Abstract
The ability to clone and manipulate DNA segments is central to molecular methods that enable expression, screening, and functional characterization of genes, proteins, and regulatory elements. We previously described the development of a novel technology that utilizes in vitro site-specific recombination to provide a robust and flexible platform for high-throughput cloning and transfer of DNA segments. By using an expanded repertoire of recombination sites with unique specificities, we have extended the technology to enable the high-efficiency in vitro assembly and concerted cloning of multiple DNA segments into a vector backbone in a predefined order, orientation, and reading frame. The efficiency and flexibility of this approach enables collections of functional elements to be generated and mixed in a combinatorial fashion for the parallel assembly of numerous multi-segment constructs. The assembled constructs can be further manipulated by directing exchange of defined segments with alternate DNA segments. In this report, we demonstrate feasibility of the technology and application to the generation of fusion proteins, the linkage of promoters to genes, and the assembly of multiple protein domains. The technology has broad implications for cell and protein engineering, the expression of multidomain proteins, and gene function analysis.
The cloning and manipulation of DNA segments, typically encoding functional elements such as promoters, genes, protein domains, or fusion tags, are central to methods of cell engineering, protein production, and gene-function analysis. The large number of available genome sequences now makes it possible to create and apply repositories of defined functional elements to conduct high-throughput, genome-wide analyses. The Gateway Cloning Technology (Hartley et al. 2000) uses in vitro site-specific recombination to clone and subsequently transfer DNA segments between vector backbones. This approach has been used to generate several large clone collections (Entry Clones), in some cases comprising the entire or nearly entire coding capacity of model genomes as open reading frames (ORFs). These ORFeomes include Caenorhabditis elegans (Walhout et al. 2000b; Reboul et al. 2001, 2003), Pseudomonas aeruginosa (LaBaer et al. 2004), and Saccharomyces cerevisiae (G. Marsischky, pers. comm.), Arabidopsis (Yamada et al. 2003; also see Atome project http://genoplante-info.infobiogen.fr/Databases/CT_Nouveaux_Outils/NO2001054/), human (clones available from several commercial sources), and an incipient collection of Drosophila ORFs (http://www.fruitfly.org/EST/gateway.shtml). A collection of sequenced, full-length Arabidopsis cDNAs in the Gateway Vector pCMV-SPORT6 will shortly be made available through INRA-Genoscope (Castelli et al. 2004). Repositories of full-length clones, some of which are in the Gateway format, are available for Xenopus (http://xgc.nci.nih.gov/), zebrafish (http://zgc.nci.nih.gov/), as well as many human, mouse, and rat genes (http://mgc.nci.nih.gov/). Subsets of these collections have been transferred into various vectors that enable highly parallel functional screens (Walhout et al. 2000a, 2002; Davy et al. 2001; Walhout and Vidal 2001; Boulton et al. 2002; Li et al. 2004) and protein expression/purification (Huang et al. 2003; Kery et al. 2003).
Gateway utilizes engineered site-specific recombination sites derived from bacteriophage λ (Hartley et al. 2000). Four types of sites are involved in two reactions as follows: the BP reaction, attB × attP → attL + attR and the LR reaction, attL × attR → attB + attP. All att sites contain a central 7-bp overlap region, defined by the integrase cleavage site that largely dictates the specificity of the recombination reaction (Landy 1989). Reactions in which unique att sites flanking a DNA of interest (DOI) such as attB1-DOI-attB2, are reacted with a vector that includes compatible sites (attP1-attP2 sites) will result in sequential recombination reactions that transfer the DOI into the selected vector backbone, now flanked by attL sites (attL1-DOI-attL2). Because the recombination events that occur within the 7-bp overlap regions are precise, the reading frame and orientation of the transferred DNA are maintained, providing for amino- and/or carboxy-terminal fusion proteins to be rapidly constructed, and conserving the integrity of the transferred DNA sequence.
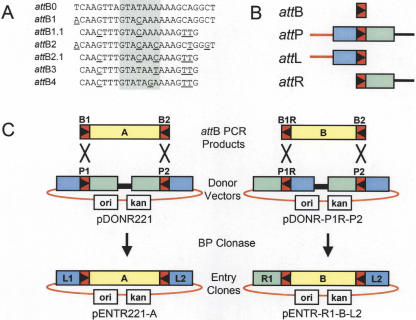
A collection of λ-based site-specific recombination sites (att sites) having unique specificities has been generated (summarized in Fig. 1A). Nucleotide differences within the 7-bp overlap region dictate the site specificity; this region, and the adjacent sequences, affect the efficiency of the recombination reaction. The sites shown were selected and characterized in a variety of experiments to maximize recombination efficiency and minimize cross-reactivity between nonidentical specificities (Cheo et al. 2002). Additional characterization of these sites has been recently published (Sasaki et al. 2004). A collection of compatible recombination sites (attB, attP, attL, and attR) for each att site specificity was constructed for use in this study. Additionally, improvements in the sequence of attB1 and attB2 (attB1.1 and attB2.1, respectively) that reduce their size, and clone with higher efficiency, have been developed (Fig. 1A). Similar changes have been incorporated into attL and attR sites, referred to as attL1.1, attR1.1, attL2.1, and attR2.1 to distinguish them from the standard attL1, attL2, attR1, and attR2 sites.
Figure 1.
Summary of att sites and BP cloning. (A) List of attB sites; attB0 is the E. coli λ bacteriophage attachment site (Landy 1989). Shaded area indicates 7-bp overlap (Int cut sites); underlined bases indicate changes from attB0. (B) Schematic diagram of recombination sites. (Red rectangles) attB sites, the orientation of the asymmetric overlap region is indicated by the triangle. (Blue and green rectangles) attL and attR arms, respectively. (Red line) vector backbone segment; (black line) ccdB counter selection gene (Bernard and Couturier 1992) and chloramphenicol acetyltransferase (cat) selection marker. (C) Examples of BP cloning of attB PCR products into Donor Vectors to generate Entry Clones. Note that reversing the orientations of both the attB1 site in the PCR product and attP1 site in the Donor Vector results in an attR1 site in the Entry Clone.
Current methods for cloning and transferring DNA segments, whether by restriction enzymes and ligase, site-specific recombination, or homologous recombination, are principally applied to single DNA segments. Manipulating multiple DNA segments simultaneously results in significant reductions in cloning efficiencies and an increase in design and execution time. In this report, we describe the extension of the Gateway technology for assembling multiple DNA segments in vitro, in an order, orientation, and reading frame-specific way. By using a collection of recombination sites of unique specificities, we demonstrate the ability to link and clone two and three DNA segments into a vector backbone with high efficiency. The approach has broad application to the engineering of proteins, pathways, and cells, and provides a highly flexible platform for functional analysis.
RESULTS
att Site Orientation Dictates Site Identity and Enables Linkage of DNA Segments
The relationship of the four att sites to each other is depicted in Figure 1B. The orientation of each site relative to vector DNA sequences determines the identity of the product sites in the selected molecules. In standard Gateway reactions, DNA segments flanked by attB1 and attB2 sites are reacted with Donor Vectors containing attP1 and attP2 sites. The attP sites are oriented such that the selected product molecules contain attL1 and attL2 sites (Entry Clones). Reversing the orientations of the attB1 site in the PCR product as well as the attP1 site in the Donor Vector (denoted as attB1R and attP1R, respectively) generates an attR1-attL2 Entry Clone (Fig. 1C). Thus, by dictating the orientation and specificity of the att sites that flank a DNA segment of interest, it is possible to generate Entry Clones with attL or attR sites on either end. Consequently, multiple Entry Clones can be combined in a single in vitro reaction, such that recombination occurs between the ends of two or more Entry Clones, or with sites present in a recipient vector (Destination Vector). In this way, DNA segments can be linked in a predefined order, orientation, and reading frame.
