Abstract
Since the advent of microarrays, vast amounts of gene expression data have been generated. However, these microarray data fail to reveal the transcription regulatory mechanisms that underlie differential gene expression, because the identity of the responsible transcription factors (TFs) often cannot be directly inferred from such data sets. Regulatory TFs activate or repress transcription of their target genes by binding to cis-regulatory elements that are frequently located in a gene's promoter. To understand the mechanisms underlying differential gene expression, it is necessary to identify physical interactions between regulatory TFs and their target genes. We developed a Gateway-compatible yeast one-hybrid (Y1H) system that enables the rapid, large-scale identification of protein-DNA interactions using both small (i.e., DNA elements of interest) and large (i.e., gene promoters) DNA fragments. We used four well-characterized Caenorhabditis elegans promoters as DNA baits to test the functionality of this Y1H system. We could detect ∼40% of previously reported TF-promoter interactions. By performing screens using two complementary libraries, we found novel potentially interacting TFs for each promoter. We recapitulated several of the Y1H-based protein-DNA interactions using luciferase reporter assays in mammalian cells. Taken together, the Gateway-compatible Y1H system will allow the high-throughput identification of protein-DNA interactions and may be a valuable tool to decipher transcription regulatory networks.
Differential gene expression is a major determinant in development. Each gene exhibits a specific temporal and spatial expression pattern and level, and as a result, each cell/tissue expresses a unique subset of proteins. Differential gene expression can be regulated at different steps, including protein synthesis as well as protein and mRNA degradation. However, it is widely appreciated that developmental gene expression patterns are predominantly established at the level of transcription regulation (Lee and Young 2000). Specifically, differential gene expression is controlled by regulatory transcription factors (TFs) that bind to cis-regulatory DNA elements (binding sites) that are often located in or near a gene's promoter. Between 5% and 10% of metazoan genes encode putative TFs (Levine and Tjian 2003). With ∼14,000-30,000 genes predicted in metazoan genomes (The C. elegans Sequencing Consortium 1998; Adams et al. 2000; Lander et al. 2001; Venter et al. 2001), this translates to hundreds or thousands of predicted TFs.
The availability of complete genome sequences and the development of functional genomic technologies enable the assessment of questions relating to differential gene expression on a genome-wide scale. For example, the use of microarrays allows the profiling of genome-wide gene expression under different experimental conditions (Schena et al. 1995). The subsequent use of clustering algorithms enables the identification of genes with similar expression profiles that may be involved in similar biological processes (Eisen et al. 1998). After aligning the promoter sequences of such genes, cis-regulatory elements can be found that may be responsible for their coexpression (Tavazoie et al. 1999). Although microarray data are informative to provide gene expression “signatures” for tissues and/or organisms under various experimental conditions, these data fail to reveal the transcription regulatory mechanisms that control differential gene expression. This is because not all functional elements can be detected by promoter alignments. In addition, the TFs that bind to elements that can be identified may not have been experimentally characterized yet. Moreover, many TFs belong to larger TF families in which members share similar DNA-binding domains and, potentially, overlapping DNA recognition elements. As a result, even though a family of TFs can sometimes be inferred from aligned promoter sequences, the exact TF member responsible for the coexpression may remain elusive. An additional explanation for the lack of inference of transcriptional mechanisms from expression profiling data is that it is not possible to discriminate between direct and indirect transcriptional effects that lead to the differences in mRNA levels. Finally, mRNA levels measured by microarrays are not only the result of transcription, but are also the result of a balance between mRNA synthesis and degradation. Thus, to gain insight into the transcriptional mechanisms that lead to differential gene expression, genes need to be experimentally “matched” with the TFs that regulate their expression. One method of doing this is by identifying protein-DNA interactions between regulatory TFs and regulatory DNA elements (e.g., cis-regulatory elements or gene promoters).
Several biochemical methods have been developed to identify protein-DNA interactions, including gel shift, DNAse I footprinting, and chromatin-immunoprecipitation (ChIP) assays (Latchman 1998; Orlando 2000). ChIP assays are particularly powerful because they allow the identification of protein-DNA interactions that occur in vivo. This approach involves the precipitation of a DNA-binding protein together with its associated DNA, and has been used to verify target gene binding by several TFs (Shang et al. 2000; Wells et al. 2002; Conkright et al. 2003). Initially, this method has been used to characterize the interactions of individual TFs with one or a few of their target genes. To increase the throughput of the assay and to allow the unbiased identification of TF-DNA interactions throughout the genome, ChIP experiments have recently been combined with microarray technologies (Ren et al. 2000; Iyer et al. 2001). This combined method has been used to map protein-DNA interactions for 106 of the 141 predicted regulatory TFs in Saccharomyces cerevisiae (Lee et al. 2002). The data obtained were used to model yeast transcription regulatory networks. It is challenging to perform ChIP assays on a large scale using intact metazoan model organisms. This is because antibodies have to be generated for each TF, or, alternatively, transgenic strains expressing epitope-tagged TFs need to be produced. Both are time-consuming and therefore only feasible for a single or handful of TFs. Also, it is difficult to detect interactions with metazoan TFs that are expressed at low levels, in a small number of cells, or during a narrow developmental time interval. In addition, with ChIPs, one usually cannot discriminate between different TF isoforms. Further, analysis of ChIPs with microarrays requires the generation of comprehensive arrays containing regulatory genomic sequences (e.g., promoters). Finally, although ChIP experiments are valuable to answer the question, “Which DNA fragments does a TF of interest bind to?”, they are less suitable to address the converse question, “Which TFs bind to a DNA fragment (e.g., promoter) of interest?”
The yeast one-hybrid (Y1H) system is a suitable method to answer the second question because it allows the identification of proteins that can bind to DNA elements of interest, including cis-regulatory elements, origins of DNA replication, and telomeres (Li and Herskowitz 1993; Lehming et al. 1994; Kim et al. 2003). The Y1H system is conceptually similar to the yeast two-hybrid system (Y2H) that is used for the detection of protein-protein interactions (Fields and Song 1989). In the Y2H system, two hybrid proteins are used. The bait protein (X) is fused to a DNA-binding domain (DB), and the prey protein (Y) is fused to a transcription activation domain (AD). When X and Y physically interact with each other, a functional TF is reconstituted and reporter gene expression is activated. In the Y1H system, a single hybrid protein, AD-Y, is used, and reporter gene expression is activated when Y interacts with the DNA bait. Although many predicted regulatory TFs contain an intrinsic AD, several TFs have a repressor domain or no activation/repressor domain at all. In addition, DNA-binding proteins that do not function in transcription (e.g., replication and DNA repair proteins) do not contain an AD. To enable the identification of a variety of DNA-binding proteins, a strong, heterologous AD is added to the prey protein. So far, the Y1H system has been mainly used with multiple copies of short DNA elements (up to 30 bp) as DNA baits. To facilitate the high-throughput, unbiased identification of protein-DNA interactions, we developed a Gateway-compatible Y1H system. This system can be used with both small (e.g., cis-regulatory elements), and with single copies of large DNA fragments (e.g., gene promoters). This is important because the use of promoters circumvents the need to identify functional cis-regulatory elements for a gene of interest.
The Gateway cloning system facilitates accurate, high-efficiency cloning of multiple DNA fragments into various vectors in vitro because it is based on recombination and does not require the use of restriction enzymes (Hartley et al. 2000; Walhout et al. 2000a). Previously, we developed a Y2H system that is compatible with the Gateway cloning system (Walhout and Vidal 2001). Combining Gateway with Y2H greatly enhanced the throughput of protein-protein interaction identification and analysis. Indeed, already >5500 protein-protein interactions have been identified using this system (Walhout et al. 2000a, 2002; Davy et al. 2001; Boulton et al. 2002; Li et al. 2004). Therefore, the simultaneous cloning of multiple DNA baits into Y1H vectors by Gateway cloning is expected to greatly enhance the throughput of the Y1H method as well.
