutinin (H) genes were detected in five domestic Vietnamese dogs with diarrhea, and two CDVs were successfully isolated from dogs positive for H genes. The complete genome of one isolate, CDV/dog/HCM/33/140816, was determined. Phylogenetic analysis showed that all Vietnamese CDVs belonged to the Asia-1 genotype. In addition, the H proteins of Vietnamese CDV strains were the most homologous to those of Chinese CDVs (98.4% to 99.3% identity). These results indicated that the Asia-1 genotype of CDV was the predominant genotype circulating among the domestic dog population in Vietnam and that transboundary transmission of CDV has occurred between Vietnam and China.
Keywords: canine distemper virus, dog, genotype
Canine distemper virus (CDV) is an infectious agent that can cause canine distemper (CD), a lethal disease, among many species of carnivores [2,3,4, 12]. Recently, a CDV epidemic was reported among non-human primates in Chinese and Japanese quarantines [21, 23]. CDV is a negative-sense, single-stranded RNA virus that is a member of the order Mononegavirales, family Paramyxoviridae, genus Morbillivirus. The genome of CDV encodes six structural proteins (nucleocapsid (N), matrix (M), fusion (F), hemagglutinin (H), phospho- (P) and large- (L) proteins) and two nonstructural proteins (C and V proteins) [5]. The H gene is one of the most variable genes in CDV and has been used to investigate the genetic relationships among the various strains [10]. At least 14 different genotypes of CDV have been reported, including Asia-1, Asia-2, Asia-3, Asia-4, Europe, European wildlife, Arctic-like, Rockborn-like, America-1, America-2, Africa, South America-1, South America-2 and South America-3 [9, 11, 22, 25, 26]. Although there has been one report of the isolation of the genotype America-1 from Vietnamese dogs in Ha Noi, Vietnam [14], information on CDVs in Vietnam is very scarce.
In this study, CDVs were detected and isolated from Vietnamese dogs with clinical symptoms and were genetically analyzed.
MATERIALS AND METHODS
Collection of fecal and eye swabs
A total of 43 fecal swabs and 10 eye swabs were collected from diarrheic dogs in one veterinary hospital in Ho Chi Minh, Vietnam, from 2013 to 2014. Their ages ranged from 2 months to 3 years. The swab samples were dissolved in 2 ml of phosphate-buffered saline and were then filtered through a 0.22-µm filter (Millipore, Carrigtwohill, Ireland) and stored at −80°C until analysis.
Viral RNA extraction: RNA was extracted from CDV-infected A72/SLAM cells using the RNeasy Mini Kit (Qiagen, Hilden, Germany) and from the filtered fecal and eye swab samples using the Viral RNA Mini Kit (Qiagen).
Reverse transcription polymerase chain reaction (RT-PCR) for H genes: To determine the H genes of the CDVs, RT-PCR was performed using QIAGEN One Step RT-PCR (Qiagen) with the primers HF (5′-AAC TTA GGG CTC AGG TAG TC-3′) and HR (5′-AGA TGG ACC TCA GGG TAT AG-3′) [8]. The amplified products were confirmed to be approximately 2 kbp in size by electrophoresis on 0.8% agarose gels and were purified using the QIAquick PCR Purification Kit (Qiagen).
Sequence analysis
The nucleotide sequences were determined using an ABI PRISM 310 Genetic Analyzer auto sequencer (Applied Biosystems, Carlsbad, CA, U.S.A.). The full length of the H gene (1,824 bp) was determined as described previously [13].
Virus isolation
For virus isolation, A72 cells expressing canine SLAM, A72/cSLAM [20], were used. The cells were maintained in Dulbecco’s minimum essential medium (DMEM; Life Technologies, St. Louis, MO, U.S.A.) containing 10% heat-inactivated fetal bovine serum (FBS; Sigma-Aldrich, St. Louis, MO, U.S.A.), 100 units/ml penicillin and 100 µg/ml streptomycin (Life Technologies). A72/cSLAM cells in 6-well plates were inoculated with the filtrates of the swab samples and then incubated at 37°C in a 5% CO2 incubator. Cells were observed daily for a cytopathic effect (CPE). If no CPE was present, the cells were blind-passaged five times. The isolates were plaque-purified three times using Crandell-Rees feline kidney cells expressing canine signaling lymphocyte activation molecule (CRFK/cSLAM) [20].