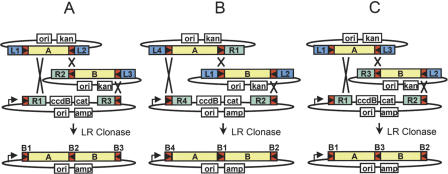
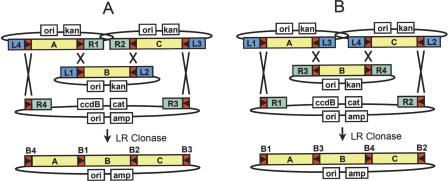
The flexibility of this approach allows several strategies for linking DNA segments to be used. For example, to take advantage of existing L1-ORF-L2 Entry Clone collections (above), new Entry Clones containing, for example, fusion or epitope tags, can be generated to allow linkage at either the 3′ end (an attR2-attL3 Entry Clone; Fig. 2A) or at the 5′ end (an attL4-attR1 Entry Clone; Fig. 2B) of a standard attL1-attL2 Entry Clone. An alternative strategy can be used to maximize compatibility with the large number of attR1-attR2 Destination vectors currently available (Fig. 2C).
Figure 2.
Overview of two-segment cloning. (A) Linkage of a 3′ element Entry Clone (R2-L3) to standard Entry Clones (L1-L2) and cloning into a Destination Vector (R1-R3). (B) Linkage of a 5′ element Entry Clone (L4-R1) to standard Entry Clone and cloning into a Destination Vector (R4-R2). (C) Alternate two-segment cloning strategy linking two Entry Clones (L1-L3 and R3-L2) and cloning into a standard Destination Vector (R1-R2).
Construction of Entry Clones Using Novel Recombination Sites
As an initial assessment of the efficiency, flexibility, and functionality of this approach, both transcription and translational fusions were generated, in which enhanced green fluorescent protein (eGFP; BD Bioscience) was fused through its N terminus, in a two-segment cloning strategy (Fig. 2), to either a yeast promoter element (transcriptional fusions) or a mammalian peptide or protein (translational fusions) reported to direct subcellular localization. In a first step, five yeast promoters including, ADH1, GPD1, TEF2, CUP1 (Funk et al. 2002) and STE2 (Hagen et al.1991) were PCR amplified from yeast genomic DNA using attB-containing promoter-specific oligonucleotides, such that the products were flanked by 5′ attB4 sites and 3′ attB1.1R sites. Reaction of these B4-B1.1R PCR products to pDONR-P4-P1R generated L4-promoter-R1.1 Entry Clones that could subsequently be linked to an L1.1-eGFP-L2.1 Entry Clone and cloned into an R4-R2 Destination Vector (note that for brevity we designate attL1 as L1, attL2 as L2, etc.). Approximately equal molar amounts of attB PCR product and pDONR-P4-P1R were mixed in 1-h BP Clonase reactions. As is typically seen for standard Gateway reactions using B1-B2-flanked PCR products, the B4-yeast promoter-B1.1R PCR products generated large numbers of colonies (Table 1). Essentially all clones examined exhibited the predicted structure as assessed by restriction endonuclease digestion of plasmid DNA (data not shown). The observed efficiencies are comparable to those of standard attB1-attB2 Gateway reactions, and suggest that the novel recombination-site specificities and organization will provide a similarly robust platform.
Table 1.
Summary of BP Reactions
| BP reaction (10 μL) | PCR product (ng) | PCR size (bp) | PCR product (fmol) | KanR (cfu)a | Correct clonesb |
|---|---|---|---|---|---|
| B4-CUP1 promoter-B1.1R × pDONR-P4-P1.1R | 10 | 332 | 45 | 86,420 | 5/5 |
| B4-STE2 promoter-B1.1R × pDONR-P4-P1.1R | 20 | 535 | 56 | 83,520 | 5/5 |
| B4-TEF2 promoter-B1.1 × pDONR-P4-P1.1R | 15 | 653 | 34 | 30,740 | 5/5 |
| B4-GPD1 promoter-B1.1R × pDONR-P4-P1.1R | 20 | 756 | 40 | 35,380 | 4/5 |
| B4-ADH1 promoter-B1.1R × pDONR-P4-P1.1R | 50 | 1535 | 49 | 10,584 | 5/5 |
| No Clonase pDONR-P4-P1.1R | 116 |
Colony forming units per milliliter transformation obtained from transformation of 1 μL BP reaction into 50 μL competent Mach 1 cells (Invitrogen) at 1 × 108 cfu/μg pUC.
Number of correct clones vs. total number of clones analyzed based on restriction enzyme analysis of miniprep DNA. (KanR) Kanamycin resistant.
To assess the feasibility of using a two-segment cloning strategy for constructing translation fusions, a set of Entry Clones was designed that could link to the 5′ (Fig. 2A) or 3′ end (Fig. 2B) of standard (L1-ORF-L2) Entry Clones, and simultaneously clone into a mammalian Destination Vector downstream of a CMV promoter. The linkage reaction is accomplished by simply mixing the two Entry Clone and Destination Vector DNAs in buffer containing LR Clonase (Invitrogen), incubating, and transforming an appropriate recipient host. We used subcellular localization signals fused to eGFP to facilitate functional confirmation of the assembled fusion constructs. These localization tags were as follows: a synthetic nuclear localization signal (NLS; Kalderon et al. 1984; Methods); amino acids 50-100 of human transferrin receptor (reported to promote localization to the plasma membrane [Jing et al. 1990]); a portion of the human mitochondrial protein succinate dehydrogenase complex subunit D (SDHD; Hirawake et al. 1999; Murphy et al. 2000); and the complete human cytoplasmic protein β-arrestin (Barak et al. 1997).
Simultaneous Cloning of Two DNA Segments Into a Vector
Two strategies were used in two-segment cloning of the mammalian localization tag-eGFP fusions. In the first strategy, Entry Clones comprising L1.1-tag-L2.1 and R2.1-eGFP-L3 were combined with an R1.1-R3 Destination Vector (Fig. 2A). The second approach utilized L4-tag-R1.1 and L1.1-eGFP-L2.1 Entry Clones reacted with an R4-R2.1 Destination Vector (Fig. 2B). In this experiment, the Destination Vector was linearized between the attR sites, adjacent to the ccdB gene. This both increases the efficiency of cloning and reduces background (see below). The results of reactions containing each of two supercoiled Entry Clones, a linearized Destination Vector, and LR Clonase are summarized in Table 2. Each reaction generated hundreds to thousands of colonies, which, upon analysis of representative clones by restriction endonuclease digestion, showed the predicted structures for each of four clones at each time point. Hence, both strategies provide high efficiency, rapid, and flexible approaches to simultaneous linking and cloning two DNA segments in an order-, orientation-, and reading frame-specific manner. We observed variable improvement in colony output with increasing incubation time; however, even at the shortest times, the desired clones were readily obtained. Other experiments have shown that colony numbers increase with time, and to maximize success, we typically incubate from 4 h to overnight. Finally, the B1.1-NLS-B2.1 PCR product failed to clone into pDONR221, and colony outputs for the L4-NLS-L1.1 reaction (Table 2) are unusually low. The low cloning efficiency of this small insert (27 bp) could reflect that a minimum distance is required between att sites for efficient recombination.
Table 2.