RESULTS AND DISCUSSION
A Gateway-Compatible Yeast One-Hybrid System
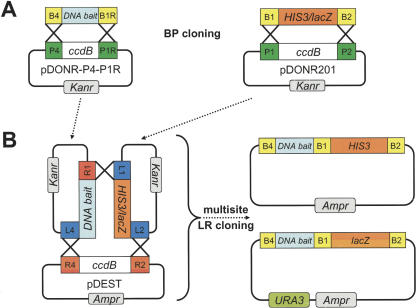
To conduct a Y1H assay, a DNA sequence of interest (the “DNA bait”) is first cloned upstream of a reporter gene to create a DNAbait::reporter construct. Here, we use the reporter genes HIS3 and lacZ. Subsequently, the DNAbait::reporter is integrated into the genome of a Y1H yeast strain by site-specific recombination into a mutant marker locus. Because the DNAbait::reporter constructs contain a wild-type marker gene, bait-containing yeast colonies can be efficiently selected. To generate a Gateway-compatible version of the Y1H system that is compatible with the Caenorhabditis elegans “promoterome” (Dupuy et al. 2004), we first cloned open reading frames (ORFs) encoding the reporters His3 and β-galactosidase (β-Gal) into a Donor vector (pDONR201; Fig. 1) by a Gateway BP reaction. This resulted in HIS3 and lacZ Entry clones. We included minimal HIS3 and CYC1 promoters in the HIS3 and lacZ reporter constructs, respectively. The reporter Entry clones can be used with promoter Entry clones (Dupuy et al. 2004) in multisite Gateway LR reactions (Cheo et al. 2004) to generate DNAbait::reporter Destination clones (Fig. 1). A generic multisite Destination vector (pDEST6; Invitrogen) was used to generate DNAbait::HIS3 fusion constructs. DNAbait::HIS3 constructs were integrated at the his3-200 locus of the Y1H yeast strain. The minimal HIS3 promoter provides sufficient levels of His3 expression for selection on minimal media lacking histidine (Sc-His). Integration of the DNAbait::reporter construct is essential to obtain reproducible results and to reduce levels of background expression (Wang and Reed 1993; data not shown). An additional vector, pDESTMW#1, was generated for Gateway multisite cloning of DNA baits together with a lacZ reporter (Fig. 1). This plasmid contains a wild-type copy of the URA3 gene, which facilitates integration of DNAbait::lacZ constructs into the ura3-52 locus of the Y1H yeast strain and selection on minimal media lacking uracil (Sc-Ura). Each DNA bait used here was fused to both HIS3 and lacZ, and the DNAbait::reporter constructs were sequentially integrated into the genome of the Y1H yeast strain and maintained on Sc-His,-Ura media. Two Y1H reporters are used in a single DNA bait yeast strain to reduce the level of false positives (Vidal 1997; see below).
Figure 1.
Creating DNAbait::reporter constructs by multisite LR cloning. (A) Gateway-cloned ORFs encoding the Y1H reporters His3 and β-galactosidase (β-Gal) were BP cloned into pDONR201 to generate HIS3 and lacZ Entry clones. DNA baits were BP cloned into pDONR-P4-P1R to generate DNA bait Entry clones. (B) The DNA baits were fused to HIS3 and lacZ by a multisite Gateway LR reaction using the DNA bait Entry clones, the Y1H reporter entry clones, and the Y1H Destination vectors to generate DNAbait::HIS3 and DNAbait::lacZ Destination clones.
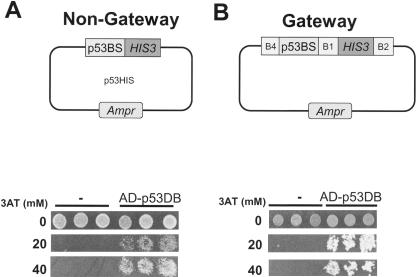
Gateway AttB Sites Do Not Influence the Y1H Readout
With the Gateway cloning system, any DNA fragment cloned into a Destination vector will be flanked by Gateway AttB recombination sites (Hartley et al. 2000; Walhout et al. 2000b). Here, DNA baits fused to HIS3 or lacZ are flanked by AttB4 and AttB1 sites (Fig. 1). We tested whether Gateway AttB recombination sites influence the readout of the Y1H system, for example, by binding an endogenous yeast TF. We compared our system to the commercially available Clontech Y1H system (BD Biosciences). As a positive control, we used the interaction between the TF p53 and its consensus DNA-binding site. Six p53-binding sites (p53BS) were fused to HIS3 and lacZ, either with or without flanking AttB Gateway recombination sites (Fig. 2). As a prey, we used the DNA-binding domain of mouse p53, fused to Gal4 AD (AD-p53DB). The induction of His3 expression by AD-p53DB was examined on media containing increasing concentrations of 3-aminotriazole (3AT). 3AT is a competitive inhibitor of the His3 enzyme. Thus, in the presence of increasing amounts of 3AT, more His3 needs to be expressed to confer growth. We found that for both DNA baits (i.e., Gateway versus “non-Gateway”), the level of background His3 expression was low and His3 expression was efficiently induced by the expression of AD-p53DB (Fig. 2). Similar results were obtained with p53-binding sites fused to lacZ (data not shown; see also Fig. 4 below). These findings demonstrate that Gateway AttB recombination sites flanking a DNA bait do not influence the detection of protein-DNA interactions in the context of the Y1H system.
Figure 2.
A Gateway-compatible Y1H system. (A) “Non-Gateway” Y1H control assay. As a DNA bait, six copies of the murine consensus p53-binding site (p53BS) fused to the HIS3 reporter gene was used with Gal4 AD alone (-) or with AD-p53DB as prey. (B) As a DNA bait, six copies of the murine consensus p53-binding site (p53BS) fused by a multisite Gateway reaction to the HIS3 reporter gene was used with Gal4 AD alone (-) or with AD-p53DB. Here, the p53BS bait is flanked by AttB4 and AttB1 Gateway recombination sites. Cells were plated on Sc-His,-Ura media with 0, 20, or 40 mM 3AT as indicated.
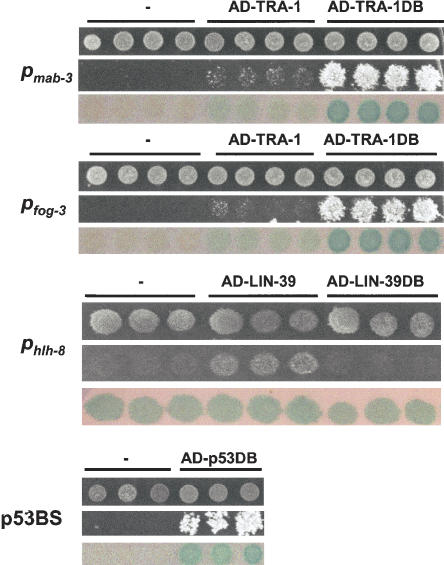
Figure 4.
TF-promoter interactions detected by the Gateway-compatible Y1H system. DNAbait::reporter strains were transformed with Gateway Destination plasmids containing either full-length TF-encoding ORFs, or the part of these ORFs that is predicted to encode the DB, fused to Gal4 AD. An empty Gateway vector containing only Gal4 AD was used as a negative control (-). The resulting yeast transformants were grown on Sc-His,-Ura,-Trp media (upper row), Sc-His,-Ura,-Trp, containing 40 mM 3AT (middle row), or spotted on YEPD plates containing a nitrocellulose filter for β-Gal assays (lower row).
Self-Activation by C. elegans Promoters
To understand the regulatory mechanisms that control differential gene expression, TFs need to be “matched” with their target genes. So far, the Y1H system has been mainly used with multiple copies of small elements as DNA baits, including cis-regulatory elements. However, the cis-regulatory DNA elements that are responsible for regulated transcription have not been identified for the vast majority of predicted genes. To facilitate the matching of TFs with their putative target genes without prior knowledge of the cis-regulatory elements these genes harbor, we tested whether single-copy gene promoters can be used as DNA baits in the Gateway-compatible Y1H system. To enable the detection of protein-DNA interactions with the Y1H assay, DNA baits need to be used that, when fused to HIS3, confer low levels of growth on media containing 3AT and, when fused to lacZ, yield white colonies. One potential difficulty with using large promoter fragments as DNA baits is that promoters are more complex in base composition than cis-regulatory elements and may thus be more likely to activate reporter gene expression in the absence of an AD-Y prey protein (e.g., by binding an endogenous yeast TF). This background expression is referred to as self-activation. We analyzed the level of self-activation by the promoters of four C. elegans genes (Pmyo-2, Pmab-3, Pfog-3, and Phlh-8). These genes were selected because trans-acting TFs that regulate their expression have been reported previously (Table 1). The ORFs encoding these TFs are available as Entry clones in the ORFeome (Reboul et al. 2003) and were transferred by a Gateway LR cloning reaction to the AD-Destination vector, pDESTAD. This vector is commonly used for Y2H screens (Walhout et al. 2000a) and is also compatible with Y1H assays.