Nucleotide sequence of the complete genome of CDV/dog/HCM/33/140816
To determine the nucleotide sequence of the complete genome of Vietnamese CDV, one representative isolate, CDV/dog/HCM/33/140816, was analyzed. The TaKaRa RNA LA PCRTM Kit (AMV) Ver. 1.1 (Takara, Otsu, Japan) was used to amplify the whole genome of this CDV strain with the following primer pairs: 53F, 5′-CTT AGG GTC AAT GAT CCT ACC-3′, and 4977R, 5′-TGG AGG GGA TCT TGT AGG GT-3′ (4,925 bp); 4398F, 5′-GTT ATC ATC AGC GAT GA TCAG-3′, and 9076R, 5′-AGA TGG ACC TCA GGG TAT AG-3′ (4,679 bp); 6947F, 5′-AAC TTA GGG CTC AGG TAG TC-3′, and 12457R, 5′- GGT TCC TTA ATG CTC TCG C-3′ (5,511 bp); and 11832F, 5′-GCA CCC ATA GGT GGT CTT AAT-3′, and 15523R, 5′- GTC TCA AGT TGA AAG AGC CAA TTC-3′ (3,692 bp). The PCR products were sequenced using specific primers, which were designed according to the overlapping strategy. The 5′- and 3′-terminal sequences were determined using the 5′ RACE System for Rapid Amplification of cDNA Ends, Version 2.0 (Invitrogen, Waltham, CA, U.S.A.). The sequence was assembled and analyzed using GENETYX® ver. 8 (Software Development Co., Tokyo, Japan).
Phylogenetic analysis
Phylogenetic relationships based on the amino acid sequences of the H proteins and the nucleotide sequence of the genome or each gene were analyzed using a distance-based (neighbor-joining) method with MEGA 6.0 software [24]. Bootstrap values were calculated based on 1,000 replicates.
RESULTS
Detection and isolation of CDV in Vietnamese dogs
A total of five CDV strains were amplified by the RT-PCR for H genes. All dogs from which CDVs were detected had diarrhea and were less than 1 year old (Table S1). The Vietnamese CDVs were named CDV/dog/HCM/01/131018, CDV/dog/HCM/24/140112, CDV/dog/HCM/29/140127, CDV/dog/HCM/33/140816 and CDV/dog/HCM/38/140827. These CDV-positive samples were inoculated into A72/cSLAM cells for virus isolation. Two CDVs, CDV/dog/HCM/33/140816 and CDV/dog/HCM/38/140827, were isolated.
Comparison of Vietnamese CDVs with other CDVs
Five H genes (1,824 bp, 607 amino acids) were determined, and the nucleotide sequences were deposited into the DNA database of Japan (DDBJ) (Accession numbers: LC159583, LC159584, LC159585, LC159586 and LC159587). All Vietnamese CDV strains possessed G at position 530 (530G) and Y at position 549 (549Y) in the H proteins. A total of nine potential asparagine-linked glycosylation sites were conserved. The percent identity between the amino acids of the H protein among the Vietnamese CDVs ranged from 98.5% to 100%; between Vietnamese CDVs and genotype Asia-1, it ranged from 96.5% to 99.5%; between Vietnamese CDVs and genotype Asia-2, it ranged from 92.9% to 93.6%; between Vietnamese CDVs and genotype Asia-3, it ranged from 86.2% to 86.7%; between Vietnamese CDVs and genotype Asia-4, it ranged from 93.7% to 94.8%; between Vietnamese CDVs and genotype America-1, it ranged from 89.6% to 91.4%; between Vietnamese CDVs and genotype America-2, it ranged from 93.9% to 96.2%; between Vietnamese CDVs and genotype Europe, it ranged from 93.4% to 95.7%; between Vietnamese CDVs and genotype European wildlife, it ranged from 94.1% to 95.4%; between Vietnamese CDVs and genotype Artic, it ranged from 92.3% to 94.1%; between Vietnamese CDVs and genotype Rockborn-like, it ranged from 94.6% to 95.9%; between Vietnamese CDVs and genotype South America, it ranged from 93.2% to 94.9%; and between Vietnamese CDVs and genotype Africa, it ranged from 93.9% to 94.7%.