Summary of LR Reactions of Two-Segment Cloning
| LR reaction (10 μL) | DNA topology | Input DNA (ng) | Insert size (bp) | DNA size (bp) | fmoles | AmpR cfua 2 h | AmpR cfu 6 h | AmpR cfu 24 h |
|---|---|---|---|---|---|---|---|---|
| L4-NLS-R1.1 | ccc | 40 | 27 | 2673 | 23 | 44 (4/4)b | 106 (4/4) | 97 (4/4) |
| L1.1-eGFP-L2.1 | ccc | 40 | 719 | 3261 | 19 | |||
| pDESTCMV42 (R4-R2.1) | linear | 134 | 7302 | 27 | ||||
| L4-Transferrin-R1.1 | ccc | 30 | 155 | 2802 | 16 | 125 (4/4) | 900 (4/4) | 1675 (4/4) |
| L1.1-eGFP-L2.1 | ccc | 40 | 719 | 3261 | 19 | |||
| pDESTCMV42 | linear | 134 | 7302 | 27 | ||||
| L1.1-Transferrin-L2.1 | ccc | 17 | 155 | 2697 | 9 | 2588 (4/4) | 2425 (4/4) | 2750 (4/4) |
| R2.1-no ATG eGFP-L3 | ccc | 67 | 713 | 3356 | 31 | |||
| pDESTCMV13 (R1.1-R3) | linear | 134 | 7302 | 27 | ||||
| L4-SDHD-R1.1 | ccc | 40 | 454 | 3001 | 20 | 462 (4/4) | 438 (4/4) | 625 (4/4) |
| L1.1-eGFP-L2.1 | ccc | 40 | 719 | 3261 | 19 | |||
| pDESTCMV42 (R4-R2.1) | linear | 134 | 7302 | 27 | ||||
| L1.1-SDHD-L2.1 | ccc | 100 | 454 | 2896 | 52 | 925 (4/4) | 675 (4/4) | 862 (4/4) |
| R2.1-no ATG eGFP-L3 | ccc | 67 | 713 | 3356 | 31 | |||
| pDESTCMV13 (R1.1-R3) | linear | 134 | 7302 | 27 | ||||
| L1.1-β-arrestin-L2.1 | ccc | 100 | 1229 | 3765 | 40 | 2667 (4/4) | 6222 (4/4) | 7111 (4/4) |
| R2.1-no ATG eGFP-L3 | ccc | 100 | 713 | 3356 | 45 | |||
| pDESTCMV13 (R1.1-R3) | linear | 200 | 7302 | 42 | ||||
| No Clonase | ||||||||
| L4-NLS-R1.1 | ccc | 40 | 27 | 2673 | 23 | 12 | ||
| L1.1-eGFP-L2.1 | ccc | 40 | 719 | 3261 | 19 | |||
| pDESTCMV42 (R4-R2.1) | linear | 134 | 7302 | 27 | ||||
| No Clonase | ||||||||
| L1.1-β-arrestin-L2.1 | ccc | 100 | 1229 | 3765 | 40 | 0 | ||
| R2.1-no ATG eGFP-L3 | ccc | 100 | 713 | 3356 | 45 | |||
| pDESTCMV13 (R1.1-R3) | linear | 200 | 7302 | 42 |
(ccc) Supercoiled, (Amp) ampicillin resistant.
Colony forming units per milliliter transformation obtained from transformation of 1 μL BP reaction into 50 μL competent DH5α cells (Invitrogen) at 1 × 108 cfu/μg pUC.
Number of correct clones vs. total number of clones analyzed based on restriction enzyme analysis of miniprep DNA. Cotransforming Entry Clone DNA was observed in 6 of 48 samples.
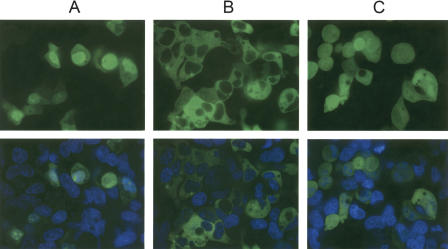
To examine whether the assembled fusion proteins were functional, HEK293 cells were transfected with three representative Expression Clones as follows: CMV promoter-B4-NLS-B1.1-eGFP-B2.1-poly adenylation signal; CMV promoter-B1.1-β-arrestin-B2.1-eGFP-B3-poly(A); and CMV promoter-B4-Transferrin receptor-B1.1-eGFP-B2.1-poly(A). As a control, we also constructed an eGFP Expression Clone [CMV promoter-B1.1-eGFP-B2.1-poly(A)]. Representative images of transfected cells stained with the nuclear dye Hoechst 33342 are shown in Figure 3. In contrast to the general whole-cell fluorescence observed with eGFP alone, the NLS-eGFP primarily localized in the nucleus, whereas the β-arrestin-eGFP protein was exclusively cytoplasmic, as predicted from published reports (Kalderon et al. 1984 and Barak et al. 1997, respectively). Additionally, CHOK1 cells were transfected with these DNAs and showed similar localization patterns (data not shown). Hence, the assembled DNA segments generated fusion proteins with the expected function. Cells transfected with the transferrin receptor-eGFP construct, although appearing to show membrane localization, rapidly rounded up, appeared unhealthy, and, therefore, were not imaged.
Figure 3.
Fluorescent, confocal, live-cell imaging of HEK-293 cells transfected with (A) NLS-eGFP, (B) β-Arrestin-eGFP fusions, or (C) untagged eGFP. Forty-eight hours post-transfection, cells were stained with Hoechst 33342 and imaged confocally at 60× magnification on a Pathway HT system (Atto Bioscience). Pseudo-colored images of a typical Z-stack section are shown. (Top) GFP channel alone; (bottom) Hoechst + GFP.
Two valuable characteristics of Gateway-cloned PCR products are worth noting; (1) reading frames, once established by primer design, always maintain register in recombination reactions, and (2) all Gateway clones and vectors of a particular site specificity, if established in the standard reading frame register, are compatible with each other (for a summary of translation products across the att sites, see Supplemental Fig. 1). Thus, the Entry Clones constructed to target proteins to particular subcellular locations can be rapidly recombined with any other set of compatible Entry Clones (ORFs) to yield constructs that are immediately useful.
Assembly of Promoter-ORF Transcriptional Fusions Using Two-Segment Cloning
In addition to the generation of fusion proteins, promoters and genes can be cloned simultaneously into a vector to profile expression patterns (Dupuy et al. 2004; Hope et al. 2004), assess promoter strength, or to optimize the level of expressed proteins. As part of a program to characterize members of the superfamily of seven-transmembrane G protein-coupled receptors (GPCRs), we have used this approach to modify and express several model GPCRs in yeast (S. cerevisiae). The STE2 gene encodes the yeast α-factor pheromone GPCR. Binding of α-factor to the Ste2 receptor induces activation of the heterotrimeric G protein, which, in turn, activates a MAP Kinase signal-transduction pathway that results in growth arrest and induction of genes in preparation for mating (Bardwell et al. 1994).
The five yeast promoter Entry Clones (ADH1, GPD1, TEF2, CUP1, and STE2) described above were used in two separate two-segment cloning reactions. First, to construct promoter-eGFP Expression Clones, each L4-promoter-R1.1 Entry Clone was reacted with an L1.1-eGFP-L2.1 and an R4-R2.1 Destination Vector. Second, to construct promoter-STE2 Expression Clones, each yeast promoter Entry Clone was reacted with an L1.1-STE2-L2.1 Entry Clone and an R4-R2.1 Destination Vector. The same Destination Vector, pDEST425.42, a LEU2, 2-micron-based vector derived from p425GAL1 (Christianson et al. 1992; Funk et al. 2002) was used in each reaction. Once again, these two-segment cloning reactions were highly efficient (data not shown).
To demonstrate functionality of the linked transcription fusions, and to estimate the level at which each of the promoters functioned, we introduced the promoter-eGFP constructs into yeast strain MBY1272 (MATa his3Δ200 leu2Δ0 met15Δ0 trp1Δ63 ura3Δ0 far1::ura3Δ sst2::ura3). Studies of GPCRs in yeast are typically conducted in a far1, sst2 background (Dowell and Brown 2002). Deletion of FAR1, a cyclin-dependent kinase inhibitor, allows growth in the presence of the yeast pheromone α-factor. Deletion of SST2, a member of the RGS GTPase-activating factors that return Gα to it's inactive state, has been shown to increase sensitivity to agonists.
