Abstract
Proteome-scale studies of protein three-dimensional structures should provide valuable information for both investigating basic biology and developing therapeutics. Critical for these endeavors is the expression of recombinant proteins. We selected Caenorhabditis elegans as our model organism in a structural proteomics initiative because of the high quality of its genome sequence and the availability of its ORFeome, protein-encoding open reading frames (ORFs), in a flexible recombinational cloning format. We developed a robotic pipeline for recombinant protein expression, applying the Gateway cloning/expression technology and utilizing a stepwise automation strategy on an integrated robotic platform. Using the pipeline, we have carried out heterologous protein expression experiments on 10,167 ORFs of C. elegans. With one expression vector and one Escherichia coli strain, protein expression was observed for 4854 ORFs, and 1536 were soluble. Bioinformatics analysis of the data indicates that protein hydrophobicity is a key determining factor for an ORF to yield a soluble expression product. This protein expression effort has investigated the largest number of genes in any organism to date. The pipeline described here is applicable to high-throughput expression of recombinant proteins for other species, both prokaryotic and eukaryotic, provided that ORFeome resources become available.
The nematode Caenorhabditis elegans is one of the best-studied multicellular model organisms (Wood 1988). Its short life span, fixed number of cells, and transparent body, together with its complete genome sequence, make it an ideal model system for investigating basic biology, especially cell differentiation and organ development (Brenner 1974; The C. elegans Sequencing Consortium 1998). Studies of this relatively simple organism have yielded many insights about the biology of higher organisms, including humans. For example, programmed cell death (apoptosis) was discovered in C. elegans (see, e.g., Ellis and Horvitz 1986).
Proteome scale studies of protein structure, function, and interactions have become a new paradigm for both investigating basic biology and developing therapeutics, as exemplified by the worldwide structural genomics initiatives and numerous proteomics projects (Burley et al. 1999; Norvell and Zapp-Machalek 2000; Zhu et al. 2001; Braun et al. 2002; Lesley et al. 2002; Adams et al. 2003). Naturally, recombinant protein expression is critical for these programs.
Under the NIH-NIGMS-sponsored Protein Structural Initiative, we selected C. elegans as our model genome to systematically express its proteins and solve their three-dimensional structures by x-ray crystallography and NMR. This effort is facilitated by the C. elegans ORFeome project, an effort that aims at cloning all predicted protein-encoding ORFs as Gateway Entry clones, which, in turn, enables a high-throughput (HTP) approach of recombinant protein expression (Reboul et al. 2003).
It is generally recognized that the production of proteins in soluble form, sufficient (milligram) quantity, and homogeneity for structural analyses is the most prodigious part of a structural genomics project (Stevens and Wilson 2001; Chambers 2002; Heinemann 2002). To express all proteins from the C. elegans genome of ∼20,000 ORFs, the traditional clone-by-clone approach is inadequate, and an automated, parallelized, HTP approach is necessary. This calls for new cloning and expression strategies amenable to automation and parallelization. The Gateway system is an ideal choice for such a genome-wide recombinant protein-expression endeavor. In a traditional approach, each ORF has to be digested with appropriate restriction enzymes, gel purified, and ligated at selected sites that may differ from protein to protein; such approaches require numerous reactions for a large number of genes and are prohibitive in parallel processing strategies. The Gateway technology revolutionized this tedious process by allowing in vitro site-specific recombination using a universal system independent of expression vector functions, host background, or candidate target gene. Once Gateway tags are added to a gene, its subsequent subcloning into an expression system requires only simple in vitro reactions that are amenable to HTP operation (Hartley et al. 2000; Walhout et al. 2000; Reboul et al. 2003).
For a genome-scale undertaking, target prioritizing (target selection) becomes an issue that is dictated by a multitude of considerations, namely, the diversity and significance of biological functions, interests of discovery science, therapeutic development, and the practical perspective of whether the ORF is experimentally tractable for structure determination by x-ray crystal-lography and NMR. For structural genomics, priority is given to novel proteins, that is, proteins without a reliable structural homolog in the Protein Data Bank (PDB; Berman et al. 2000). A traditional approach is to select protein targets by bioinformatics analysis and then obtain soluble expression using all methods available. This type of target selection, however, has drawbacks of criteria bias and prediction inaccuracy. This is evident from the following line of arguments. Firstly, more than 30% of the ORFs code for proteins of completely unknown function, and only about 50% have a definite function assigned from our analysis of the sequence-alignment notation. Therefore, any selection criteria based on existing knowledge would be inadequate due to lack of prior data. Secondly, in some reported studies, 50% or more selected targets were dropped prematurely due to solubility problems, in spite of having been predicted by bioinformatics analysis to be soluble and amenable to structural elucidation (Heinemann 2002). To produce large numbers of proteins from the C. elegans genome, we utilized a two-tiered approach for target screening and production. Our target-selection strategy is analogous to the shotgun approach used in human genome sequencing. We systematically express the C. elegans ORFs using HTP methods in 0.6 mL scale with one selected expression system, currently one expression vector, and one E. coli strain, and determine the solubility of expressed proteins under limited conditions. Soluble proteins are then produced in large quantity for structure determination. The combined approach is essentially a comprehensive target-selection strategy.
For HTP protein expression, we developed a robotic pipeline to systematically process all cloned ORFs from the C. elegans ORFeome (Reboul et al. 2003). Here, we describe the pipeline and report the protein expression data on 10,167 unique C. elegans ORFs.
RESULTS
An Integrated Robotic Pipeline
We developed a robotic pipeline on the basis of an approach of step-wise automation on an integrated robotic platform that is versatile for handling multistep processes for HTP recombinant protein expression. We optimized and miniaturized the basic protocols and developed a novel transformation platform, including a robotic device for heat-shock in 96-well plate, as well as an ElectroTip for automated electroporation (Finley et al. 2004). Because of our shotgun approach of target selection and the limited number of soluble proteins expressed in heterologous expression systems, we developed an automated ELISA (enzyme-linked immunosorbent assay) for solubility profiling. The method is applied successfully in analyzing individual ORFs in 96-well plate formats for total expression level and percentage of soluble proteins. Whereas various experimental and technological details have been previously described (Finley et al. 2004), our emphasis here is on genome-wide HTP protein expression for C. elegans.