Complete sequence of a Vietnamese CDV
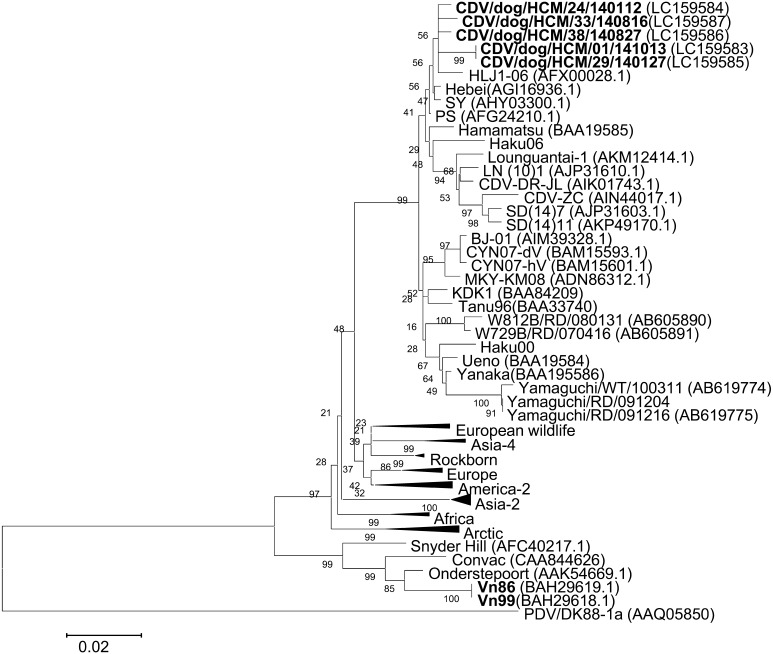
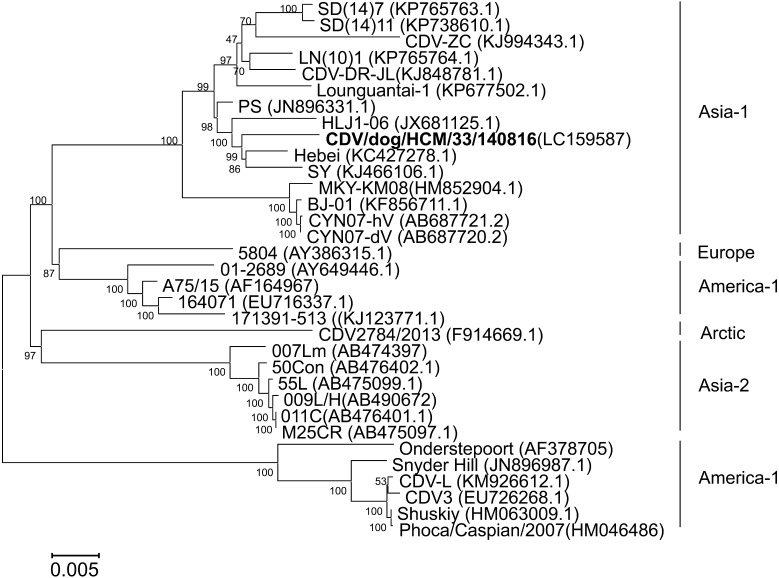
CDV/dog/HCM/33/140816 was isolated from the eye swabs of dogs with diarrhea and was plaque-purified three times. The complete genome was determined and deposited into the DDBJ (accession number: LC159587). The genome of the CDV/dog/HCM/140816 strain is 15,690-bp long and consists of six structure genes in the order of 3′-N-P-M-F-H-L-5′. The open reading frames (ORF) of N, P, M, F, H and L were located at nucleotides 108 to 1,679, 1,081 to 3,324, 3,432 to 4,439, 4,935 to 4,623, 7,079 to 8,902 and 9,030 to 15,584, respectively. The percent identities between the complete sequences of CDV/dog/HCM/33/140816 and 32 other CDVs deposited in GeneBank ranged from 92.4% to 98.7% and that with the Hebei strain showed the highest sequence similarity (Table S2). Phylogenetic trees were constructed based on H gene (Fig. 1) and the whole genome (Fig. 2). The results showed that the Vietnamese CDV/dog/HCM/33/140816 was close to Chinese CDVs, Hebei, SY, PS and HLJ1-06.
Fig.1.