Table 1.
Tested TF-Promoter Interactions
| Bait | Interacting TFs | References |
|---|---|---|
| Pfog-3 | TRA-1 | Chen and Ellis 2000 |
| Pmab-s | TRA-1 | Yi et al. 2000 |
| Pmyo-2 | CEH-22, PHA-4, DAF-3 | Kalb et al. 1998; Okkema and Fire 1994; Thatcher et al. 1999 |
| Phlh-8 | LIN-39, MAB-5 | Koh et al. 2002; Liu and Fire 2000 |
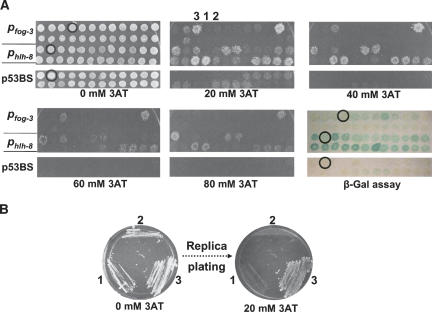
Transcription start sites have not been mapped for most C. elegans genes. However, 5′-untranslated regions are generally short. Thus, we define “promoters” as the intergenic sequences upstream of the predicted start of each ORF, with a maximal length of 2.5 kb. This should include the proximal promoter that likely has the highest density of cis-regulatory elements. The four promoters were Gateway-cloned as part of the promoterome cloning project (Dupuy et al. 2004). Each DNA bait was fused to the HIS3 and lacZ reporter genes by a multisite Gateway LR reaction (Fig. 1; Cheo et al. 2004). Successful cloning of DNAbait::reporter constructs was verified by PCR (data not shown), after which the constructs were used to generate yeast DNA bait strains. For each DNA bait strain, 24 independent yeast colonies were examined for self-activation by a β-Gal assay and by growth on Sc-His,-Ura media containing increasing concentrations of 3AT. Self-activation by the four promoters was compared with self-activation by p53BS (Fig. 3A; Pfog-3 and Phlh-8 are shown as examples). As with protein baits in the Y2H system (Walhout and Vidal 2001), various degrees of self-activation were observed for individual DNA baits and between different DNA baits. The variation in self-activation for individual DNA baits is caused by differences in the number of integration events of each promoter::reporter construct as the level of self-activation observed is higher when multiple copies are integrated compared with a single integration event (data not shown). Interestingly, individual p53BS::reporter-containing colonies also exhibit variable levels of self-activation on 3AT-containing selective media (Fig. 3A). This indicates that self-activation is not restricted to large promoter baits.
Figure 3.
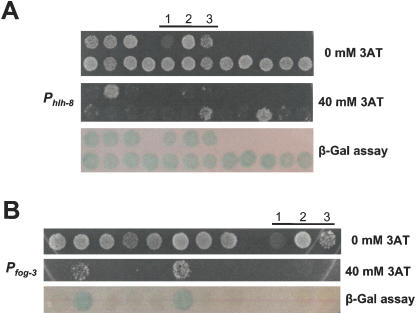
Self-activation by C. elegans promoters. (A) The Y1H Destination plasmids containing the Gateway-cloned promoters Pfog-3 or Phlh-8 were integrated into the genome of YM4271 yeast. Twenty-four double integrant colonies were picked and spotted either onto (1) Sc-His,-Ura media plus increasing amounts of 3AT to determine self-activation of the HIS3 reporter, and onto (2) YEPD plates containing a nitrocellulose filter for a β-Gal assay. Circles indicate colonies that were selected for downstream Y1H experiments. (B) To determine if individual colonies exhibit phenotypic variations after clonal expansion, three Pfog-3 colonies, varying in their extent of self-activation (low = 1, moderate = 2, and high = 3), were picked and restreaked on Sc-His,-Ura media after which they were replica-plated onto selective 20 mM 3AT-containing media.
We selected colonies with minimal levels of self-activation with both reporters for Pmab-3 and Pfog-3 and with low HIS3 self-activation for Phlh-8 (Fig. 3A). Because all Pmyo-2-containing colonies exhibited high levels of self-activation with both reporters (data not shown), a single colony was randomly selected for downstream Y1H experiments. Next, we determined whether colony self-activation phenotypes are stable, that is, whether they exhibit phenotypic variations after several clonal expansions. Three Pfog-3::reporter-containing colonies that vary in the extent of HIS3 self-activation (low, moderate, and high; Fig. 3B) were picked, restreaked, and replica-plated onto selective 3AT-containing media. Phenotypic variations in propagated colonies were not observed, indicating that DNAbait::reporter-containing yeast strains that do not exhibit self-activation are relatively stable (Fig. 3B). Finally, we tested whether omission of the minimal promoter driving HIS3 expression would reduce the extent of self-activation. We found that yeast cells transformed with a DNAbait::HIS3 construct without a minimal HIS3 promoter failed to grow on Sc-His media (data not shown). This suggests that C. elegans promoters cannot drive basal levels of gene expression in yeast. We hypothesize that this is because of different architectural requirements of metazoan and yeast promoters, such as differences in distance requirements between the transcription start site and other basal promoter elements (e.g., the TATA box). Taken together, although some promoter baits can self-activate reporter gene expression, a yeast colony that exhibits minimal levels of self-activation can usually be selected for subsequent use in Y1H assays.
Y1H-Based TF-Promoter Interactions
Only a small number of C. elegans TF-promoter interactions have been described. We tested whether several of these could be detected with the Gateway-compatible Y1H system. DNAbait::reporter strains were generated for each promoter and transformed with the appropriate AD-TF-coding Destination clones. Subsequently, protein-DNA interactions were examined on Sc-His,-Ura,-Trp +3AT plates and by β-Gal assays. For three of the four promoters, we were able to confirm previously reported interactions (Fig. 4; see also Fig. 6 below). We detected binding of TRA-1 to both Pmab-3 and Pfog-3, but the growth induction on selective 3AT media, as well as the β-Gal expression, was modest compared with the positive control (p53DB binding to p53BS; Fig. 4). One explanation for this difference is the fact that the number of consensus binding sites for TRA-1 within Pmab-3 and Pfog-3 is relatively low (three and one, respectively, vs. six p53 consensus sites in the p53BS control). Other possibilities include the need of TRA-1 to dimerize with a specific partner to bind DNA firmly, or the interference of a TRA-1 protein domain with transcription activation. Because TRA-1 functions as a transcriptional repressor in vivo, it likely contains a repressor domain (Chen and Ellis 2000). Therefore, we examined if the removal of sequences outside the TRA-1 DB improves the Y1H read-out. The DB-encoding part of tra-1 was cloned and fused to Gal4 AD using Gateway BP and LR cloning, and transformed into Pmab-3- and Pfog-3-containing yeast strains. As illustrated in Figure 4, Pmab-3 or Pfog-3 bait strains that express AD-TRA-1DB grow better on selective media than strains expressing full-length AD-TRA-1. The increased induction of β-Gal expression by AD-TRA-1DB corroborated these observations (Fig. 4). These findings suggest that a regulatory protein domain of TRA-1 may interfere with AD reporter activation.
Figure 6.