Using our HTP pipeline, we are able to process 384 unique C. elegans ORFs every 3 wk, starting with the ORFeome collection of Entry clones (Reboul et al. 2003). A Gateway LR reaction is used to transfer individual ORFs into a destination vector for protein expression in Escherichia coli, followed by solubility profiling, production of soluble proteins in the 10 mg range, and finally structure determination by x-ray crystallography and/or NMR methodologies. This throughput can be further increased in an assembly-line fashion by staggering new plates in the process as demands require.
The pipeline developed using C. elegans ORFs is applicable to HTP recombinant protein expression for other species, provided that ORFeome resources become available. We have tested the pipeline on 350 genes from a bacterial genome arrayed on four 96-well plates. Small-scale protein expression was verified by both ELISA and SDS-PAGE, and the results are >98% consistent between the two methods (C.-H. Luan, S.H. Qiu, R.J. Gray, P.S. Horanyi, Z.-J. Liu, J. Zhou, M. Luo, and B.C. Wang, unpubl.).
Statistics of Protein Expression of C. elegans
Table 1 summarizes the results of protein expression for 118 plates of C. elegans ORFs. To achieve proteome-scale analysis, expression and solubility data were obtained using the same conditions for all genes. From the 10,167 unique ORFs, small-scale expression in E. coli was observed for 4854, of which 1536 were in a soluble form as determined by a new ELISA method (C-H. Luan, S.H. Qiu, R.J. Gray, B.J. Finley, and M. Luo, in prep.), which identified candidates that have an expression level not less than ∼2 μg/mL-expressed target protein. This accounts for ∼15% of the ORFs tested. Among those soluble proteins identified in the small-scale screen that are novel targets for structural genomics and that have a projected yield in large-scale production suitable for structure determination by NMR and x-ray crystallography, 590 have been confirmed so far by 1 L scale-up expression. Of these, 197 proteins were produced and purified without further yield-optimization for structure determination. A summary of these efforts and up-to-date results can be obtained from the Structural Genomics for C. elegans (SGCE) Web site (http://sgce.cbse.uab.edu).
Table 1.
Summary of C. elegans Recombinant Protein Expressiona
| No. of genes | % | |
|---|---|---|
| Genes processedb | 10,167 | |
| Protein expression observedc | 4854 | 47.7%d |
| Soluble expressionc | 1536 | 15.1%d |
| Soluble expression (1 L scale-up)e | 590 |
For up-to-date results: http://sgce.cbse.uab.edu.
Unique ORFs from 118 plates of 96-well each containing 88 or 94 genes.
0.6 ml expression.
Percentage of genes processed.
1 liter scale-up expression to-date on part of the soluble proteins identified in small scale screen.
Reproducibility of Protein Expression in 96-Well Format
Because of the cost and effort involved in the recombinant protein expression using our approach, it is not feasible or practical to repeat the experiments routinely on each plate to be confident about the expression and solubility of all tested proteins. Therefore, it is important to have confidence in the reproducibility of the screening results. The data in Figure 1 demonstrate the reproducibility of our multistep protein-expression process. These data are from two experiments of small-scale expression, purification, and solubility profiling for Plate 19 from the same expression clones and experimental conditions. In the first experiment, nine ORF clones expressed soluble protein in E. coli. In the second experiment, the same nine ORFs displayed soluble expression in almost the same order of solubility ranking, except for one. As shown in Figure 1, 19-A7 was ranked as the third most soluble ORF in the first experiment, whereas it was the ninth most soluble in the repeat experiment. This means that if only the second experiment were performed, 19-A7 may not have been identified as a candidate for further effort. There are 32 proteins expressed in the first experiment and 32 in the second experiment. Among them, 31 are common to both experiments. Each experiment missed one protein having mid-level expression in the other experiment. Taking into account the fact that each experiment involves multiple steps, these data demonstrate reliable reproducibility in terms of classifying candidates on the basis of protein expression and solubility profiling for the multi-ORF, multistep experiments, and are certainly satisfactory for a high-throughput screening approach.
Figure 1.
Reproducibility of protein expression in 96-well format. The data are from two experiments of protein expression, purification, and solubility profiling for plate 19. The data demonstrate good reproducibility for identifying soluble proteins in the multigene, multistep experiments. The data on left in 96-well format are ELISA readings of OD (optical density) at 405 nm for the supernatant (soluble fraction) of the expression, each well for one ORF. The maximum OD reading is 4.0. Color coding describes the level of protein expression, where red is for high, orange for medium, and yellow for low. On the right, listed with OD for each soluble protein, is σ which is calculated as the ratio of OD reading for the supernatant (soluble fraction, data shown) to that for pellet (insoluble fraction, data not shown) from the same expression experiment of an ORF. The OD and σ values combined serve as the solubility signature of a protein.
Temperature Dependence of Total Expression and Soluble Protein Expression
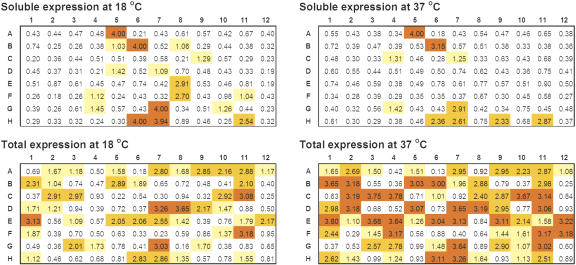
The temperature dependence of protein expression and solubility was optimized as described (Finley et al. 2004). In our HTP production mode, each ORF was routinely tested at two temperatures, 18 and 37°C, in order to find suitable expression conditions. The general observation is that more soluble proteins are obtained at 18°C, whereas more proteins are expressed at 37°C. A representative example of the results is shown for one multi-ORF plate in Figure 2. For all ORFs analyzed, 3779 were found expressed at 18°C, in contrast to 4195 at 37°C; of these, 1404 were soluble at 18°C, in contrast to 902 soluble at 37°C. When a soluble protein is found at both temperatures, the yield is usually higher at 18 than at 37°C. This is shown in Figure 3 by the 1 L expression data for 76-F10 at the two temperatures. The total expression, however, is higher at 37°C.