Phylogenetic tree based on the amino acid sequences of the H gene (607 amino acids). The Vietnamese CDV strains isolated in this and previous studies are shown in bold. Genbank accession numbers are shown in parentheses. The accession numbers of reference strains in collapsed branches (triangles) are as follows. European wildlife: H06Ny11 (ABK35780), H06Ny13 (ABK35782), Danish Mink (CAA87688) and German ferret (CAA59358). Asia-4: NJ (11) 2 (AHN52227), NJ (12) 4 (AHN52229) and NJ (12) 5 (AHN52230). Rockborn: Rockborn (ADN96002), Rockborn Candur (ADU4476) and lesser panda (AAD54601). Europe: 5804 (AAQ49703), Dog Turkey (AAM11476), DK91A (AAQ05829) and 2544 (CAB01252). America-2: 16407 (ACD92997), 171391-513 (AHM26181.1), A75/17 (AAD49703.1), 01-2689 (AAT94553.1), Black panther A-92 (CAA90879) and America dog (CAA87691). Africa: 4L70214 (ACS36478), 21L (ACS36479) and 1bn (ACS36498). Arctic: H05Bp7F (ABK35776), H05Bp10S (ABK35779), H06Bp2S (ABK35771), CDV2784/2013 (AHF81428.1), Greenlandic dog (CAA87689) and PDV2 (CAA59357). Asia-2: 50Con (BAN16508.1), 50Con (BAN16508.1), 55L (BAN16489.1), 5L (BAN16489.1), 55L (BAN16489.1), 55L (BAN16489.1), 26D (BAB39166), 98-002 (BAA84208) and HM3 (BAB39167). The nucleotide sequences of Haku00 and Haku06 were reported by Hirama et al. (2004). Scale bar indicates the number of amino acid substitutions per site.
Fig. 2.
Phylogenetic tree based on the nucleotide sequences of the CDV genomes. The Vietnamese CDV strain, CDV/dog/HCM/33/140816, isolated in this study is shown in bold. GenBank accession numbers are shown in parentheses.
DISCUSSION
Although CD has been recognized since the 1950s and vaccines against CDV have been available for controlling the virus, CDV remains a threat to many species of animals. CD outbreaks in both domestic and wild animals have been reported in many countries [1, 6, 7, 9, 11, 15, 16, 19, 22, 25, 26]. CDVs have been categorized into many genotypes based on the H protein, and the genotypes were consistent with the geographic distribution of the viruses [17, 18]. Therefore, the genotypes make it clear where the animals were infected with CDV. In Vietnam, only genotype America-1 had been detected in domestic dogs in 2009 [14]. In this study, all of the five Vietnamese CDVs identified belonged to genotype Asia-1, indicating that this genotype is dominant among the domestic dog population in Vietnam as in other Asian countries. This is the first report of the isolation of genotype Asia-1 in Vietnam. Genotype Asia-1 of CDV was first reported in Japan in 1991 [18]. Since then, this genotype has spread to many Asian countries, including Taiwan in 2003 [7, 16], Korea in 2007 [1], Thailand in 2013 [22] and China in 2004 to 2015 [6, 15, 26]. Recently, some CD outbreaks of genotype Asia-1 had caused the deaths of tigers and monkeys [19, 23].
Phylogenetic analysis revealed that all Vietnamese CDVs in this study are closely related to the Hebei (Mink, 2008), SY (Raccoon dog, 2012), PS (Dog, 2010) and HLJ1-06 (Fox, 2006) strains found in China (Figs. 1, 2 and S1). These results suggested that there might be cross-border transmission of CDV between China and Vietnam.
Another genotype of CDV, America-1, has also been reported to have caused CD outbreaks in Vietnam [14]. However, the Vietnamese CDVs analyzed in the present study possessed low identity with genotype America-1 (89.6% to 91.4% identity in the amino acid sequences of H genes). Therefore, a vaccine against CDV genotype America-1 might not completely protect domestic dogs from CDV infection in Vietnam. Further investigations will be required to clarify the efficacy of a Vietnamese vaccine.
In this study, two Vietnamese CDV strains were successfully isolated, and their whole genomes were analyzed. Further investigations of the biological characteristics of Vietnamese CDVs would be useful for the diagnosis of CD and for vaccine development in Vietnam.
Supplementary
Acknowledgments
We would like to thank the veterinarians in participating animal hospitals for sample collection. This study was funded by Japan Society for the Promotion and Science (JSPS) KAKENHI (grant number 15H04599) and Japan Agency for Medical Research and Development (AMED).