Populations of freshly transformed yeast colonies were assayed for GFP fluorescence in the presence and absence of α-factor over a 16-h period. In the absence of α-factor, each culture generated increasing fluorescence over time (Fig. 4A). The levels of fluorescence observed paralleled the relative promoter strengths reported previously by others (Pillai et al. 2001; Funk et al. 2002). In the presence of α-factor, levels of fluorescence were generally lower for all promoters, with the exception of the STE2 promoter, which was modestly induced and demonstrated higher levels of GFP fluorescence than all of the other promoter fusions. Hence, the two-segment cloning approach can be used to efficiently link and clone, in parallel, collections of promoters to ORFS for functional evaluation.
Figure 4.
Functional analysis of transcriptional fusions. (A) Promoter-eGFP fusions and (B) promoter-STE2 receptor fusions in yeast cells co-transformed with the FUS1-yeGFP reporter. Relative GFP fluorescence was measured over a 16-h period. Yeast strains were assayed in the presence (filled symbols) or absence (open symbols) of 500 nM α-factor as described in the Methods. Promoter fusions are as follows: STE2 (Red), ADH1 (yellow), GPD1 (blue), TEF2 (green), and CUP1 (black). Yeast strains containing the CUP1 promoter fusions were assayed in the presence (black circles) and absence (black squares) of 100 μM copper sulfate.
The promoter of the yeast pheromone-inducible FUS1 gene, when fused to a reporter gene, has been shown to function as a response indicator of α-factor binding to Ste2 receptor (Hagen et al. 1991). Using a two-segment cloning strategy, we linked the FUS1 promoter to yeGFP (an optimized eGFP, in which the first 15 codons were modified to conform to yeast codon bias, see primer sequences in Supplemental Table 1) in a TRP1, 2-micron-based Destination Vector (pDEST424.42). Using the FUS1-yeGFP reporter, and the yeast promoter-STE2 Expression Clones, we attempted to optimize functional Ste2 receptor expression levels. Yeast strain MBY1273 (MBY1272 ste2) was cotransformed with each of the promoter-STE2 Expression Clones and the FUS1-yeGFP reporter construct. Populations of freshly transformed yeast cells were assayed for GFP fluorescence in the presence and absence of α-factor. Cultures to which α-factor had been added showed significant increases in fluorescence (3×-6×) relative to those lacking α-factor (Fig. 4B). In contrast to the promoter-eGFP results, where the Ste2 promoter showed highest expression levels, functional activity of Ste2 receptor was highest when expressed from the ADH1 promoter, although all promoters provided receptor activities within a twofold range of one another. The results demonstrate the capability and utility of applying two-segment cloning to protein expression optimization, and the characterization of promoters and ORFs.
Defined Replacement of att-Flanked Segments Within Multi-Segment Constructs
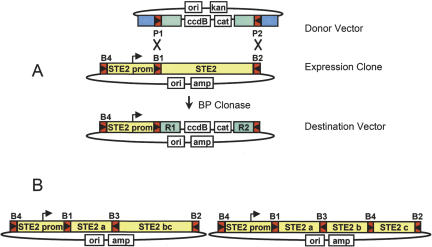
A unique advantage of Gateway multi-site technology is that once assembled, it is possible to access and replace one or more segments of a construct flanked by unique recombination sites. On the basis of indications from the promoter-eGFP data (Fig. 4A), we decided to use the STE2 promoter for expression and functional analysis of various forms of the Ste2 receptor. The STE2 Expression Clone construct had the structure B4-STE2 promoter-B1.1-STE2-B2.1 in the pEXP425 vector backbone (ampicillin resistance). By replacing the STE2 ORF with a counter-selection cassette encoding ccdB, we could rapidly convert this Expression Clone to a Destination Vector. This would enable the subsequent insertion of one, two, three, or higher order constructs downstream of the promoter, and demonstrate the concept of segment exchange. For this, we simply reacted the Expression Clone with pDONR221 (P1-P2) in a BP reaction as schemed in Figure 6A, below. Plating Escherichia coli transformants on ampicillin and chloramphenicol selected the desired Destination Vector product with the structure B4-STE2 promoter-R1-cat-ccdB-R2. Because ccdB inhibits growth of most standard E. coli cells (Bernard and Couturier et al. 1992), we used the ccdB-resistant strain DB3.1 (Invitrogen). As a consequence, unreacted pDONR221 (kanamycin + chloramphenicol resistant) can cotransform cells also containing the desired product. To minimize this possibility, the Expression Clone molecule was used in a 2:1 molar excess. Cloning efficiencies using various topologies of the two molecules were examined, and the results are summarized in Table 3. Thousands of colonies are obtained when both molecules are supercoiled; however, the background (no clonase) is similarly high, and only 1/6 of the clones examined (restriction endonuclease digestion) had the desired structure. Reduced recombination efficiencies have been seen previously in standard Gateway B × P reactions when both molecules are supercoiled (D. Cheo, unpubl.). Linearizing either or both molecules substantially reduced background colonies and increased the percentage of desired clones. The ability to rapidly replace existing segments of an assembled construct with other single or multi-segment Entry Clones (below) substantially expands the flexibility and potential for generating high-order, multi-segment constructs.
Figure 6.
STE2 Expression Constructs (A) Reaction of the B1-B2 sites in a multi-segment Expression Clone with a Donor Vector (P1-P2) to generate a STE2 promoter Destination Vector. (B) Diagram of STE2 receptor constructs resulting from 2 and 3 segment cloning into the Destination Vector shown in (A).
Table 3.
Defined Replacement of att-Flanked Segments With a BP Reaction
| BP reaction (10μL) | DNA topology | Input DNA (ng) | DNA size (bp) | fmoles | AmpR CmR cfu 20 ha |
|---|---|---|---|---|---|
| pEXP425-[B4-STE2 promoter-B1.1-STE2-B2.1] | ccc | 300 | 8925 | 50 | 3206 |
| pDONR221 (P1-ccdB CmR-P2) | ccc | 75 | 4759 | 24 | (1/6)b |
| No Clonase | 928 | ||||
| pEXP425-[B4-STE2 promoter-B1.1-STE2-B2.1] | ccc | 300 | 8925 | 50 | 55 |
| pDONR221 (P1-ccdB CmR-P2) | linear | 75 | 4759 | 24 | (4/6) |
| No Clonase | 0 | ||||
| pEXP425-[B4-STE2 promoter-B1.1-STE2-B2.1] | linear | 300 | 8925 | 50 | 562 |
| pDONR221 (P1-ccdB CmR-P2) | ccc | 75 | 4759 | 24 | (5/6) |
| No Clonase | 8 | ||||
| pEXP425-[B4-STE2 promoter-B1.1-STE2-B2.1] | linear | 300 | 8925 | 50 | 94 |
| pDONR221 (P1-ccdB CmR-P2) | linear | 75 | 4759 | 24 | (6/6) |
| No Clonase | 0 |
Colony forming units per milliliter transformation obtained from transformation of 1 μL reaction into 50 μL competent DB3.1 cells at 1 × 108 cfu/μg pUC.
Number of correct clones vs. total number of clones analyzed based on restriction enzyme analysis of miniprep DNA. (ccc) Supercoiled; (AmpR) ampicillin resistant; (CmR) chloramphenicol resistant.
Assembling and Cloning Three DNA Segments
Introducing an additional recombination-site specificity to link and clone three DNA segments in a predefined order, orientation, and reading frame register can further extend the multi-site strategies thus far described. As seen above, any site specificity can be used at any position, typically dictated by the sites present in pre-existing ORF or Destination Vector collections (we have not yet exhaustively characterized the sites for effects of the 8 amino acid translation products on expression levels or function. Properties of particular site specificities may also influence site placement decisions). The three-segment assembly scheme is depicted in Figure 5. In this case, three Entry Clone DNAs are constructed, such that the sites flanking the centrally positioned DNA segment recombine with the internal sites of the other two Entry Clones. As with two-segment cloning, the outermost sites are configured to recombine with sites on the Destination Vector. The schemes shown could utilize existing ORFeome collections (L1-ORF-L2; Fig. 5A) or existing Destination Vectors (R1-ccdB-R2; Fig. 5B).