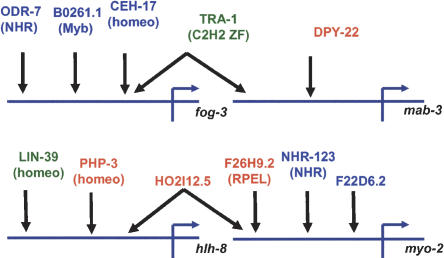
Graphic representation of the protein-promoter interactions detected. Proteins in green were detected by targeted Y1H experiments, proteins in red were identified by AD-cDNA library screens, and proteins in blue by AD-TF minilibrary screens. The TF family is indicated between parentheses under the respective ORF name. Homeo = homeodomain, Myb = Myb-like DNA binding, ZF = zinc finger, NHR = nuclear hormone receptor type zinc finger, RPEL = RPEL repeat.
LIN-39 and MAB-5 have previously been shown to bind Phlh-8 in vivo; however, there is ambiguity whether these TFs bind alone, or whether they require the association with the cofactor CEH-20 (Liu and Fire 2000). Our Y1H analysis demonstrates that LIN-39 is able to bind Phlh-8 without the presence of CEH-20 (Fig. 4). However, we could not detect an interaction between Phlh-8 and MAB-5 (data not shown). Because TRA-1DB provided a stronger Y1H readout than full-length TRA-1, we examined whether LIN-39 also exhibits stronger binding to its target when only its DB was fused to Gal4 AD. Interestingly, the LIN-39-Phlh-8 interaction was no longer detected with LIN-39 DB (Fig. 4). There are several explanations for this observation. First, removal of protein sequences outside the LIN-39 DB may have prevented correct folding of the truncated fusion protein, resulting in an inability of the fusion protein to bind DNA. Second, sequences outside DB may be necessary for LIN-39 to homodimerize or bind to an endogenous yeast dimerization partner, which may be essential for adequate DNA binding.
Pmyo-2 exhibits high levels of self-activation in the context of the Y1H system, that is, Pmyo-2 drives sufficient His3 expression to confer growth on Sc-His,-Ura media containing up to 80 mM 3AT (data not shown). Therefore, known protein-DNA interactions involving Pmyo-2 were difficult to detect. However, we were able to identify novel potential interactions with this promoter using a Y1H screening approach (see below).
Identification of Novel Protein-Promoter Interactions
To test whether we could identify protein-DNA interactions using a screening method, we performed Y1H screens with the four C. elegans promoter baits and two prey libraries: an AD-cDNA library and an AD-TF minilibrary (see below). The AD-cDNA library has already been used to generate several protein-protein interactions maps with the Gateway-compatible Y2H system (Walhout et al. 2000a; Davy et al. 2001; Boulton et al. 2002; Li et al. 2004). Yeast transformation reactions were performed as described (Walhout and Vidal 2001) and plated on Sc-His,-Ura, -Trp media containing 3AT. The amount of 3AT used varied depending on the level of self-activation exhibited by each DNA bait. After incubating for 7 d at 30°C, colonies were picked. Subsequently, growth on media containing 3AT was retested, and, in addition, β-Gal expression was tested. ORFs encoding the interactors present in colonies positive for both reporters were amplified k;1from yeast by PCR and sequenced as described (Walhout and Vidal 2001). Several putative protein-promoter interactions were identified and are summarized in Figure 6 below. Using Pmab-3 as a bait, we found a single putative interactor, DPY-22. This protein is homologous to the human transcriptional mediator TRAP230 and contains a predicted nucleic acid-binding domain (Harris et al. 2003). DPY-22 inhibits WNT-dependent ray formation in the C. elegans male tail, and its expression partly overlaps with that of MAB-3 (Zhang and Emmons 2000). MAB-3 is a TF that promotes ray differentiation (Shen and Hodgkin 1988), which suggests that the interaction between Pmab-3 and DPY-22 may be relevant in vivo.
The Pmyo-2- and Phlh-8-containing reporter strains exhibit high levels of background β-Gal expression (Fig. 3A; data not shown). Thus, activation of lacZ cannot be used as a secondary assay with these promoters. However, we retested 3AT-positive interactions of both promoters by PCR/gap-repair (Fig. 5; Walhout and Vidal 2001; data not shown). Seven out of 67 Pmyo-2 positives were retested, of which four are potentially biologically relevant (i.e., they contain a predicted DB or were found multiple times; see below). One potential Pmyo-2 interactor, F26H9.2, contains a predicted RPEL DNA-binding domain (Bateman et al. 2004). The three other positives all belong to a family of three genes that encode proteins with extensive homology: H02I12.5, Y62E10A.14, and Y17G9B.9. Two positives corresponded to H02I12.5, and one to Y62E10A.14. There is no functional annotation available for these proteins. Interestingly, H02I12.5 self-activates as a DB-bait in the context of the Y2H system, which suggests that H02I12.5 may function in transcription regulation (Walhout et al. 2002). Intriguingly, H02I12.5 was also identified as an interactor with Phlh-8 (Fig. 6). hlh-8 encodes a helix-loop-helix domain-containing TF that is involved in muscle development (Corsi et al. 2000), and MYO-2 is a myosin heavy chain isoform expressed in the pharynx muscle (Jantsch-Plunger and Fire 1994). This implies that H02I12.5 may function in muscle development as well. To test the specificity of the interactions of H02I12.5 with both promoters, we transformed an AD-H02I12.5 plasmid into the other DNAbait::reporter strains and examined Y1H interaction phenotypes. H02I12.5 binding to Pfog-3, Pmab-3, or p53BS could not be detected, indicating that the interactions of this protein with Pmyo-2 and Phlh-8 are relatively specific (data not shown). Another putative Phlh-8 interactor identified is the homeodomain-containing TF PHP-3. Because php-3 and hlh-8 exhibit similar expression profiles across 553 microarray experiments (Kim et al. 2001), the interaction between PHP-3 and Phlh-8 may be relevant in vivo. In summary, using the AD-cDNA library, we were able to identify novel potential protein-DNA interactions with three of the four promoter baits. Importantly, two of the four promoters exhibit relatively high levels of self-activation but could nonetheless be used to identify putative protein partners.
Figure 5.
Examples of retest of Phlh-8 and Pfog-3 positives. (A) Phlh-8 retest. (B) Pfog-3 retest. Double integrant yeast cells were transformed with an empty vector (containing Gal4 AD) and PCR products containing sequences that encode potential interaction partners as described previously (Walhout and Vidal 2001). Transformants were grown on Sc-His,Ura,-Trp plates with 40 mM 3AT or spotted on YEPD plates containing a nitrocellulose filter for β-Gal assays. Controls: 1 = no DNA, 2 = circular (uncut) empty AD vector, 3 = linear (cut) AD vector.
In a random search using ∼50 TFs, we found that most TF-encoding genes are covered by a relatively small number of ESTs (i.e., <5). Because the AD-cDNA library is not normalized, many colonies would have to be screened to identify such underrepresented TFs. Previously, we have generated an AD-ORF minilibrary to identify protein-protein interactions with proteins encoded by germ-line-enriched genes (Walhout et al. 2002). To increase the likelihood of retrieving TFs that are present in low proportions in the AD-cDNA library, we generated a C. elegans AD-TF minilibrary. Entry clones containing predicted TF-encoding ORFs (B. Deplancke and A.J.M. Walhout, in prep.) were obtained from the ORFeome (Reboul et al. 2003). The TF-encoding ORFs were fused to Gal4 AD by a high-throughput Gateway LR reaction. The AD-TF minilibrary contains ∼65% of all predicted TF-encoding full-length ORFs in the correct orientation and frame, and in roughly equimolar amounts. Because this minilibrary has a highly reduced complexity compared with the AD-cDNA library, fewer colonies have to be screened. We screened each promoter bait versus the AD-TF minilibrary, and each double positive was retested by PCR/gap repair (Fig. 5; Table 2). Interestingly, although no positives were found for Pfog-3 with the AD-cDNA library, five double positives were found with the AD-TF minilibrary (Table 2). The first Pfog-3 interactor, B0261.1, is a novel protein with a predicted Myb-like DNA-binding domain. The second interactor, ODR-7, is an olfactory-specific member of the nuclear hormone receptor superfamily (Sengupta et al. 1994). The remaining interactors correspond to CEH-17, a homeodomain-containing TF that has been implicated in axonal development (Pujol et al. 2000). Y1H screens with the AD-TF minilibrary yielded two additional putative Pmyo-2 interactors. F22D6.2 is a zinc-finger-containing protein with unassigned function or anatomic expression pattern. NHR-123 is a novel nuclear hormone receptor that has not been studied in C. elegans. However, NHR-123 exhibits significant homology with the human vitamin D3 receptor (VDR). Interestingly, VDR regulates the expression of several genes involved in muscle development, including myosin heavy chain genes (Endo et al. 2003). Because myo-2 encodes a myosin heavy chain, the NHR-123-Pmyo-2 interaction may constitute an “interolog,” that is, an evolutionarily conserved interaction (Walhout et al. 2000a). The usefulness of the AD-TF minilibrary is illustrated by the observation that no ESTs have been identified for ceh-17, four for nhr-123, and two for odr-7. The identification of nonoverlapping interactors (Fig. 6) indicates that both libraries should be used in future Y1H screens.