REFERENCES
- 1.An D. J., Yoon S. H., Park J. Y., No I. S., Park B. K.2008. Phylogenetic characterization of canine distemper virus isolates from naturally infected dogs and a marten in Korea. Vet. Microbiol. 132: 389–395. doi: 10.1016/j.vetmic.2008.05.025 [DOI] [PubMed] [Google Scholar]
- 2.Appel M. J. G., Summers B. A.1995. Pathogenicity of morbilliviruses for terrestrial carnivores. Vet. Microbiol. 44: 187–191. doi: 10.1016/0378-1135(95)00011-X [DOI] [PubMed] [Google Scholar]
- 3.Appel M. J., Yates R. A., Foley G. L., Bernstein J. J., Santinelli S., Spelman L. H., Miller L. D., Arp L. H., Anderson M., Barr M., Susan K. P., Brian S. A.1994. Canine distemper epizootic in lions, tigers, and leopards in North America. J. Vet. Diagn. Invest. 6: 277–288. doi: 10.1177/104063879400600301 [DOI] [PubMed] [Google Scholar]
- 4.Beineke A., Puff C., Seehusen F., Baumgärtner W.2009. Pathogenesis and immunopathology of systemic and nervous canine distemper. Vet. Immunol. Immunopathol. 127: 1–18. doi: 10.1016/j.vetimm.2008.09.023 [DOI] [PubMed] [Google Scholar]
- 5.Carvalho O. V., Botelho C. V., Ferreira C. G. T., Scherer P. O., Soares-Martins J. A., Almeida M. R., Silva Júnior A.2012. Immunopathogenic and neurological mechanisms of canine distemper virus. Adv. Virol. 2012: 163860. doi: 10.1155/2012/163860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng Y., Wang J., Zhang M., Zhao J., Shao X., Ma Z., Zhao H., Lin P., Wu H.2015. Isolation and sequence analysis of a canine distemper virus from a raccoon dog in Jilin Province, China. Virus Genes 51: 298–301. doi: 10.1007/s11262-015-1236-3 [DOI] [PubMed] [Google Scholar]
- 7.Chulakasian S., Lee M. S., Wang C. Y., Chiou S. S., Lin K. H., Lin F. Y., Hsu T. H., Wong M. L., Chang T. J., Hsu W. L.2010. Multiplex Amplification Refractory Mutation System Polymerase Chain Reaction (ARMS-PCR) for diagnosis of natural infection with canine distemper virus. Virol. J. 7: 122. doi: 10.1186/1743-422X-7-122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demeter Z., Lakatos B., Palade E. A., Kozma T., Forgách P., Rusvai M.2007. Genetic diversity of Hungarian canine distemper virus strains. Vet. Microbiol. 122: 258–269. doi: 10.1016/j.vetmic.2007.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Espinal M. A., Díaz F. J., Ruiz-Saenz J.2014. Phylogenetic evidence of a new canine distemper virus lineage among domestic dogs in Colombia, South America. Vet. Microbiol. 172: 168–176. doi: 10.1016/j.vetmic.2014.05.019 [DOI] [PubMed] [Google Scholar]
- 10.Gámiz C., Martella V., Ulloa R., Fajardo R., Quijano-Hernandéz I., Martínez S.2011. Identification of a new genotype of canine distemper virus circulating in America. Vet. Res. Commun. 35: 381–390. doi: 10.1007/s11259-011-9486-6 [DOI] [PubMed] [Google Scholar]
- 11.Guo L., Yang S. L., Wang C. D., Hou R., Chen S. J., Yang X. N., Liu J., Pan H. B., Hao Z. X., Zhang M. L., Cao S. J., Yan Q. G.2013. Phylogenetic analysis of the haemagglutinin gene of canine distemper virus strains detected from giant panda and raccoon dogs in China. Virol. J. 10: 109. doi: 10.1186/1743-422X-10-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harder T. C., Kenter M., Vos H., Siebelink K., Huisman W., van Amerongen G., Örvell C., Barrett T., Appel M. J. G., Osterhaus A. D. M. E.1996. Canine distemper virus from diseased large felids: biological properties and phylogenetic relationships. J. Gen. Virol. 77: 397–405. doi: 10.1099/0022-1317-77-3-397 [DOI] [PubMed] [Google Scholar]
- 13.Kameo Y., Nagao Y., Nishio Y., Shimoda H., Nakano H., Suzuki K., Une Y., Sato H., Shimojima M., Maeda K.2012. Epizootic canine distemper virus infection among wild mammals. Vet. Microbiol. 154: 222–229. doi: 10.1016/j.vetmic.2011.07.006 [DOI] [PubMed] [Google Scholar]