Figure 2.
Temperature dependence of total and soluble protein expression in 96-well format. More soluble proteins were found at 18°C; more proteins were expressed at 37°C. For this plate, the total protein expression was about 40%, whereas the soluble expression was ∼12%. The data are OD readings from 96-well format ELISA for ORFs in plate 51. Color coding is the same as in Figure 1.
Figure 3.
Comparison of expression vector pDEST17.1 vs. pET15G for protein expression. Protein expression of the genes in the four plates was carried out using pDEST17.1 (plots on left) and pET15G (plots on right). The number of soluble proteins was increased in all cases by using pET15G. The data are represented as histograms of OD readings at 405 nm from ELISA vs. ORFs in the 96-well plate.
Expression Vector Engineering
The expression vector, pDEST17.1 (Invitrogen, http://www.invitrogen.com), was less favorable for these studies, because the His6-tag on the expressed proteins is not amenable to proteolytic cleavage. It is desirable to have the option of removing the tag for protein crystallization. In addition, the yield of soluble proteins obtained with pDEST17.1 is relatively low. To overcome these problems, we engineered two vectors using two pET-vectors (Novagen, http://www.novagen.com), each encoding a thrombin cleavage site within the peptide sequence LVPRGS. We generated Gateway-compatible versions of these two pET vectors as follows: pET15G, which encodes the 21 amino acid sequence MGSSHHHHHHSS GLVPRGSQS in the pET15b backbone vector, and pET21G, which uses the pET21b backbone with the 29 amino acid sequence MASMTGGQQMG SSHHHHHSSGLVPRG SQS. For the pET21G vector, an additional fusion tag, T7 tag with epitope sequence MASMTGGQQMG, is included in the N-terminal sequence upstream of the His6-tag and thrombin cleavage site. The T7 tag promotes protein expression, as observed in a number of experiments in our study (data not shown).
Figure 3 compares the protein expression results obtained using pDEST17.1 and pET15G for ORFs in three 96-well plates. The total number of bacterial transformants with heterologous protein expression, as well as those with soluble expression increased in all cases when using pET15G. The same soluble proteins obtained by using pDEST17.1 were also obtained when pET15G was used.
To improve soluble protein production, we examined three bacterial strains, BL21(DE3), BL21-AI, and BL21(DE3)pLysS (Invitrogen) in combination with the three destination vectors, pDEST17.1, pET15G, and pET21G. The most successful combination uses either the pET15G or pET21G vector and the E. coli strain BL21-AI, as suggested by the 0.6-mL scale expression data in Table 2, whereas pDEST17.1 is most effective when coupled with BL21(DE3)pLysS.
Table 2.
Comparison of Expression by Different Vector and Bacterial Strain Combinationsa
| pET15G with BL21-AI | pET15G with BL21(DE3)pLysS | pET21G with BL21-AI | pET21G with BL21(DE3)pLysS | |
|---|---|---|---|---|
| No. of proteins expressed | 70 | 25 | 65 | 11 |
| % ORFs in plate | 79.5% | 28.4% | 73.9% | 12.5% |
From expression data for the 88 ORFs in plate 18.
Scale-Up of the Small-Scale Expression
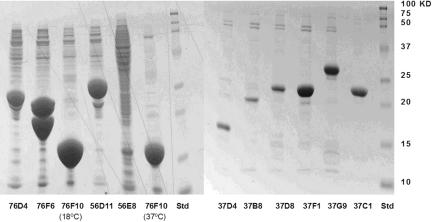
Before attempting large-scale production (six 1-Liter scale), we perform 1 L-scale expression on the soluble candidates identified in the small-scale screening. Figure 4 exemplifies the correspondence between the results of soluble proteins of 0.6 mL and 1 L expressions. The soluble protein bands in the SDS-polyacrylamide gels are consistent with ELISA data in small-scale screening. Approximately 85% of the proteins in 1-L expression have the correct molecular size. The majority of the rest either have molecular weight lower than the theoretical value or have multiple bands. These could presumably be due to incorrect gene annotation, truncation in expression, or degradation after expression.
Figure 4.
SDS-polyacrylamide gels showing the 1-L scale-up expressions of 12 soluble proteins. The expression levels in the gel are typical for the C. elegans proteins expressed in E. coli. The bands for 76D4, 76F6, 56D11, and 76F10 represents a high-level expression with yield greater than 10 mg per liter of bacterial cell culture (mg/L). The bands for 37F1, 37G9, and 37C1 represent a medium level expression from 6 to 10 mg/L, whereas the bands for 37D4, 37B8, and 37D8 represent a low level expression from 3 to 6 mg/L.
For large-scale expression, we have used both the conventional LB (Luria Broth) medium and the medium developed by F.W. Studier at Brookhaven National Laboratory, in which auto-induction of expression occurs close to saturation of bacterial growth. There is no need to monitor culture densities or add inducer at the proper time. In our experiments, the autoinduction medium led to higher levels of protein expression than LB medium in 80% of cases, depending on the specific protein. Multistep purification is performed using Ni-affinity filtration-, and ion-exchange-chromatography, taking into account the size and charge of the individual proteins.