Figure 5.
Overview of three-segment cloning. (A) Linkage of a 5′ Entry Clone, a standard (L1-L2) Entry Clone, and a 3′ Entry Clone and a Destination Vector (R4-R3). (B) Alternate three-segment cloning strategy using a standard Gateway Destination Vector (R1-R2).
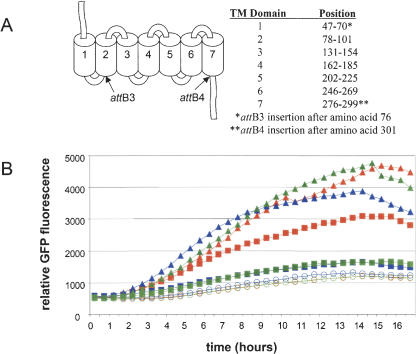
As part of an effort to understand further the determinants defining 7-transmembrane receptor-agonist interactions, we have dissected the Ste2 receptor and several mammalian receptors (to be reported elsewhere) to allow rapid combinatorial assembly of putative agonist-binding domains, following from work described previously for the study of olfactory receptors (Krautwurst et al. 1998). As a first step, we identified locations, on the basis of protein structural predictions (Parrish et al. 2002), within the Ste2 receptor, which would potentially accommodate insertion of the att-site translation products, and divided the receptor into three parts (Ste2a, Ste2b, and Ste2c, according to the scheme depicted in Fig. 5B for DNA segments A, B, and C, respectively). Three positions surrounding the first cytoplasmic domain were chosen as a junction between Ste2a and Ste2b (corresponding to amino acids 76, 81, and 95; Fig. 7A, below). A single position (amino acid 301) near the predicted seventh transmembrane/cytoplasmic transition served as the junction between Ste2b and Ste2c. STE2-specific primers, appended with the appropriate attB sequences, were used to generate three PCR-products for the STE2a segment, from which three L1.1-STE2a-L3 Entry Clones were constructed (essentially as described above). Similarly three R3-STE2b-R4 Entry Clones, one L4-STE2c-L2.1 Entry Clone, and an Entry Clone containing an intact STE2bc segment (R3-STE2bc-L2.1) were generated. These Entry Clones were used in two- and three-segment cloning reactions with the B4-STE2 promoter-R1-R2 Destination Vector (Fig. 6A). As seen in Table 4, both two- and three-segment cloning reactions using Entry Clones with the STE2a/b junction at amino acid 76 generated similar numbers of colonies. Expression Clones isolated from these reactions (Fig. 6B) displayed predicted restriction patterns in 10 of 10 and 8 of 10 samples, respectively. Ste2 receptors containing att-site insertions at amino acids' positions 81 and 95 (Fig. 7) were ultimately found to be nonfunctional in assembled STE2 Expression Clones. Because each of the STE2 Expression Clone constructs generated from two- and three-segment cloning reactions were sequence validated, we speculate that the insertion of attB3 at positions 81 and 95 disrupts transmembrane domain II. Consequently, the studies below focused only on the Ste2a/Ste2b junction at amino acid 76.
Figure 7.
Functional analysis of STE2 receptor constructs (A) Ste2 transmembrane topology (Parrish et al. 2002) and att site locations. (B) Response to α-factor treatment of yeast strains containing STE2 receptor constructs cotransformed with the FUS1-yeGFP reporter. The STE2 receptor constructs were expressed using the STE2 promoter. Relative GFP fluorescence was measured over a 16-h period. Yeast strains were assayed in the presence of 500 nM (triangles), 50 nM (squares), or absence (circles) of α-factor as described in the Methods. Symbols are as follows: wild-type Ste2 receptor (red), Ste2 receptor containing att3 (blue), and Ste2 receptor containing both attB3 and attB4 (green).
Table 4.
Summary of LR Reactions of Two- and Three-Segment Cloning
| LR reaction (10 μL) | DNA topology | Input DNA (ng) | DNA size (bp) | fmoles | ampR cfu 5 ha | ampR cfu 20 ha |
|---|---|---|---|---|---|---|
| L1.1-STE2a-L3 | linear | 50 | 2768 | 27 | ||
| R3-STE2bc-L2.1 | linear | 50 | 3713 | 20 | 71 | 238 (10/10)b |
| pDEST425-B4-STE2 promoter | linear | 150 | 9844 | 23 | ||
| R1.1-ccdB-R2.1 | ||||||
| L1.1-STE2a-R3 | linear | 50 | 2768 | 27 | ||
| L3-STE2b-L4 | linear | 50 | 3340 | 22 | 149 | 330 (8/10) |
| R4-STE2c-L2.1 | linear | 50 | 2930 | 26 | ||
| pDEST425-B4-STE2 promoter | linear | 150 | 9844 | 23 | ||
| R1.1-ccdB-R2.1 | ||||||
| No Clonase | 3 |
Colony forming units per milliliter transformation obtained from transformation of 1 μL reaction into 50 μL DH5α competent cells at 1 × 108 cfu/μg pUC.
Number of correct clones vs. total number of clones analyzed based on restriction enzyme analysis of miniprep DNA.
Functional Analysis of Multi-Segment STE2 Receptors To determine whether the assembled STE2 was functional, three Expression Clone constructs consisting of pEXP425-STE2 promoter-STE2, pEXP425-STE2 promoter-STE2a-STE2bc, and pEXP425-STE2 promoter-STE2a-STE2b-STE2c, were cotransformed with the pEXP424-FUS1-yeGFP reporter into yeast strain MBY1273. Populations of freshly transformed yeast cells were assayed for GFP fluorescence as before, in the presence and absence of α-factor (Fig. 7B). At the higher concentration of α-factor (500 nM), all three receptors were observed to respond to approximately similar levels, validating that the assembled Ste2 receptors maintained all DNA segments in the proper reading frame, orientation, and order, and that translation across the novel att sites generated a functional fusion protein with kinetic and dynamic properties similar to that of the wild-type intact protein. At lower α-factor concentrations (50 nM), both attB-containing receptors showed reduced levels of response relative to an intact Ste2 receptor. As removal of attB4 failed to improve the response, either the position and/or presence of attB3 appears to be having an observable effect under certain conditions. Additional experiments are in progress to further assess these possibilities.
DISCUSSION
Using an expanded set of site specificities that mediate λ-based site-specific recombination (Gateway Cloning), we have demonstrated the concerted assembly and cloning of multiple DNA segments into a plasmid vector backbone. Because the site-specific recombination mechanism is precise, each DNA segment of the multiple segment construct is linked and cloned in a predefined order, orientation, and translational reading frame. By dictating the orientation of the attB sites that flank a DNA of interest, as well as the orientation of attP sites within Donor Vectors, Entry Clones can be generated in BP reactions such that attL or attR sites can be appended at either end of the cloned insert. The proper arrangement of flanking attL and attR sites in a set of multiple Entry Clones enables their linkage and cloning into a Destination Vector in a highly efficient LR reaction.
Novel att-site specificities were identified by examining all possible single-base changes within the 7-bp overlap of the attL and attR core regions (Cheo et al. 2002). Alterations within the first three positions of the seven-base overlap created new specificities that did not appreciably cross-react with noncognate partner sites. With four base possibilities at each of three positions, at least 64 different specificities are possible. We demonstrate here the use of three and four att sites in the cloning of two and three DNA segments. Thus far, we have successfully used six unique att-site specificities in a single LR reaction to mediate the cloning of five DNA segments into a Destination Vector (D.L. Cheo, in prep.).
Nucleotide sequence changes within attB sites (outside of the seven-base overlap) that increased cloning efficiencies in BP reactions with cognate attP sites were also identified (Fig. 1). Nucleotides at the ends of the attB site (e.g., in the attB1 and attB2 site) were found to be unnecessary and were eliminated to effectively shorten the attB sequence to 21 bp. These changes were incorporated into each of the attB sites used in this study (attB1.1, attB2.1, attB3, and attB4). Because each of the site specificities are interchangeable with respect to the position within the assembled construct, and as the site can accommodate numerous other base changes in flanking regions, substantial flexibility exists for dictating the sequence of the sites and their resulting translation products to minimize or impart particular functional attributes.