- 14.Lan N. T., Yamaguchi R., Kien T. T., Hirai T., Hidaka Y., Nam N. H.2009. First isolation and characterization of canine distemper virus in Vietnam with the immunohistochemical examination of the dog. J. Vet. Med. Sci. 71: 155–162. doi: 10.1292/jvms.71.155 [DOI] [PubMed] [Google Scholar]
- 15.Li W., Li T., Liu Y., Gao Y., Yang S., Feng N., Sun H., Wang S., Wang L., Bu Z., Xia X.2014. Genetic characterization of an isolate of canine distemper virus from a Tibetan Mastiff in China. Virus Genes 49: 45–57. doi: 10.1007/s11262-014-1062-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang C. T., Chueh L. L., Lee K. H., Huang H. S., Uema M., Watanabe A., Miura R., Kai C., Liang S. C.2008. Phylogenetic Analysis and Isolation of Canine Distemper Viruses in Taiwan. Taiwan Vet. J. 34: 198–210. [Google Scholar]
- 17.Martella V., Cirone F., Elia G., Lorusso E., Decaro N., Campolo M., Desario C., Lucente M. S., Bellacicco A. L., Blixenkrone-Møller M., Carmichael L. E., Buonavoglia C.2006. Heterogeneity within the hemagglutinin genes of canine distemper virus (CDV) strains detected in Italy. Vet. Microbiol. 116: 301–309. doi: 10.1016/j.vetmic.2006.04.019 [DOI] [PubMed] [Google Scholar]
- 18.Mochizuki M., Hashimoto M., Hagiwara S., Yoshida Y., Ishiguro S.1999. Genotypes of canine distemper virus determined by analysis of the hemagglutinin genes of recent isolates from dogs in Japan. J. Clin. Microbiol. 37: 2936–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagao Y., Nishio Y., Shiomoda H., Tamaru S., Shimojima M., Goto M., Une Y., Sato A., Ikebe Y., Maeda K.2012. An outbreak of canine distemper virus in tigers (Panthera tigris): possible transmission from wild animals to zoo animals. J. Vet. Med. Sci. 74: 699–705. doi: 10.1292/jvms.11-0509 [DOI] [PubMed] [Google Scholar]
- 20.Nakano H., Kameo Y., Andoh K., Ohno Y., Mochizuki M., Maeda K.2009. Establishment of canine and feline cells expressing canine signaling lymphocyte activation molecule for canine distemper virus study. Vet. Microbiol. 133: 179–183. doi: 10.1016/j.vetmic.2008.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qiu W., Zheng Y., Zhang S., Fan Q., Liu H., Zhang F., Wang W., Liao G., Hu R.2011. Canine distemper outbreak in rhesus monkeys, China. Emerg. Infect. Dis. 17: 1541–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Radtanakatikanon A., Keawcharoen J., Charoenvisal N. T., Poovorawan Y., Prompetchara E., Yamaguchi R., Techangamsuwan S.2013. Genotypic lineages and restriction fragment length polymorphism of canine distemper virus isolates in Thailand. Vet. Microbiol. 166: 76–83. doi: 10.1016/j.vetmic.2013.05.015 [DOI] [PubMed] [Google Scholar]
- 23.Sakai K., Nagata N., Ami Y., Seki F., Suzaki Y., Iwata-Yoshikawa N., Suzuki T., Fukushi S., Mizutani T., Yoshikawa T., Otsuki N., Kurane I., Komase K., Yamaguchi R., Hasegawa H., Saijo M., Takeda M., Morikawa S.2013. Lethal canine distemper virus outbreak in cynomolgus monkeys in Japan in 2008. J. Virol. 87: 1105–1114. doi: 10.1128/JVI.02419-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S.2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. doi: 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yi L., Xu H., Wang J., Cheng Y., Zhang H., Yan X., Cheng S.2013. Complete Nucleotide Sequence of Canine Distemper Virus CDV-PS, Isolated from Dogs in China. Genome Announc. 1: e00232–e13. doi: 10.1128/genomeA.00232-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao J. J., Yan X. J., Chai X. L., Martella V., Luo G. L., Zhang H. L., Gao H., Liu Y. X., Bai X., Zhang L., Chen T., Xu L., Zhao C. F., Wang F. X., Shao X. Q., Wu W., Cheng S. P.2010. Phylogenetic analysis of the haemagglutinin gene of canine distemper virus strains detected from breeding foxes, raccoon dogs and minks in China. Vet. Microbiol. 140: 34–42. doi: 10.1016/j.vetmic.2009.07.010 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.