Bioinformatics Analysis
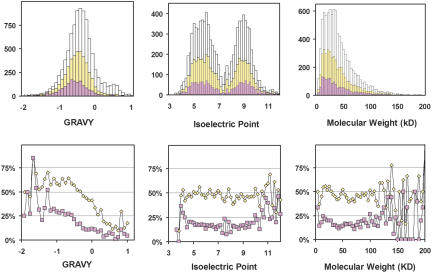
We have generated a database containing a variety of biochemical properties and predictions calculated from the sequences of each of the C. elegans ORFs and have correlated these predictions with the protein expression and solubility results obtained from our HTP screen of 10,167 bacterial-expressed ORFs (accessible via http://sgce.cbse.uab.edu). From this analysis, 34 parameters were correlated to expression and solubility using the linear correlation coefficient (LCC), and the most prominent ones are given in Table 3 with signal peptide, GRAVY (Grand Average of Hydropathicity, an indicator for average hydrophobicity of a protein), and transmembrane helices on the top of the list. Because signal peptide and transmembrane helices are hydrophobic in nature, the conclusion is that hydrophobicity is the most important indicator for heterologous expression and solubility of eukaryotic proteins in E. coli. A negative LCC of -0.20 between GRAVY and the soluble expression indicates that solubility is inversely correlated to the hydrophobicity of the protein. To a lesser extent, the number of cysteines in a protein adversely affects its expression and solubility. Correlations with molecular weight, rare codon count, and isoelectric point that we had previously observed with a far less number of samples are no longer present. The data are presented as histograms and plots of percentage expressed and percentage soluble versus GRAVY, isoelectric point, and MW in Figure 5. The GRAVY of the center of the distribution for total proteins expressed is lower than that for the 10,167 genes studied, which is close to the genome average of -0.4. The GRAVY of the center of the distribution for the soluble protein expressions is even lower. The molecular weights of the expressed proteins range from 6 KD to >100 KD, with 2%-3% having MW >100 KD. The majority of the expressed proteins are in the range of from10 to 60 KD, the same as the overall molecular weight distribution of the proteins encoded by the 10,167 ORFs. The most significant trend noted by SGI's MineSet decision tree software (http://sgi.com) is that a homologous structure deposited in the PDB implies solubility, which is also suggested by the positive LCC values in Table 3.
Table 3.
Maximum Correlation of Expression and Solubility to Molecular Properties
| LCCa
|
||
|---|---|---|
| Expression | Solubility | Parameter |
| −0.31 | −0.17 | Signal Peptideb |
| −0.24 | −0.20 | GRAVY (hydrophobicity)c |
| −0.24 | −0.11 | Transmembrane Helicesd |
| −0.14 | −0.09 | Number of cysteines |
| −0.17 | −0.08 | Anchor peptide |
| −0.12 | −0.06 | Prokaryotic membrane lipoprotein lipid attachment sitee |
| 0.16 | 0.12 | PDB identity |
Linear correlation coefficients (LCC). A cut-off of 0.10 is used for the absolute value of LCC for expression in reporting analysis results in the table.
Signal Peptides (http://www.cbs.dtu.dk/services/SignalP/) (Nielsen et al. 1997).
GRAVY (Grand Average of Hydropathicity) value for a peptide or protein is calculated as the sum of hydropathy values of all of the amino acids, divided by the number of residues in the sequence using the EXPASY site (http://ca.expasy.org/tools/protparam.html) (Kyte and Doolittle 1982; Gasteiger et al. 2003).
Transmembrane Helices (http://www.cbs.dtu.dk/services/TMHMM-2.0/) (Krogh et al. 2001).
Prokaryotic membrane lipoprotein lipid attachment site predicted by PROSITE (Bairoch et al. 1997).
Figure 5.
Histograms (top) of GRAVY, molecular weight, and isoelectric point for the 10,167 ORFs studied (white), expressed (yellow), and soluble (pink), and percentage plots (bottom) of the expressed and soluble ORFs over the studied, respectively, vs. the three parameters. Correlation between protein expression and GRAVY is apparent in the plots. The lack of a correlation to isoelectric point and molecular weight is indicated by the flatness of the curves in the percentage plots.
One of the missions of structural genomics is to prioritize those sequences with a protein family (Pfam) domain whose structure is unknown. We analyzed our expression results with respect to Pfam domains found in each ORF tested. A summary of these results is shown in Table 4. The Pfam domains associated with the highest percentage of soluble expressions (RRM_1, Motile_Sperm, Helicase_C, adh_short, Histone) all have well-known structural domains. Conversely, the targets more interesting to structural genomics (the 7TM chemoreceptor, the Neurotransmitter-gated ion-channel, and the Collagen triple helix repeat) prove difficult to express. Our data are consistent with previous observations for bacterial expression of eukaryotic proteins (Braun et al. 2002), especially with respect to Ras-like proteins, but less so with proteins containing protein kinase or MATH domains. One explanation for these differences is that Braun et al. (2002) used denaturing conditions for analyzing expression, whereas we have focused on obtaining soluble proteins using nondenaturing conditions for purification.
Table 4.
Expression/Solubility Data for the Most Common Pfam Domainsa
| Countb | %Exp | %Sol | Domain length | Pfam | Description |
|---|---|---|---|---|---|
| 60 | 78.3 | 23.3 | 104 | Motile-Sperm | Major sperm protein domain |
| 34 | 70.6 | 17.6 | 111 | Histone | Core histone H2A/H2B/H3/H4 |
| 72 | 69.4 | 25.0 | 71 | RRM-1 | RNA recognition motif |
| 38 | 68.4 | 2.6 | 70 | GST-N | GST, N-terminal |
| 44 | 65.9 | 18.2 | 247 | Adh-short | Short chain dehydrogenase |
| 48 | 64.6 | 4.2 | 163 | Ras | Ras family |
| 63 | 57.1 | 3.2 | 38 | WD40 | WD domain, G-beta repeat |
| 49 | 44.9 | 18.4 | 80 | Helicase-C | Helicase conserved C-terminal |
| 38 | 44.7 | 10.5 | 56 | Homeobox | Homeobox domain |
| 35 | 40.0 | 2.9 | 208 | Metallophos | Calcineurin-like phosphoesterase |
| 56 | 39.3 | 5.4 | 106 | BTB | BTB/POZ domain |
| 236 | 39.0 | 6.4 | 275 | Pkinase | Protein kinase domain |
| 47 | 31.9 | 4.3 | 114 | MATH | MATH domain |
| 35 | 31.4 | 2.9 | 117 | DUF290 | Transthyretin-like family |
| 65 | 27.7 | 10.8 | 77 | zf-C4 | Zinc finger, C4 type |
| 68 | 26.5 | 0 | 45 | F-box | F-box domain |
| 56 | 25.0 | 0 | 142 | FTH | FTH domain |
| 48 | 22.9 | 10.4 | 232 | Y-phosphatase | Protein-tyrosine phosphatase |
| 41 | 22.0 | 9.8 | 195 | Hormone-recep | Ligand-binding domain of nuclear hormone receptor |
| 46 | 10.9 | 0 | 212 | Neur-chan-LBD | Neurotransmitter-gated ion-channel |
| 39 | 7.7 | 0 | 294 | 7tm-5 | 7TM chemoreceptor |
| 41 | 7.3 | 0 | 202 | Neur-chan-memb | Neurotransmitter-gated ion-channel |
| 105 | 6.7 | 1.9 | 59 | Collagen | Collagen triple helix repeat |
| 70 | 2.9 | 1.4 | 61 | Col-cuticle-N | Nematode cuticle collagen N-terminal |
| 57 | 1.8 | 0 | 37 | ShTK | ShTK domain |
The list is sorted on % Expressed. %Exp stands for %Expressed, and %Sol for %Soluble.