Table 2.
Overview of Y1H Screens
| DNA bait | Total no. of colonies screened | Picked colonies | Double positives | Retest by gap repair | Relevant hits |
|---|---|---|---|---|---|
| AD-cDNA library | |||||
| Pfog-3 | 1.2 × 105 | 60 | 4 | — | 0 |
| Pmab-3 | 1.8 × 105 | 48 | 4 | — | 1 |
| Pmyo-2 | 1.5 × 105 | 67 | —a | 7 | 4 |
| Phlh-8 | 1.1 × 106 | 15 | —a | 3 | 3 |
| AD-TF mini-library | |||||
| Pfog-3 | 1.4 × 106 | 45 | 22 | 5 | 5 |
| Pmab-3 | 6.2 × 105 | 48 | 0 | — | — |
| Pmyo-2 | 1.2 × 106 | 41 | —a | 2 | 2 |
| Phlh-8 | 5.6 × 105 | 44 | —a | 0 | — |
The β-Gal test could not be performed for these promoters because of high lacZ reporter self-activation.
False Negatives and False Positives
As with any system, our approach will not identify all possible interactors (i.e., false negatives). We can detect three out of seven reported protein-DNA interactions with single-copy promoter baits (∼40%). Although the sample set is small, this suggests that the rate of false negatives (i.e., missed protein-DNA interactions) is similar to that of the Gateway-compatible Y2H system (Walhout et al. 2000a). There are several explanations for missed protein-DNA interactions. First, AD-TF ORFs that are underrepresented or absent in the AD-cDNA library will be missed. Second, the AD-TF minilibrary is not suitable to identify DNA-binding proteins that can only provide a positive Y1H readout in a truncated form. Third, the AD-TF minilibrary is composed of ∼65% of all predicted worm TFs. Thus, the remaining 35% of TFs will be missed. Recently, >2000 C. elegans ORFs have been Gateway-cloned and added to the ORFeome resource (Lamesch et al. 2004). In the future, it should be possible to retrieve the putative TF-encoding ORFs from this set and include them in the AD-TF minilibrary. Fourth, elements that are located at the 5′-end of promoters may be missed because the yeast transcriptional machinery may not mediate long-range interactions. Finally, TFs that require heterodimerization or posttranslational modifications for their DNA binding will be missed. It is important to note, however, that in addition to detecting known interactions, we could detect TFs from different TF families, indicating that the Gateway-compatible Y1H system can be used to identify different classes of protein-DNA interactions (Fig. 6).
In addition to missing interactions, we may also identify irrelevant proteins (i.e., false positives). We aim to limit the number of false positives by only considering clones that score positive in both reporter assays and/or by retesting interactions using gap repair (Table 2; Walhout and Vidal 2001). Although it is difficult to estimate the residual level of false positives, it is relatively straightforward to discriminate interactors with a high likelihood of relevance in the Y1H system, compared with Y2H protein-protein interactions. This is because we are particularly interested in putative regulatory TFs and other predicted DNA-binding proteins. For each prey obtained with the AD-cDNA library, we therefore examined whether it contains a predicted DNA binding domain using the Pfam (Bateman et al. 2004) and Interpro (Mulder et al. 2003) databases. Because not all DBs may have been correctly predicted, clones that were found multiple times and that were positive with both reporters (if possible) were also considered potentially relevant. All the interactions depicted in Figure 6 fulfill these criteria.
Validation of Y1H Interactions
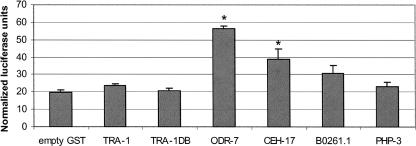
To validate Y1H-based protein-DNA interactions, we performed transient cotransfections and luciferase reporter assays in mouse 3T3 cells. We focused on interactions detected with Pfog-3 as all the corresponding TF-encoding ORFs are available as Entry clones in the C. elegans ORFeome (Reboul et al. 2003). Pfog-3 was cloned into a Gateway-compatible luciferase reporter vector that contains a minimal SV40 promoter (a kind gift from Glenn Maston, UMass Medical School). The ORFs encoding putative Pfog-3 interactors were subcloned into an N-terminal GST-tag-containing vector that allows high-level protein expression in mammalian cells. Each transfection was performed in triplicate and independently repeated three times. Results of a representative experiment are shown in Figure 7. Both ODR-7 and CEH-17 significantly induced luciferase expression compared with negative controls, confirming that these TFs can bind to Pfog-3. No significant inductions were observed for B0261.1, TRA-1, and TRA-1DB. The lack of induction by TRA-1 may be because TRA-1 functions as a repressor in C. elegans (Chen and Ellis 2000). It is important to note that no heterologous transcription activation domain was added to the TFs for this assay. Thus, the data obtained suggest that both ODR-7 and CEH-17 may function as transcriptional activators in vivo.
Figure 7.
Validation of Y1H-based protein-DNA interactions. Results of a representative experiment validating Y1H TF-Pfog-3 interactions by transient transfections and luciferase assays are shown. Negative controls include an empty GST-tag-containing vector and GST-php-3. Values are means ± SEM of three replicates. (*) P < 0.05 vs. negative controls.
In summary, we have developed a Gateway-compatible Y1H system that allows the identification of protein-DNA interactions using not only multiple copies of small (i.e., repeats of DNA elements of interest), but also single copies of large (i.e., gene promoters) DNA fragments. The system is compatible with the novel C. elegans promoterome resource (Dupuy et al. 2004) and is amenable to high-throughput settings. The Gateway-compatible Y1H system may be a valuable tool to dissect transcription regulatory networks in C. elegans and, potentially, other metazoan organisms.
METHODS
Cloning and Vector Preparation
ORFs encoding the Y1H reporters His3 and β-galactosidase (β-Gal) were obtained by PCR using the vectors pHISi-1 and pLacZi (BD Biosciences) as template DNA, respectively. The primers pHISiFW: 5′-GAATTCCCGGGGAGCTCA-3′ and pHISiRV: 5′-CTCGGGGACACCAAATATGG-3′ were used for amplification of the HIS3 ORF, and pLacZiFW: 5′-AAGCTTGAATTCGAGCTCG GTA-3′ and pLacZiRV: 5′-TTACGCGAAATACGGGCAGACAT-3′ for amplification of the lacZ ORF. To facilitate Gateway BP cloning of the ORFs into the Entry vector pDONR201, AttB1 and AttB2 tails were added to the 5′-end of both FW and RV primers, respectively. B1-HIS3-B2 and B1-lacZ-B2 PCR products were cloned into pDONR201 as described (Walhout et al. 2000b; Reboul et al. 2001). Individual L1-HIS3-L2 Entry clones were sequenced using pDONR201 FW and RV primers to retrieve a wild-type HIS3 Entry clone. The lacZ ORF is too large (3.3 kb) to sequence in a single run. Therefore, a lacZ Entry clone was selected on the basis of its functionality in a positive control Y1H experiment (see below). Reporter Entry clones can be used in a multisite LR reaction (Cheo et al. 2004) together with L4-DNAbait-R1 Entry clones and R4-ccdB-R2 cassette-containing Destination vectors to create B4-DNAbait-B1-reporter-B2 Y1H Destination clones. L4-DNAbait-R1 Entry clones were generated by performing a PCR reaction with the specific primers for each DNA target, that is, Pfog-3, Pmab-3, Pmyo-2, and Phlh-8, and genomic C. elegans DNA as a template (Dupuy et al. 2004). Subsequently, the resulting PCR products were cloned into the Entry vector pDONR-P4-P1R as described (Dupuy et al. 2004). To generate an L4-DNAbait-R1 Entry clone containing the murine p53 consensus binding site (p53BS), FW and RV primers were designed containing six p53 consensus binding site repeats tailed with AttB4 and AttB1R tails, respectively. These primers were annealed and the 5′-ends of the double-stranded product were filled in by a PCR reaction. The resulting PCR products were cloned into pDONR-P4-P1R by a Gateway BP reaction.