To maintain the previously defined Gateway reading frame through attB1 and attB2 (to maximize clone compatibility), we have added two bases at the 3′ end of every upstream (left) attB primer, and one base at the 3′ end of every downstream (right) attB primer. We have defined these bases as GA and C, which encode glycine and alanine, respectively, when translated (see Supplemental Fig. 1B). These sequences also serve to define the left and right ends of every insert within clones and PCR products. We have taken advantage of these sequence tags to develop an attB conversion PCR methodology. With a complete set of left and right attB primers, we can successfully create new Entry Clones of any type or site specificity from any att-site-flanked DNA segment using a simple 5-cycle PCR reaction, followed by BP cloning.
Whereas the reactions are generally quite robust, several parameters can affect the efficiency with which the desired multi-segment constructs are obtained in an LR reaction. These include the topology of the reacting molecules, molar ratios of the input DNAs, and incubation time. Regarding topology, the use of supercoiled Entry Clones eliminates the need to identify and digest restriction sites absent from the insert, as well as the need to inactivate residual restriction enzyme activity prior to the LR reaction. Linearization of the Destination Vector reduces cotransformation by unreacted Destination Vector when selecting Expression Clone products (cotransformation can reduce colony output due to growth inhibition by ccdB). Cloning efficiencies are typically lower if all supercoiled molecules are used. We find the reactions to be fairly tolerant to relative differences (three- to fourfold) in molar ratios of the molecules, and gel-based intensity estimates are usually acceptable. Exceeding 200 fmole of total DNA/10 μL reaction can negatively affect reaction efficiencies. In contrast to standard Gateway reactions, which often generate substantial amounts of desired products within 30 min to 1 h, multi-segment assemblies have the highest probabilities of success after reaction times of 2 h or longer. Finally, we have not yet characterized the effect of insert size or the upward size limitations on the assembly of DNA segments by these methods. We routinely use two- and three-segment cloning strategies for segments ranging from 3 to 6 Kb into Destination Vectors of from 6 to 8 Kb, generating Expression Clones of from 13 to 18 Kb with success rates similar to those reported here.
Once assembled, a multi-segment construct can be further modified using Donor Vectors having cognate attP sites in a BP reaction. With the appropriate selections, novel multi-segment Entry Clones and Destination Vectors can be isolated from the same BP reaction. In this study, we have applied this approach to the conversion of a two-segment Expression Clone construct (B4-promoter-B1.1-ORF-B2.1) by replacing the ORF segment with the ccdB cat cassette from pDONR221 to generate a B4-promoter-R1-ccdB-R2 Destination Vector (Fig. 6A). Subsequent use of this Destination Vector in a three-segment cloning generated an Expression Clone containing four segments. This approach further increases the flexibility with which larger multi-segment assemblies can be constructed and manipulated.
Numerous strategies can be used for the design of Entry Clone collections that can facilitate gene function analysis, protein expression, protein purification, genetic screens (Deplancke et al. 2004), and the like. For a given strategy (e.g., Figs. 2, 5), collections of Entry Clones can be generated, sequence validated, and used in a combinatorial way to rapidly generate (typically in 2 or 3 d) customized multi-segment Expression Clone constructs. We have validated this approach here for linking collections of promoters to collections of ORFs, and for the optimization of protein expression and promoter characterization. We have also demonstrated here the use of multiple attB insertions to dissect proteins for functional analysis or assembly of protein domains (using a 7-transmembrane G-protein-coupled receptor as a model). Whereas the assemblies described here were cloned into a single landing site in a vector backbone, this need not be so. For example, we have generated Entry Clones comprised of individual elements of plasmid vectors (i.e., selectable markers, promoters, origins of replication, protein fusion tags, etc.; Cheo et al. 2002). Combining selected Entry Clones in a single LR reaction generated custom-designed vector systems, in which the assembled segments comprised a replicon with various desirable features. A similar approach could be extended to assemble segments of adenovirus, baculovirus, artificial chromosomes, and small genomes. Numerous other application areas can be easily envisaged, including the expression of multi-subunit enzymes, the generation of siRNA and gene-targeting constructs, the assembly of multi-gene biochemical pathways, and the construction of highly complex combinatorial libraries. The system is potentially amenable to high-throughput laboratory automation methodologies and will likely provide a suitable platform for complex cell and protein-engineering challenges.
METHODS
PCR
All PCR reactions were prepared using 20 pmoles of each primer (see Supplemental Table 1 for all primer sequences) and Platinum Taq DNA Polymerase Hiqh Fidelity (Invitrogen) in 50-μL volumes. Yeast promoters and STE2 ORF were amplified from yeast genomic DNA (Invitrogen). The β-arrestin, SDHD, and TFRC mammalian localization tags were PCR amplified using a human leukocyte cDNA library (Invitrogen) as template. PCR products encoding N-terminal fusion tags lacked in-frame stop codons. The NLS tag was amplified using annealed and extended primers as template. Full-length attB gene-specific primers were used to amplify β-arrestin. Partial (15 bp) attB gene-specific primers were used in primary PCR amplifications of SDHD, TFRC, and NLS. For amplification of full-length attB PCR products of SDHD, TFRC, and NLS, one-tenth (5 μL of 50 μL) of the primary PCR reactions were used as template in 5-cycle secondary PCR reactions containing the appropriate attB adapter primers. Using this adapter attB PCR approach, partial attB PCR products were converted to full attB PCR products. In addition, for SDHD and TFRC, partial attB sites were simultaneously converted to full attB sites with alternative orientation and specificity.
BP Clonase Reactions
BP reactions were prepared in 10 μL using 1-5 μL of unpurified PCR product brought to 5 μL with TE and containing 1 μL of the appropriate Donor Vector at 150 ng/μL (50 femtomoles), 2 μL of 5× BP reaction buffer and 2 μL of BP Clonase (Invitrogen). The reactions were incubated at room temperature for 1-4 h, and 1 μL (without a proteinase K treatment) was used to transform 50 μL of chemically competent Mach 1 E. coli cells (Invitrogen). Kanamycin-resistant colonies were selected on LB agar containing 50 μg/mL kanamycin at 37°C. Total colony outputs per milliliter were normalized to 1 × 108 CFU/μg of pUC19.
Destination Vector Construction
The attR reading frame cassettes for the mammalian CMV Destination Vectors (pDESTCMV12, pDESTCMV13, and pDESTCMV42) were prepared by PCR amplification of a standard Gateway reading frame cassette using SacII- and XbaI-containing attR-specific oligos (see Supplemental Table 1 for primer sequences). PCR products were digested with SacII and XbaI and ligated between SacII-XbaI sites of pQBI-B23sgBFP (Quantum Biotechnologies). The attR reading frame cassettes of the yeast Destination Vectors pDEST424.42 and pDEST425.42 were prepared by PCR amplification of a standard Gateway reading frame cassette with SacI and XhoI containing attR-specific oligos. The PCR product was digested with SacI and XhoI and ligated between the SacI-XhoI sites of p424GAL1 and p425GAL1 (Funk et al. 2002).
Multi-Segment LR Reactions
For two-segment cloning of mammalian localization-eGFP protein fusions, two supercoiled Entry Clones and a linear Destination Vector were combined in a 30-μL reaction containing 6 μL of 5× LR reaction buffer and 6 μL of LR Clonase. Entry Clone DNA concentrations were estimated from Ethidium Bromide-stained agarose gels containing known amounts of supercoiled pBR322. Reactions were incubated for 2, 6, and 24 h at room temperature. A total of 10 μL aliquots were removed at each time point and treated with 1 μL of proteinase K (2 mg/mL) for 10 min at 37°C. A 1-μL sample of each reaction was introduced by transformation into chemically competent DH5α E. coli (Invitrogen), and ampicillin-resistant colonies were selected on LB agar containing 100 μg/mL ampicillin. For all other multi-segment cloning reactions, equal amounts of each DNA were added to reactions, totaling ∼100 femtomoles per 10 μL of reaction. The two- and three-segment STE2 Expression Clones were generated by combining approximately equal molar ratios of the Entry Clones and Destination Vector (see Fig. 6A,B) incubated with LR Clonase (5 or 20 h at room temperature), after which 2 μL (of 20 μL) were introduced into E. coli DH5α cells by transformation.