The 33 count is 2% of total 1689 distinct pfams in the studied sequences. The list contains all with count >2% of the 1689 distinct Pfams in the studied sequences.
GRAVY and Expression
Our bioinformatics analysis of the expression data for the 10,167 ORFs suggests that overall hydrophobicity is the most important factor for an ORF to yield a soluble expression product. This provides significant experimental validation for using hydrophocity in bioinformatics analysis. This conclusion is demonstrated by 0.6-mL scale expression data for 87 genes in one plate, which was not included in the data set used in the analysis. Figure 6 lists the genes by GRAVY value, annotated with the presence of a signal peptide and transmembrane helices, and color coded with expression data.
Figure 6.
Expression data in correlation to GRAVY, Signal peptide, and Transmembrane helices as demonstrated on 87 ORFs for plate 11041. The genes are listed in two panels ordered by GRAVY value from low to high in the left panel and continued in the same manner in the right panel. The break point for the two panels was chosen for easy presentation. (Right) Higher GRAVY values. The expression data are color coded with gray for no expression, white for expression but not soluble, yellow for low level soluble, orange for mid level soluble, and red for high level soluble. (SP) Signal peptide or anchor; (TM) number of transmembrane helices.
No expression was observed for the 19 genes with the highest GRAVY values. The more negative the GRAVY value becomes, the more likely that an ORF exhibits soluble protein expression. Nine of the 11 soluble proteins, including the three with the highest total protein yield, have GRAVY values less than -0.4, the average GRAVY value for the C. elegans genome. Although protein expression was observed for the majority of the genes in that range, they are not all soluble. Even for those with the lowest GRAVY, solubility varies. In summary, the observation is that low GRAVY implies expressability. Soluble expression, however, depends on other factors and can not be accurately predicted by bioinformatics methods alone. Thus, for the foreseeable future, empirical screening appears to be the only reliable way to identify those ORFs that can be expressed in a soluble form.
DISCUSSION
In the genome-wide protein expression effort, we have analyzed >10,167 ORFs, comprising nearly half of the predicted C. elegans ORFeome. By comparison, Zhu and colleagues expressed 5800 yeast genes, accounting for 93% of the genome (Zhu et al. 2001), using a homologous expression system in yeast. Other reported heterologous protein expression efforts have involved selected sets of genes up to ∼1000 (Christendat et al. 2000; Braun and LaBear 2003). Importantly, the protein expression results presented here are based on essentially a diverse and unfiltered collection of both previously characterized and uncharacterized genes from an entire metazoan genome.
Our current HTP protein expression pipeline uses a single expression vector, pET15G with a His6-tag, in combination with one E. coli host strain. Protein expression was observed on 47.7% of the 10,167 ORFs studied, by comparison to the success rates of 50% in a study of 65 genes (Reboul et al. 2003) and 65% in a study of 167 genes (Chance et al. 2002), both selected from the same ORFeome and expressed under similar conditions using E. coli and vectors with an N-terminal His6-tag. In the small-scale screen, ∼15% of the ORFs tested express soluble protein. This rate is consistent with the results in similar studies to express eukaryotic proteins in E. coli using a poly-His purification tag (Braun et al. 2002; Chance et al. 2002). Various solubility expression rates have been reported in the literature. The majority of the data available, however, has been generated using preselected subsets of genes, as opposed to our unbiased screening of all ORFs.
A number of factors contribute to whether or not any given gene expresses soluble protein in an E. coli-based heterologous system. The first is the biological properties of the target gene. The bioinformatics analysis of our expression data indicates that the most significant trend was that a homologous structure deposited in the PDB implied solubility. However, sequences with a PDB homolog are the lowest priority for structural genomics efforts. We had prioritized our plates by giving a higher priority to the plates that contain fewer ORFs having PDB homologs. Therefore, the half of the genome for which we report data herein is enriched for those ORFs encoding fewer PDB homologs. This may affect the soluble rate in a negative way. On the other hand, our current approach is not directed at expressing folded membrane proteins, which constitute 30% of a typical proteome (Christendat 2000; Heinemann 2002). Membrane proteins account for 31% of the C. elegans genome, but only 23% of the ORFs tested in this study. We had also prioritized our plates with higher priority to those with fewer transmembrane helices. Fewer membrane proteins in the current set may affect the soluble rate in a positive way.
A second factor is protein expression conditions. To achieve a proteome-scale analysis, expression and solubility data were obtained using the same conditions for all genes. Generally, expression solubility can be improved by optimization of expression conditions for each clone. HTP operation in 96-well format precludes such individual ORF-based optimization.
The third factor in the limitation resulted from the current single ORF-based approach, where each ORF is placed in one well and each protein expression construct contains only one ORF of interest. As previously observed (Adams et al. 2003), a major problem with single ORF-expression strategies is that they intrinsically select for cytoplasmic, homomeric, unmodified, and/or cofactorless recombinant proteins. If a protein requires its partner to fold properly and to be soluble, then, the one-ORF-at-a-time approach will likely not be successful.
The fourth factor is simply expressing eukaryotic proteins in a prokaryotic host. We are expressing eukaryotic proteins that are known in their native environment to undergo posttranslational modification, whereas E. coli lacks posttranslational modification and other properties of the eukaryotic system. One of the consequences is that eukaryotic multidomain proteins cannot be expressed in E. coli in soluble form.