Two different Destination vectors were used to generate DNAbait::reporter fusions. First, a generic R4-ccdB-R2 cassette-containing Destination vector (pDEST6; Invitrogen) was used to generate DNAbait::HIS3 fusion constructs. DNAbait::HIS3 constructs were verified by PCR using M13 FW and RV primers. We generated a novel Y1H Destination vector, pDEST-MW#1, for multisite LR cloning of DNA baits together with the lacZ reporter by cloning an R4-ccdB-R2 cassette into the HindIII and AfeI sites of the URA3-gene-containing pLacZi plasmid, thereby disrupting the endogenous lacZ gene (BD Biosciences). The URA3 gene is used as a marker for selection of integration of bait::lacZ constructs. The integrity of DNAbait::lacZ constructs was verified by PCR using FW (5′-GTTCGGAGATTACCGAATCAA-3′) and RV (5′-GCCCGGATAAACGGAACTGGAA-3′) pDEST-MW#1 primers, which anneal 100 bp upstream of and 100 bp downstream from the R4-ccdB-R2 cassette, respectively. Because the lacZ ORF is 3.3 kb long, DNAbait::lacZ amplicons are frequently larger than 4 kb, which decreases the overall PCR efficiency. A new RV primer (5′-ATGCGCTCAGGTCAAATTCAGA-3′) was therefore designed, which anneals 592 bp downstream from the β-Gal ATG.
Entry clones containing ORFs encoding TFs that have been reported to bind Pfog-3, Pmab-3, Pmyo-2, or Phlh-8 were retrieved from the ORFeome (Reboul et al. 2003), and used together with pDESTAD to fuse the ORF to Gal4 AD via a conventional Gateway LR reaction as described (Walhout and Vidal 2001). To determine the success of LR cloning, a PCR reaction was performed with the AD-TF Destination clones as a template and with AD and TERM primers (Walhout and Vidal 2001). To generate a Gateway-compatible Y1H positive control, the sequence encoding the DB of the mouse p53 transcription factor (p53DB) was obtained by PCR using pGAD53m (BD Biosciences) as a DNA template and with AttB1-tailed Mp53BDFW 5′-TCCCTGTCACCGAGAC CCCTG-3′ and AttB2-tailed Mp53BDRV: 5′-TCAGTCTGAGT CAGGCCCCA-3′ primers. Subsequently, the PCR product was cloned by a Gateway BP reaction into pDONR201. The resulting p53DB Entry clone was used in a Gateway LR reaction together with pDESTAD to generate pAD-p53DB.
The DNA-binding domains of relevant TFs were predicted using Pfam (Bateman et al. 2004). The corresponding DNA sequences were retrieved by PCR using the B1-tailed FW 5′-TTACGAATCAAACAGCACCAAA-3′ and B2-tailed RV: 5′-CGATCCCGATCCACCGAATCCTCCA-3′ primers for tra-1 and the B1-tailed FW 5′-CACGCGGCGAGAAGCGACAACGA-3′ and the B2-tailed RV 5′-AGAATTGATTGAAAAGTGGGAACCGGA-3′ primers for lin-39, after which the PCR products were Gateway BP cloned into pDONR201. The resulting Entry vectors were used in a Gateway LR cloning reaction together with pDESTAD to fuse the DBs to Gal4 AD as described.
Generation of the AD-TF Minilibrary
To generate the AD-TF minilibrary, we first identified which of the ∼1000 predicted TF-encoding ORFs (B. Deplancke and A.J.M. Walhout, in prep.) are available in the ORFeome using the Worfdb database (Vaglio et al. 2003). A total of 675 TF-ORF-containing Entry clones were transformed into DH5α bacteria and purified in an automated manner using a Biorobot 8000 (QIAGEN). Subsequently, all TF-ORFs were subcloned by high-throughput Gateway LR cloning into pDESTAD to generate AD-TF fusions. These fusions can be used both in Y1H and in Y2H experiments. Destination clones were analyzed by PCR to examine if they contain an insert of the expected size (data not shown). The AD-TF minilibrary was generated by mixing AD-TF glycerol stocks in equal amounts and purifying mixed plasmid DNA from the pooled clones using CsCl2-based plasmid purification.
Yeast Manipulations
The Y1H strain YM4271 was obtained from BD Biosciences. To integrate reporter vectors into the YM4271 genome, DNAbait::reporter constructs were linearized. DNAbait::HIS3 constructs were cut with AflII or XhoI. Subsequently, DNAbait::HIS3 fusions were integrated at the his3-200 locus, and integrants were selected on Sc-His media. To generate double yeast integrants, DNAbait::lacZ constructs were linearized with NcoI or ApaI, and integrated at the ura3-52 locus of the DNAbait::HIS3-containing strains. Double integrants were selected on Sc-His,-Ura media. For each digestion and subsequent transformation, ∼300 ng of reporter plasmid was used. The high-throughput transformation protocol was performed as described in Walhout and Vidal (2001), except that the cells were grown until they reached an OD600 of 0.7 instead of 0.6, and that the heat-shock time at 42°C was extended to 20 min. Plates were incubated for 2-3 d at 30°C. Each reporter integration yielded between 20 and 200 colonies. Integration was verified by PCR on yeast genomic DNA using pDEST-specific primers (data not shown).
For each DNA bait, 24 double integrant colonies were picked and spotted (1) onto Sc-His,-Ura media with increasing concentrations of 3AT (0, 20, 40, 60, and 80 mM) to test for HIS3 reporter self-activation, and (2) on YEPD plates containing a nitrocellulose filter to analyze lacZ reporter self-activation using a β-Gal assay. For each DNA bait, a colony that exhibited minimal self-activation with both reporters was picked for use in subsequent Y1H experiments. Y1H screens were performed using the lowest bait-dependent 3AT concentration from which HIS3 reporter self-activation was sufficiently suppressed and varied between 40 and 100 mM 3AT.
For individual AD-TF transformations, 100 ng of AD-TF Destination clone DNA was transformed into the relevant DNAbait::reporter strain. Transformants were spotted on Sc-His, -Ura,-Trp media to generate a masterplate. After incubating for 2 dat30°C, colonies were replica-plated, as described (Vidal 1997), to Sc-His,-Ura,-Trp plates with increasing concentrations of 3AT (20-100 mM) and to YEPD for β-Gal assays. Plates were monitored daily and were incubated for 7-8 d.
To screen baits versus a worm AD-cDNA library (Walhout et al. 2000b), 25 μg of library DNA was used per bait. Transformation reactions were plated directly onto Sc-His,-Ura,-Trp media with the appropriate 3AT concentration (see above). For each screen, a 100- and a 1000-fold dilution was plated onto Sc-His, -Ura,-Trp to estimate the transformation efficiency. Potential positives were picked after 6-8 d of incubation at 30°C, spotted both onto an Sc-His,-Ura,-Trp masterplate as well as an Sc-His, -Ura,-Trp +3AT plate for reanalysis of potential positives. The latter plate was incubated for 4-5 d at 30°C, whereas the masterplate was replica-plated after 2 d to a YEPD plate containing a nitrocellulose filter for the β-Gal assay. To screen DNA baits versus the worm AD-TF minilibrary, the transformation protocol that was used for AD-cDNA library screens was followed, except that 5 μg of library DNA was used.