Live-Cell Confocal Fluorescent Imaging of Mammalian Cells
A 96-well special optics coverglass plate (Nunc catalog no. 164588) was treated with 50 μL of a 1% Matrigel solution (BD Bioscience catalog no. 354234) in serum-free Optimem DMEM (Invitrogen) for 30 min at 37°C. The matrigel was removed, and 293H cells were seeded at a density of 5 × 104 per well. The cells were allowed to attach to the plate at 37°C in 5% CO2 for 3 h. Each well of cells was transfected with 0.9 μL of Lipofectamine 2000 and 200 ng plasmid DNA as per the manufacturer's instructions (Invitrogen). After 24 h, cells were labeled with 1 μg/mL Hoechst 33342 (Molecular Probes) in DMEM medium at 37°C, 5% CO2 for 15 min. Cells were washed twice in HBSS and imaged on a Pathway HT (Atto Bioscience) using an Olympus Plan-Apo 60× oil objective having a 150-μM working distance and a 1.40 numerical aperture. Z-stacks were obtained using AttoVision software on the Pathway HT (Atto Bioscience).
Yeast Strain Construction
A parental yeast strain BY1126 (MATa his3Δ200 leu2Δ0 met15Δ0 trp1Δ63 ura3Δ0; Brachmann et al. 1998) was used to successively knock-out the FAR1 and SST2 loci using URA3-containing PCR products designed with 40-bp regions of homology (Baudin et al. 1993) immediately upstream and downstream of the FAR1 and SST2 ORFs. Competent BY1126 cells were transformed with 200 ng of FAR1-oligo-flanked URA3 PCR product, and Ura+ transformants were plated on SD-Ura selective medium. Resulting colonies were checked phenotypically for failure to growth arrest in the presence of α-factor, and verified by PCR analysis. The far1::URA3 strain was plated on 0.2% 5-fluoroorotic acid (5FOA) to select for isolates in which URA3 had excised. The resulting strain, BY1126 far1Δura3 was transformed with 200 ng of PCR product encoding SST2-flanked URA3, and colonies were again selected on SD-Ura plates. Following PCR validation, this strain (BY1126 far1Δ sst2::URA3 = MBY1272) was transformed with 200 ng of ste2::G418 PCR product (generated using the ste2::G418 knockout strain BY5645 [Open Biosystems] as template DNA [Wach et al. 1994; Giaever et al. 2002]) and colonies were selected on 400 μg/mL G418 YPAD medium. The resulting strain was confirmed by PCR and failure to respond to α-factor, and designated MBY1273.
Functional Assays in Yeast Cells
Expression Clones encoding the yeast promoter-eGFP transcriptional fusions were introduced by transformations into yeast strain MBY1272. Competent yeast cells (10 μL) were transformed with 1 μL of each Expression Clone miniprep DNA (∼100 ng) using the EZ Yeast Transformation Kit (Zymo Research). The entire transformation mixture was plated on synthetic-defined dropout medium containing glucose, but lacking leucine (SD-Leu dropout, Q-Bio Gene) agar, and incubated at 30°C for 2-3 d. Yeast strain MBY1273 (15 μL competent cells) was cotransformed with 1 μL of each of the yeast promoter-STE2 constructs along with 1 μL of the FUS1-yeGFP Expression Clone and plated on SD-leu-trp dropout agar. Populations of freshly transformed yeast colonies representative of the entire transformation mixture were suspended in 2 mL of the appropriate dropout liquid medium (above) and adjusted to OD 0.8 with medium. Equal volumes (190 μL) of normalized culture were placed in a 96-well plate in the presence or absence of varying concentrations of α-factor (10 μL diluted in water) and assayed for fluorescence (485 nm excitation/520 nm emission) over a 16-h period at 28°C. Yeast strains harboring constructs with the CUP1 promoter were also treated with 100 μM CuSO4). Measurements of GFP fluorescence were made on a Fluostar Optima plate reader (BMG Labtechnologies).
Acknowledgments
We gratefully acknowledge the many practical and conceptual contributions of our colleagues at Life Technologies/Invitrogen and Atto Bioscience. We thank Dr. Danny Dhanasekaran for valuable guidance on manipulation of GPCRs, and Drs. Mindy Goldsborough and Maria DeBernardi for assistance with imaging. Portions of this work were supported by contracts N66001-02-C-8027 and N66001-03-C-8027 from the Defense Advance Research Projects Agency.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2512204.
References
- Barak, L.S., Ferguson, S.S., Zhang, J., and Caron, M.G. 1997. A β-arrestin/green fluorescent protein biosensor for detecting G protein-coupled receptor activation. J. Biol. Chem. 272: 27497-27500. [DOI] [PubMed] [Google Scholar]
- Bardwell, L., Cook, J.G., Inouye, C.J., and Thorner, J. 1994. Signal propagation and regulation in the mating pheromone response pathway of the yeast Saccharomyces cerevisiae. Dev. Biol. 166: 363-379. [DOI] [PubMed] [Google Scholar]
- Baudin, A., Ozier-Kalogeropoulos, O., Denouel, A., Lacroute, F., and Cullin, C. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21: 3329-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard, P. and Couturier, M. 1992. Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. J. Mol. Biol. 226: 735-745. [DOI] [PubMed] [Google Scholar]
- Boulton, S.J., Gartner, A., Reboul, J., Vaglio, P., Dyson, N., Hill, D.E., and Vidal, M. 2002. Combined functional genomic maps of the C. elegans DNA damage response. Science 295: 127-131. [DOI] [PubMed] [Google Scholar]
- Brachmann, C.B., Davies, A., Cost, G.J., Caputo, E., Li, J., Hieter, P., and Boeke, J.D. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115-132. [DOI] [PubMed] [Google Scholar]
- Castelli, V., Aury, J-M., Jaillon, O., Wincker, P., Clepet, C., Menard, M., Cruaud, C., Schachter, V., Temple, G., Caboche, M., et al. 2004. Whole-genome sequence comparisons and “Full-length” cDNA sequences: A combined approach to evaluate and improve Arabidopsis genome annotation. Genome Res. 14: 406-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheo, D.L., Brasch, M.A., Temple, G.F., Hartley, J.L., and Byrd, D.R.N. 2002. Use of multiple recombination sites with unique specificity in recombinational cloning. Patent US20020007051A1.