The above limitations also point to avenues for future improvements. There are various approaches for improving soluble expression that are feasible in an HTP environment. For example, the use of Multi-fusion tags has been demonstrated in several studies to improve the overall soluble expression rate. Using four fusion tags, 128 human proteins were expressed in E. coli with a combined soluble expression of 83% (Braun et al. 2002), whereas an 80% soluble expression is achieved for 40 genes using eight fusion protein constructs (Shih et al. 2002).
E. coli is notorious for driving overexpressed foreign proteins into inclusion bodies, therefore rendering them insoluble. Many proteins in our experiments have high-expression yields, but low solubility. Refolding is a practical approach after treatment of inclusion bodies to solubilize the proteins.
We are exploring a number of these approaches, namely, potential new fusion tags, coexpression of potential partner ORFs, eukaryotic expression systems, and HTP refolding to increase soluble protein production. Our work reported with the E. coli expression system, however, demonstrates the ability to develop automation strategies and methods for HTP recombinant protein expression. The pipeline thus developed can be readily adapted to expressions using different vectors and/or host combinations, a eukaryotic expression system, or coexpression of two or more ORFs simultaneously. In addition, the availability of an evolving ORFeome resource (Lamesch et al. 2004) cloned in the Gateway system will allow us to easily adapt to any new vector required to increase the success rate of protein production.
HTP methods are clearly an important tool for structural proteomics. Our integrated robotic pipeline streamlines the complex experimental procedures and makes it possible to carry out protein expression for thousands of genes in a timely and reproducible manner. The effort of the largest recombinant protein expression for >10,167 genes from a single organism has yielded a significant number of novel targets for structural characterization. Furthermore, the efforts have given scientific insights on using bacterial hosts to express eukaryotic proteins, as well as providing enough data to critically consider the often anecdotal results of recombinant protein expression.
METHODS
There are three aspects in developing a robotic pipeline for HTP recombinant protein expression, that is, cloning of expression constructs and optimization of the related basic molecular biology protocols, miniaturization of the protocols, and automation of bench processes.
Cloning of Expression Constructs
Our basic molecular biology approach is based on the Gateway cloning and expression technology (Hartley et al. 2000; Walhout et al. 2000).
The focus of our experiments was to adapt the Gateway technology to our general pipeline by selecting and engineering compatible vectors and host cell lines for DNA plasmid miniprep and protein expression, and by converting and developing protocols that are amenable to automation. The E. coli expression system based on bacteriophage T7 RNA polymerase developed by Studier (Studier et al. 1990) is the basis of recombinant protein expression used here. For an expression host, E. coli is still the first choice because of its low cost, easy set up, and availability of large number of expression vectors, despite the fact that E. coli lacks the machinery for features unique to eukaryotic protein expression. After experimenting with the expression vector pDEST17.1, we developed two new Gateway-compatible vectors, pET15G and pET21G, based on pET15b and pET21b (Novagen) to make use of the tightly controlled, high-level gene expression of pET vectors in E. coli. There are a number of E. coli strains to choose from, in combination with different expression vectors. We examined multiple combinations for DNA plasmid miniprep and for protein expression and selected E. coli strain DH5α (Invitrogen) for production of plasmid minipreps and strains BL21 (DE3) and BL21-AI in combination with pET15G for protein expression.
Miniaturization of Basic Protocols
Miniaturization of basic molecular biology protocols requires adjusting the processes and conditions used for conventional tube and/or flask-based culture methods to 96-well microplate-based methods. For HTP operation, all chemical and biological reactions are carried out in 96-well format in order to utilize common robotic liquid handlers. We investigated and optimized conditions for miniaturization of bacterial growth, DNA plasmid propagation, bacterial transformation, protein expression, and purification in 96-well plates (Finley et al. 2004).
Automation of Miniaturized Protocols
Because molecular biology protocols often require strict temperature controls and shaking, centrifugation, or incubation for a prolonged period, we devised a strategy of step-wise automation on an integrated robotic platform and successfully automated nearly all aspects of recombinant protein expression.
Our integrated robotic platform is centered on the Beckman/Sagian core system, including a Biomek FX and Biomek 2000 liquid handlers (Beckman-Coulter), a DNA Engine Tetrad Cycler (MJ Research), an ELX-405UV plate washer (Bio-Tek), a SpectraMax UV/Vis (Molecular Devices), plate reader, a Polarstar (BMG Lab-Technologies) fluorescence plate reader, four temperature-controlled shaker-incubators, a centrifuge with microplate carrier, and a BioRobot 9600 (QIAGEN, http:www1.qiagen.com). The integrated robotic system is supported by its system software and in-house programs for specific applications.
An automated method for DNA plasmid miniprep was developed on Biomek FX configured with a vacuum manifold, a plate shaker, a 96-well pipetting tool, along with a labware gripping tool for plate movement. A semiautomated method was developed on a BioRobot 9600 robot, which has a dynamically controlled vacuum manifold, but the movement of plates requires hands-on operation. A manual vacuum device by Eppendorf having four filter plate positions was also used for plasmid miniprep.
To automate the bacterial transformation using the standard heat-shock method, which requires strict time and temperature control, we created a novel heat-shock station and software control for the Beckman core robotic system, whereby four complete 96-well plates may be transformed in an automatic fashion in ∼2 h, as described in Finley et al. (2004). In addition, a novel ElectroTip for automated electroporation was developed.
Small-scale protein purification was automated on a Biomek FX robot with two vacuum manifolds. The pellets of bacterial cell cultures were harvested and lysed by lysozyme using robot manipulations, followed by centrifugation to separate the supernatant from pellets, and then Ni affinity purification using Ni-NTA (QIAGEN) beads in 96-well filter plates. The purification was coupled to an automated ELISA in 96-well format for protein expression analysis. The current manual centrifugation can be integrated into the automated process by purchasing a robotic-compatible centrifuge.