To sequence ORFs encoding potential interactors or for gap-repair-based phenotypic retesting, yeast colony PCR was performed as described previously (Walhout and Vidal 2001). Gap repair was also performed as described (Walhout and Vidal 2001). For sequencing purposes, PCR products were purified using Millipore purification kits (Millipore) adapted for use on a Qiagen Biorobot 8000 (QIAGEN). Sequencing and BLAST comparisons to identify the sequence encoding a potential interactor were performed as described previously (Reboul et al. 2001).
Transient Transfection and Luciferase Assays
Pfog-3 was cloned by a Gateway BP reaction into pDONR201 as described (Walhout et al. 2000b), and was subcloned by Gateway LR cloning into a Gateway-compatible luciferase reporter vector containing a minimal SV40 promoter (a gift from Glenn Maston, UMass Medical School). ORFs encoding Pfog-3 interactors were retrieved from the ORFeome (Reboul et al. 2003) and subcloned by Gateway LR cloning into pDEST27 (Invitrogen), which contains an N-terminal GST tag and allows high-level protein expression in mammalian cells. Mouse 3T3 fibroblasts were transiently transfected with the promoter- and interactor-containing clones, together with a Renilla reporter vector (pRL-SV40) to normalize for transfection efficiency, following instructions provided with the Effectene transfection kit (QIAGEN). Each transfection was performed in triplicate. An empty pDEST27 vector and GST-php-3 were included as negative controls. Then, 48 h after transfection, cell lysates were prepared using the Dual-Glo Luciferase assay kit (Promega) and firefly and Renilla luciferase activities were measured by a Victor Light Luminescence Counter (Perkin-Elmer). Firefly luciferase activity was normalized to Renilla luciferase activity. Each experiment was repeated three times. Luciferase results are expressed as mean ± SEM of three replicates. Differences in the extent of luciferase induction were analyzed using the paired Student's t-test function of Microsoft Excel. Statistical significance was set at P < 0.05.
Acknowledgments
The authors thank Nele Gheldof, Job Dekker, Claude Gazin, and members of the Walhout lab for helpful discussions and critical reading of the manuscript. We thank Glenn Maston for providing the Gateway-compatible luciferase reporter vector, and Michael Brasch for helpful discussions. This work was supported by the NCI 4 R33 CA097516-02 grant awarded to M.V. and A.J.M.W. as part of a promoterome consortium that also includes Ian Hope (University of Leeds, UK). B.D. was supported by a Henri Benedictus-BAEF fellowship in Biomedical Engineering of the King Baudouin Foundation and the Belgian American Educational Foundation.
Footnotes
[The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: G. Maston.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2445504.
References
- Adams, M.D., Celniker, S.E., Holt, R.A., Evans, C.A., Gocayne, J.D., Amanatides, P.G., Scherer, S.E., Li, P.W., Hoskins, R.A., Galle, R.F., et al. 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185-2195. [DOI] [PubMed] [Google Scholar]
- Bateman, A., Coin, L., Durbin, R., Finn, R.D., Hollich, V., Griffiths-Jones, S., Khanna, A., Marshall, M., Moxon, S., Sonnhammer, E.L., et al. 2004. The Pfam protein families database. Nucleic Acids Res. 32: D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton, S.J., Gartner, A., Reboul, J., Vaglio, P., Dyson, N., Hill, D.E., and Vidal, M. 2002. Combined functional genomic maps of the C. elegans DNA damage response. Science 295: 127-131. [DOI] [PubMed] [Google Scholar]
- The C. elegans Sequencing Consortium. 1998. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science 282: 2012-2018. [DOI] [PubMed] [Google Scholar]
- Chen, P. and Ellis, R.E. 2000. TRA-1A regulates transcription of fog-3, which controls germ cell fate in C. elegans. Development 127: 3119-3129. [DOI] [PubMed] [Google Scholar]
- Cheo, D.L., Titus, S.A., Byrd, D.R.N., Hartley, J.L., Temple, G.F., and Brasch, M.A. 2004. Concerted assembly and cloning of multiple DNA segments using in vitro site-specific recombination: Functional analysis of multi-segment expression clones. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Conkright, M.D., Guzman, E., Flechner, L., Su, A.I., Hogenesch, J.B., and Montminy, M. 2003. Genome-wide analysis of CREB target genes reveals a core promoter requirement for cAMP responsiveness. Mol. Cell 11: 1101-1108. [DOI] [PubMed] [Google Scholar]
- Corsi, A.K., Kostas, S.A., Fire, A., and Krause, M. 2000. Caenorhabditis elegans twist plays an essential role in non-striated muscle development. Development 127: 2041-2051. [DOI] [PubMed] [Google Scholar]
- Davy, A., Bello, P., Thierry-Mieg, N., Vaglio, P., Hitti, J., Doucette-Stamm, L., Thierry-Mieg, D., Reboul, J., Boulton, S., Walhout, A.J., et al. 2001. A protein-protein interaction map of the Caenorhabditis elegans 26S proteasome. EMBO Rep. 2: 821-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy, D., Li, Q.-R., Deplancke, B., Boxem, M., Hao, T., Lamesch, P., Sequerra, R., Bosak, S., Doucette-Stamm, L., Hope, I.A., et al. 2004. A first version of the Caenorhabditis elegans promoterome. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Eisen, M.B., Spellman, P.T., Brown, P.O., and Botstein, D. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. 95: 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo, I., Inoue, D., Mitsui, T., Umaki, Y., Akaike, M., Yoshizawa, T., Kato, S., and Matsumoto, T. 2003. Deletion of vitamin D receptor gene in mice results in abnormal skeletal muscle development with deregulated expression of myoregulatory transcription factors. Endocrinology 144: 5138-5144. [DOI] [PubMed] [Google Scholar]
- Fields, S. and Song, O. 1989. A novel genetic system to detect protein-protein interactions. Nature 340: 245-246. [DOI] [PubMed] [Google Scholar]
- Harris, T.W., Lee, R., Schwarz, E., Bradnam, K., Lawson, D., Chen, W., Blasier, D., Kenny, E., Cunningham, F., Kishore, R., et al. 2003. WormBase: A cross-species database for comparative genomics. Nucleic Acids Res. 31: 133-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley, J.L., Temple, G.F., and Brasch, M.A. 2000. DNA cloning using in vitro site-specific recombination. Genome Res. 10: 1788-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer, V.R., Horak, C.E., Scafe, C.S., Botstein, D., Snyder, M., and Brown, P.O. 2001. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409: 533-538. [DOI] [PubMed] [Google Scholar]
- Jantsch-Plunger, V. and Fire, A. 1994. Combinatorial structure of a body muscle-specific transcriptional enhancer in Caenorhabditis elegans. J. Biol. Chem. 269: 27021-27028. [PubMed] [Google Scholar]
- Kalb, J.M., Lau, K.K., Goszczynski, B., Fukushige, T., Moons, D., Okkema, P.G., and McGhee, J.D. 1998. pha-4 is Ce-fkh-1, a fork head/HNF-3α,β,γ homolog that functions in organogenesis of the C. elegans pharynx. Development 125: 2171-2180. [DOI] [PubMed] [Google Scholar]
- Kim, S.K., Lund, J., Kiraly, M., Duke, K., Jiang, M., Stuart, J.M., Eizinger, A., Wylie, B.N., and Davidson, G.S. 2001. A gene expression map for Caenorhabditis elegans. Science 293: 2087-2092. [DOI] [PubMed] [Google Scholar]
- Kim, S.H., Hwang, S.B., Chung, I.K., and Lee, J. 2003. Sequence-specific binding to telomeric DNA by CEH-37, a homeodomain protein in the nematode Caenorhabditis elegans. J. Biol. Chem. 278: 28038-28044. [DOI] [PubMed] [Google Scholar]
- Koh, K., Peyrot, S.M., Wood, C.G., Wagmaister, J.A., Maduro, M.F., Eisenmann, D.M., and Rothman, J.H. 2002. Cell fates and fusion in the C. elegans vulval primordium are regulated by the EGL-18 and ELT-6 GATA factors—Apparent direct targets of the LIN-39 Hox protein. Development 129: 5171-5180. [DOI] [PubMed] [Google Scholar]
- Lamesch, P., Milstein, S., Hao, T., Rosenburg, J., Li, N., Sequerra, R., Bosak, S., Doucette-Stamm, L., Vandenhaute, J., Hill, D.E., et al. 2004. C. elegans ORFeome version 3.1: Increasing the coverage of ORFeome resources with improved gene predictions. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Latchman, D.S. 1998. Eukaryotic transcription factors. Academic Press, San Diego, CA.