- Christianson, T.W., Sikorski, R.S., Dante, M., Shero, J.H., and Hieter, P. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110: 119-122. [DOI] [PubMed] [Google Scholar]
- Davy, A., Bello, P., Thierry-Mieg, N., Vaglio, P., Hitti, J., Doucette-Stamm, L., Thierry-Mieg, D., Reboul, J., Boulton, S., Walhout, A.J., et al. 2001. A protein-protein interaction map of the Caenorhabditis elegans 26S proteasome. EMBO. Rep. 2: 821-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplancke, B., Dupuy, D., Vidal, M., and Walhout, A.J.M. 2004. A Gateway-based high-throughput yeast one-hybrid system. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Dowell, S.J. and Brown, A.J. 2002. Yeast assays for G-protein-coupled receptors. Receptors Channels. 8: 343-352. [PubMed] [Google Scholar]
- Dupuy, D., Li., Q., Deplancke, B.D., Boxem, M., Hao, T., Lamesch, P., Hope, I.A., Hill, D., Walhout, A.J.M., and Vidal, M. 2004. A first version of the Caenorhabditis elegans promoterome. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Funk, M., Niedenthal, R., Mumberg, D., Brinkmann, K., Ronicke, V., and Henkel, T. 2002. Vector systems for heterologous expression of proteins in Saccharomyces cerevisiae. Methods Enzymol. 350: 248-257. [DOI] [PubMed] [Google Scholar]
- Giaever, G., Chu, A.M., Ni, L., Connelly, C., Riles, L., Veronneau, S., Dow, S., Lucau-Danila, A., Anderson, K., Andre, B., et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387-391. [DOI] [PubMed] [Google Scholar]
- Hagen, D.C., McCaffrey, G., and Sprague Jr., G.F. 1991. Pheromone response elements are necessary and sufficient for basal and pheromone-induced transcription of the FUS1 gene of Saccharomyces cerevisiae. Mol. Cell Biol. 11: 2952-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley, J.L., Temple, G.F., and Brasch, M.A. 2000. DNA cloning using in vitro site-specific recombination. Genome Res. 10: 1788-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirawake, H., Taniwaki, M., Tamura, A., Amino, H., Tomitsuka, E., and Kita, K. 1999. Characterization of the human SDHD gene encoding the small subunit of cytochrome b (cybS) in mitochondrial succinate-ubiquinone oxidoreductase. Biochim. Biophys. Acta 1412: 295-300. [DOI] [PubMed] [Google Scholar]
- Hope, I.A., Stevens, J., Garner, A., Hayes, J., Cheo, D.L., Brasch, M.A., and Vidal, M. 2004. Feasibility of genome-scale construction of promoter::reporter gene fusions for expression in Caenorhabditis elegans using a multi-site Gateway recombination system. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Huang, R.Y., Boulton, S.J., Vidal, M., Almo, S.C., Bresnick, A.R., and Chance, M.R. 2003. High-throughput expression, purification, and characterization of recombinant Caenorhabditis elegans proteins. Biochem. Biophys. Res. Commun. 307: 928-934. [DOI] [PubMed] [Google Scholar]
- Jing, S., Spencer, T., Miller, K., Hopkins, C.R., and Trowbridge, I.S. 1990. Role of human transferrin receptor cytoplasmic domain in endocytosis: Localization of a specific sequence for internalization. J. Cell. Biol. 110: 283-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon, D., Roberts, B.L., Richardson, W.D., and Smith, A.E. 1984. A short amino acid sequence able to specify nuclear location. Cell 39: 499-509. [DOI] [PubMed] [Google Scholar]
- Kery, V., Savage, J.R., Widjaja, K., Blake, B.K., Conklin, D.R., Ho, Y.S., Long, X., von Rechenberg, M., Zarembinski, T.I., and Boniface, J.J. 2003. Expression screen by enzyme-linked immunofiltration assay designed for high-throughput purification of affinity-tagged proteins. Anal. Biochem. 317: 255-258. [DOI] [PubMed] [Google Scholar]
- Krautwurst, D., Yau, K.W., and Reed, R.R. 1998. Identification of ligands for olfactory receptors by functional expression of a receptor library. Cell 95: 917-926. [DOI] [PubMed] [Google Scholar]
- LaBaer, J., Qiu, J., Anumanthan, A., Mars, W., Zuo, D., Murthy, T.V.S., Taycher, H., Halleck, A., Hainsworth, E., Lory, S., et al. 2004. The Pseudomonas aeruginosa PA01gene repository. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Landy, A. 1989. Dynamic, structural, and regulatory aspects of λ site-specific recombination. Annu. Rev. Biochem. 58: 913-949. [DOI] [PubMed] [Google Scholar]
- Li, S., Armstrong, C.M., Bertin, N., Ge, H., Milstein, S., Boxem, M., Vidalain, P.O., Han, J.D.J., Chesneau, A., Hao, T., et al. 2004. A map of the interactome network of the metazoan C. elegans. Science 303: 540-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, R.F., Boland, M.V., and Velliste, M. 2000. Towards a systematics for protein subcelluar location: Quantitative description of protein localization patterns and automated analysis of fluorescence microscope images. Proc. Int. Conf. Intel. Syst. Mol. Biol. 8: 251-259. [PubMed] [Google Scholar]
- Parrish, W., Eilers, M., Ying, W., and Konopka, J.B. 2002. The cytoplasmic end of transmembrane domain 3 regulates the activity of the Saccharomyces cerevisiae G-protein-coupled α-factor receptor. Genetics 160: 429-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai, B., Sampath, V., Sharma, N., and Sadhale, P. 2001. Rpb4, a non-essential subunit of core RNA polymerase II of Saccharomyces cerevisiae is important for activated transcription of a subset of genes. J. Biol. Chem. 276: 30641-30647. [DOI] [PubMed] [Google Scholar]
- Reboul, J., Vaglio, P., Tzellas, N., Thierry-Mieg, N., Moore, T., Jackson, C., Shin-i, T., Kohara, Y., Thierry-Mieg, D., Thierry-Mieg, J., et al. 2001. Open-reading frame sequence tags (OSTs) support the existence of at least 17,300 genes in C. elegans. Nat. Genet. 27: 1-5. [DOI] [PubMed] [Google Scholar]
- Reboul, J., Vaglio, P., Rual, J.F., Lamesch, P., Martinez, M., Armstrong, C.M., Li, S., Jacotot, L., Bertin, N., Janky, R., et al. 2003. C. elegans ORFeome version 1.1: Experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat. Genet. 34: 35-41. [DOI] [PubMed] [Google Scholar]
- Sasaki, Y., Sone, T., Yoshida, S., Yahata, K., Hotta, J., Chesnut, J.D., Honda, T., and Imamoto, F. 2004. Evidence for high specificity and efficiency of multiple recombination signals in mixed DNA cloning by the Multisite Gateway system. J. Biotechnol. 107: 233-243. [DOI] [PubMed] [Google Scholar]
- Wach, A., Brachat, A., Pohlmann, R., and Philippsen, P. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10: 1793-1808. [DOI] [PubMed] [Google Scholar]
- Walhout, A.J.M. and Vidal, M. 2001. High-throughput yeast two-hybrid assays for large-scale protein interaction mapping. Methods 24: 297-306. [DOI] [PubMed] [Google Scholar]
- Walhout, A.J.M., Sordella, R., Lu, X., Hartley, J.L., Temple, G.F., Brasch, M.A., Thierry-Mieg, N., and Vidal, M. 2000a. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science 287: 116-122. [DOI] [PubMed] [Google Scholar]
- Walhout, A.J.M., Temple, G.F., Brasch, M.A., Hartley, J.L., Lorson, M.A., van den Heuvel, S., and Vidal, M. 2000b. GATEWAY recombinational cloning: Application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 328: 575-592. [DOI] [PubMed] [Google Scholar]
- Walhout, A.J., Reboul, J., Shtanko, O., Bertin, N., Vaglio, P., Ge, H., Lee, H., Doucette-Stamm, L., Gunsalus, KC., Schetter, A.J., et al. 2002. Integrating interactome, phenome, and transcriptome mapping data for the C. elegans germline. Curr. Biol. 12: 1952-1958. [DOI] [PubMed] [Google Scholar]
- Yamada, K., Lim, J., Dale, J.M., Chen, H., Shinn, P., Palm, C.J., Southwick, A.M., Wu, H.C., Kim, C., and Nguyen, M. 2003. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302: 842-846. [DOI] [PubMed] [Google Scholar]
- http://genoplante-info.infobiogen.fr/Databases/CT_Nouveaux_Outils/NO2001054; Arabidopsis Atome project.
WEB SITE REFERENCES
- http://www.fruitfly.org/EST/gateway.shtml; Drosophila ORFs.
- http://xgc.nci.nih.gov; Xenopus ORFs.
- http://zgc.nci.nih.gov; zebrafish ORFs.
- http://mgc.nci.nih.gov; human, mouse and rat ORFs.