Streamlined Process
Following is an overall view of the general approach, and the robotic pipeline we developed for recombinant protein expression using the Gateway technology and the C. elegans ORFeome (Reboul et al. 2003). The streamlined process consists of the following major steps: (1) generating expression clones, (2) expression screening and solubility profiling, and (3) 1-L scale-up confirmation and large-scale production. As shown in Figure 7 and described in detail below, this multistep process includes the LR reaction, plasmid miniprep, bacterial transformation, colony selection, protein expression, protein purification, expression level and solubility profiling by ELISA, and large-scale production. If a cloned ORFeome is not available, two additional steps are required preceding that described above.
Figure 7.
Schematic for the HTP protein expression pipeline. The rectangles represent material, the arrows represent process, and the octagon represents data.
Construction of the Expression Clones
Our starting material for the HTP protein expression was the ORFeome collection of full-length C. elegans ORFs (Reboul et al. 2003) in the form of Entry clone. Expression clones were prepared by using the LR reaction to subclone the ORFs into an expression vector, either pDEST17.1, pET15G, or pET21G.
The product of the LR reactions were transformed into competent E. coli DH5α cells and selected on 12-well ampicillin plates. Plasmid miniipreps were produced and used to transform E. coli expression hosts. The reason for using this additional step instead of introducing the LR reaction product directly into hosts for protein expression was to reduce the outgrowth of faulty recombinant reactions (Finley et al. 2004).
Protein Expression and Solubility Profiling
The expression clones were then used to transform the protein expression E. coli strain to obtain stable stocks. Then, bacterial plating, colony picking, and bacterial culture were carried out for protein expression. Here, a colony selection was performed after the transformation to improve expression yield for the recombinant protein.
For any proteome-scale protein expression in E. coli, the percentage of soluble proteins found by using a single host and expression vector combination usually is <25%. We carried out protein expression in two steps, a small-scale screen of 0.6 mL in 96-well plates at two temperatures, typically 18 and 37°C, to profile the protein expression level and identify soluble candidates for scale-up expression, which is at 1-L or 6-L scales for protein production.
Small-scale protein expression was carried out in 2-mL 96-well block plates. After bacterial growth, cell pellets were lysed by freezing overnight at -80°C and then thawed at room temperature for 15 min before adding 500 μL lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, 1 mg/mL lysozyme at pH 8.0). After mixing, cell lysis continued by shaking for ∼30 min at 1000 rpm in Vortemp shakers. Immediately after lysis, plates were spun at 4000 rpm for 30 min and a Beckman Biomek FX robot was used to separate the supernatant from the pellet by slowly aspirating 300 μL lysis buffer from the top of the solution for soluble protein purification. Then 500-μL 9M Urea (100 mM NaH2PO4, 10 mM Tris·Cl at pH 8.0) is added to the remaining pellet, followed by mixing and shaking at 1000 rpm in Vortemp shakers for ∼30 min to dissolve the pellet. Resulting solutions were purified using Ni-NTA resin in 96-well filter plates (QIAGEN). Native elution buffer containing 250 mM Imidazole (50 mM NaH2PO4, 300 mM NaCl at pH 8.0) was used to elute soluble proteins from the supernatant plate, and 8 M urea denaturing buffer (100 mM NaH2PO4, 10 mM Tris-Cl at pH 4.5) was used to elute insoluble proteins in the pellet plate. Thus, the soluble and insoluble proteins were analyzed from the same bacterial growth plate.
Solubility profiling uses a fully automated ELISA based on a new profiling concept (C.-H. Luan, S.H. Qui, R.J. Gray, B.J. Finley, and M. Luo, in prep.). The multi-data-set ELISA was analyzed by using in-house software to score solubility and to assist in determining optimal conditions for large-scale expression. The analysis also provides information to assist refolding decisions for those not yielding enough soluble proteins. The method has a success rate of >95% in expression of C. elegans proteins in E. coli as judged by 1-L scale-up expression of the soluble candidates identified in small-scale screen. ELISA, however, does not show whether a protein expressed has the correct molecular size. Therefore, orthogonal methods, such as SDS-PAGE and DNA or protein sequencing were used in a 1-L confirmation stage of the soluble candidates identified in a small-scale screen.
In using the multidata-set ELISA, each bacterial culture plate was separated into four plates for analysis, one for supernatant without purification, one for supernatant with purification, one for pellet without purification, and one for pellet with purification. The corroboration of results across plates reduces the error in detection by ELISA. Each gene plate was expressed at two temperatures, 37°C and 18°C. Thus, each gene was associated with eight ELISA data sets, effectively increasing the accuracy of the solubility profiling.
Because of the concern that the scale up from a 0.6-mL to a 1-L format is not always successful, the soluble candidates from the small-scale screen were expressed in 1-L cultures for confirmation and pilot study of large-scale purification. The 1-L expression was performed using two liter flasks in a temperature-controlled incubation shaker. To accommodate the individuality of each protein while in a high throughput experimental operation, a small number of purification conditions are designed and used to select optimal purification condition for large-scale production.
Scale-Up Protein Expression and Purification After profiling expression level, solubility, and optimal expression conditions for each protein, individual clones were selected for production in six 1-L cultures. The soluble proteins were purified by use of the standard protocols with affinity, ion-exchange, and size exclusion chromatography to obtain homogenous protein preparations. The purified proteins were then concentrated and used in crystallization trials. Insoluble proteins were subject to in vitro refolding, if necessary.
Acknowledgments
We acknowledge funding from NIH (NIGMS 1P50-GM62407), usage of a robotic system purchased by funds from NSF (EPSCOR), and partial support to C.-H. Luan from NASA's cooperative agreement (NCC8-126) to the Center for Biophysical Sciences and Engineering. We thank ResGen for providing a number of the Entry clones during the initial phase of this work.
Footnotes
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2520504.