- Lee, T.I. and Young, R.A. 2000. Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 34: 77-137. [DOI] [PubMed] [Google Scholar]
- Lee, T.I., Rinaldi, N.J., Robert, F., Odom, D.T., Bar-Joseph, Z., Gerber, G.K., Hannett, N.M., Harbison, C.T., Thompson, C.M., Simon, I., et al. 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298: 799-804. [DOI] [PubMed] [Google Scholar]
- Lehming, N., Thanos, D., Brickman, J.M., Ma, J., Maniatis, T., and Ptashne, M. 1994. An HMG-like protein that can switch a transcriptional activator to a repressor. Nature 371: 175-179. [DOI] [PubMed] [Google Scholar]
- Levine, M. and Tjian, R. 2003. Transcription regulation and animal diversity. Nature 424: 147-151. [DOI] [PubMed] [Google Scholar]
- Li, J.J. and Herskowitz, I. 1993. Isolation of the ORC6, a component of the yeast origin recognition complex by a one-hybrid system. Science 262: 1870-1874. [DOI] [PubMed] [Google Scholar]
- Li, S., Armstrong, C.M., Bertin, N., Ge, H., Milstein, S., Boxem, M., Vidalain, P.O., Han, J.D., Chesneau, A., Hao, T., et al. 2004. A map of the interactome network of the metazoan C. elegans. Science 303: 540-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. and Fire, A. 2000. Overlapping roles of two Hox genes and the exd ortholog ceh-20 in diversification of the C. elegans postembryonic mesoderm. Development 127: 5179-5190. [DOI] [PubMed] [Google Scholar]
- Mulder, N.J., Apweiler, R., Attwood, T.K., Bairoch, A., Barrell, D., Bateman, A., Binns, D., Biswas, M., Bradley, P., Bork, P., et al. 2003. The InterPro Database, 2003 brings increased coverage and new features. Nucleic Acids Res. 31: 315-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkema, P.G. and Fire, A. 1994. The Caenorhabditis elegans NK-2 class homeoprotein CEH-22 is involved in combinatorial activation of gene expression in pharyngeal muscle. Development 120: 2175-2186. [DOI] [PubMed] [Google Scholar]
- Orlando, V. 2000. Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem. Sci. 25: 99-104. [DOI] [PubMed] [Google Scholar]
- Pujol, N., Torregrossa, P., Ewbank, J.J., and Brunet, J.F. 2000. The homeodomain protein CePHOX2/CEH-17 controls antero-posterior axonal growth in C. elegans. Development 127: 3361-3371. [DOI] [PubMed] [Google Scholar]
- Reboul, J., Vaglio, P., Tzellas, N., Thierry-Mieg, N., Moore, T., Jackson, C., Shin-i, T., Kohara, Y., Thierry-Mieg, D., Thierry-Mieg, J., et al. 2001. Open-reading frame sequence tags (OSTs) support the existence of at least 17,300 genes in C. elegans. Nat. Genet. 27: 1-5. [DOI] [PubMed] [Google Scholar]
- Reboul, J., Vaglio, P., Rual, J.F., Lamesch, P., Martinez, M., Armstrong, C.M., Li, S., Jacotot, L., Bertin, N., Janky, R., et al. 2003. C. elegans ORFeome version 1.1: Experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat. Genet. 34: 35-41. [DOI] [PubMed] [Google Scholar]
- Ren, B., Robert, F., Wyrick, J.J., Aparicio, O., Jennings, E.G., Simon, I., Zeitlinger, J., Schreiber, J., Hannett, N., Kanin, E., et al. 2000. Genome-wide location and function of DNA binding proteins. Science 290: 2306-2309. [DOI] [PubMed] [Google Scholar]
- Schena, M., Shalon, D., Davis, R.W., and Brown, P.O. 1995. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270: 467-470. [DOI] [PubMed] [Google Scholar]
- Sengupta, P., Colbert, H.A., and Bargmann, C.I. 1994. The C. elegans gene odr-7 encodes an olfactory-specific member of the nuclear receptor superfamily. Cell 79: 971-980. [DOI] [PubMed] [Google Scholar]
- Shang, Y., Hu, X., DiRenzo, J., Lazar, M.A., and Brown, M. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103: 843-852. [DOI] [PubMed] [Google Scholar]
- Shen, M.M. and Hodgkin, J. 1988. mab-3, a gene required for sex-specific yolk protein expression and a male-specific lineage in C. elegans. Cell 54: 1019-1031. [DOI] [PubMed] [Google Scholar]
- Tavazoie, S., Hughes, J.D., Campbell, M.J., Cho, R.J., and Church, G.M. 1999. Systematic determination of genetic network architecture. Nat. Genet. 22: 281-285. [DOI] [PubMed] [Google Scholar]
- Thatcher, J.D., Haun, C., and Okkema, P.G. 1999. The DAF-3 Smad binds DNA and represses gene expression in the Caenorhabditis elegans pharynx. Development 126: 97-107. [DOI] [PubMed] [Google Scholar]
- Vaglio, P., Lamesch, P., Reboul, J., Rual, J.F., Martinez, M., Hill, D., and Vidal, M. 2003. WorfDB: The Caenorhabditis elegans ORFeome database. Nucleic Acids Res. 31: 237-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter, J., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304-1351. [DOI] [PubMed] [Google Scholar]
- Vidal, M. 1997. The reverse two-hybrid system. In The yeast two-hybrid system (eds. P. Bartels and S. Fields), pp. 109-147. Oxford University Press, New York.
- Walhout, A.J.M. and Vidal, M. 2001. High-throughput yeast two-hybrid assays for large-scale protein interaction mapping. Methods 24: 297-306. [DOI] [PubMed] [Google Scholar]
- Walhout, A.J.M., Sordella, R., Lu, X., Hartley, J.L., Temple, G.F., Brasch, M.A., Thierry-Mieg, N., and Vidal, M. 2000a. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science 287: 116-122. [DOI] [PubMed] [Google Scholar]
- Walhout, A.J.M., Temple, G.F., Brasch, M.A., Hartley, J.L., Lorson, M.A., van den Heuvel, S., and Vidal, M. 2000b. GATEWAY recombinational cloning: Application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 328: 575-592. [DOI] [PubMed] [Google Scholar]
- Walhout, A.J.M., Reboul, J., Shtanko, O., Bertin, N., Vaglio, P., Ge, H., Lee, H., Doucette-Stam, L., Gunsalus, K.C., Schetter, A.J., et al. 2002. Integrating interactome, phenome, and transcriptome mapping data for the C. elegans germline. Curr. Biol. 12: 1952-1958. [DOI] [PubMed] [Google Scholar]
- Wang, M.M. and Reed, R.R. 1993. Molecular cloning of the olfactory neuronal transcription factor Olf-1 by genetic selection in yeast. Nature 364: 121-126. [DOI] [PubMed] [Google Scholar]
- Wells, J., Graveel, C.R., Bartley, S.M., Madore, S.J., and Farnham, P.J. 2002. The identification of E2F1-specific target genes. Proc. Natl. Acad. Sci. 99: 3890-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, W., Ross, J.M., and Zarkower, D. 2000. Mab-3 is a direct tra-1 target gene regulating diverse aspects of C. elegans male sexual development and behavior. Development 127: 4469-4480. [DOI] [PubMed] [Google Scholar]
- Zhang, H. and Emmons, S.W. 2000. A C. elegans mediator protein confers regulatory selectivity on lineage-specific expression of a transcription factor gene. Genes & Dev. 14: 2161-2172. [DOI] [PMC free article] [PubMed] [Google Scholar]