References
- Adams, M.W.W., Dailey, H.A., DeLucas, L.J., Luo, M., Prestegard, J.H., Rose, J.P., and Wang, B.C. 2003. The Southeast collaboratory for structural genomics: A high-throughput gene to structure factory. Acc. Chem. Res. 36: 191-198. [DOI] [PubMed] [Google Scholar]
- Bairoch, A., Bucher, P., and Hofmann, K. 1997. The PROSITE database, its status in 1997. Nucleic Acids Res. 25: 217-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, I.N., and Bourne, P.E., 2000. The Protein Data Bank. Nucleic Acids Res. 28: 235-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, P. and LaBaer, J. 2003. High throughput protein production for functional proteomics. Trends Biotechnol. 21: 383-388. [DOI] [PubMed] [Google Scholar]
- Braun, P., Hu, Y., Shen, B., Halleck, A., Koundinya, M., Harlow, E., and LaBar, J. 2002. Proteome-scale purification of human proteins from bacteria. Proc. Natl. Acad. Sci. 99: 2654-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, S. 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley, S.K., Almo, S.C., Bonanno, J.B., Capel, M., Chance, M.R., Gaasterland, T., Lin, D., Sali, A., Studier, F.W., and Swaminathan, S. 1999. Structural genomics: Beyond the human genome project. Nat. Genet. 23: 151-157. [DOI] [PubMed] [Google Scholar]
- The C. elegans Sequencing Consortium. 1998. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science 282: 2012-2018. [DOI] [PubMed] [Google Scholar]
- Chambers, S.P. 2002. High-throughput protein expression for the post-genomic era. Drug Discov. Today 7: 759-765. [DOI] [PubMed] [Google Scholar]
- Chance, M.R., Bresnick, A.R., Burley, S.K., Jiang, J.-S., Lima, C.D., Sali, A., Almo, S.C., Bonanno, J.B., Buglino, J.A., Boulton, S., et al. 2002. Structural genomics: A pipeline for providing structures for the biologist. Protein Sci. 11: 723-738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christendat, D., Yee, A., Dharamsi, A., Kluger, Y., Savchenko, A., Cort, J.R., Booth, V., Mackereth, C.D., Saridakis, V., Ekiel, I., et al. 2000. Structural proteomics of an archaeon. Nat. Struct. Biol. 7: 903-909. [DOI] [PubMed] [Google Scholar]
- Ellis, H.M. and Horvitz, H.R. 1986. Genetic control of programmed cell death in the nematode C. elegans. Cell 44: 817-829. [DOI] [PubMed] [Google Scholar]
- Finley, B.J., Qiu, S.H., Luan, C.-H., and Luo, M. 2004. Structural genomics for Caenorhabditis elegans: High throughput protein expression analysis. Protein Expr. Purif. 34: 49-55. [DOI] [PubMed] [Google Scholar]
- Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R.D., and Bairoch, A. 2003. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31: 3784-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley, J.L., Temple, G.F., and Brasch, M.A. 2000. DNA cloning using in vitro site-specific recombination. Genome Res. 10: 1788-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann, U. 2002. Establishing a structural genomics platform: The Berlin-based Protein Structural Factory. Gene Funct. Disease 3: 25-32. [Google Scholar]
- Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E.L.L. 2001. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305: 567-580. [DOI] [PubMed] [Google Scholar]
- Kyte, J. and Doolittle, R.F. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157: 105-132. [DOI] [PubMed] [Google Scholar]
- Lamesch, P., Milstein, S., Hao, T., Rosenberg, J., Li, N., Sequerra, R., Bosak, S., Doucette-Stamm, L., Vandenhaute, J., Hill, D.E., et al. 2004. C. elegans ORFeome version 3.1: Increasing the coverage of ORFeome resources with improved gene predictions. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Lesley, S.A., Kuhn, P., Godzik, A., Deacon, A.M., Mathews, I., Kreusch, A., Spraggon, G., Klock, H.E., McMullan, D., Shin, T., et al. 2002. Structural genomics of the Thermotoga maritime proteome implemented in a high-throughput structure determination pipeline. Proc. Natl. Acad. Sci. 99: 11664-11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, H., Engelbrecht, J., Brunak, S., and von Heijne, G. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10: 1-6. [DOI] [PubMed] [Google Scholar]
- Norvell, J.C. and Zapp-Machalek, A. 2000. Structural genomics programs at the US National Institute of General Medical Sciences. Nat. Struct. Biol. 7: 931. [DOI] [PubMed] [Google Scholar]
- Reboul, J., Vaglio, P., Rual, J.-F., Lamesch, P., Martinez, M., Armstrong, C.M., Li, S., Jacotot, L., Bertin, N., Janky, R., et al. 2003. C. elegans ORFeome version 1.1: Experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat. Genet. 34: 35-41. [DOI] [PubMed] [Google Scholar]
- Shih,Y.-P., Kung, W.-M., Chen, J.-C., Yeh, C.-H., Wang, A. H.-J., and Wang, T.-F. 2002. High-throughput screening of soluble recombinant proteins. Protein Sci. 11: 1714-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, R.C. and Wilson, I.A. 2001. Industrializing structural biology. Science 293: 519-520. [DOI] [PubMed] [Google Scholar]
- Studier, F.W., Rosenberg, A.H., Dunn, J.J., and Dubendorff, J.W. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185: 60-89. [DOI] [PubMed] [Google Scholar]
- Walhout, A.J., Temple, G.F., Brasch, M.A., Hartley, J.L., Lorson, M.A., van den Heuvel, S., and Vidal, M. 2000. GATEWAY recombinational cloning: Application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 328: 575-592. [DOI] [PubMed] [Google Scholar]
- Wood, W.B. 1988. The nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York.
- Zhu, H., Bilgin, M., Bangham, R., Hall, D., Casamayor, A., Bertone, P., Lan, N., Jansen, R., Bidlingmaier, S., Houfek, T., et al. 2001. Global analysis of protein activities using proteome chips. Science 293: 2101-2105. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://sgce.cbse.uab.edu; Structural Genomics of C. elegans.
- http://sgi.com; MineSet decision tree software.
- http://ww.invitrogen.com; Gateway Cloning and Expression Technologies—Invitrogen.
- http://ww1.qiagen.com; Qiagen.
- http://ww1.novagen.com; Novagen.
- http://gce.cbse.uab.edu; SGCE.
- http://www.cbs.dtu.dk/services/SignalP/; Signal Peptides.
- http://www.cbs.dtu.dk/services/TMHMM-2.0/; Tramsmembrane Helices.
- http://ca.expasy.org/tools/protparam.html; EXPASY (for MW, pI, GRAVY).