Summary
Juicer and Juicebox described by Durand et al. (2016) are two new tools for fast and reliable processing of Hi-C data; providing approaches for read processing, multiple normalization schemes, feature annotation, and dynamic browsing of chromatin contacts, thus reducing arduous Hi-C analysis into an easy yet flexible pipeline.
The study of chromatin 3-D organization is a rapidly growing field, yet the time and expertise needed to analyze chromatin conformation data can be overwhelming. The Hi-C method utilizes proximity ligation and high-throughput sequencing to provide genome-wide chromatin contact data (Lieberman-Aiden et al., 2009). However, due to its fairly recent development and complicated nature of Hi-C, data analysis has traditionally required a well-versed computational biologist to extract meaningful observations. To ease this computational burden, Durand et al. in Cell Systems (Durand et al., 2016a, 2016b) have provided two companion tools, Juicer and Juicebox, which dramatically improve the ease, speed, and customization of Hi-C data analysis.
The analysis of chromatin organization grows ever more detailed, and the sheer amount of sequence processing and analysis required can become overwhelming. To this point, kilobase-resolution chromatin contact maps for the human genome require billions of Hi-C sequencing reads (Rao et al., 2014). The large read number is due both to the 2-D point-to-point nature of the Hi-C matrix and to the linear distance decay of chromatin interactions. As high-throughput sequencing becomes less expensive, these read amounts become more attainable and high resolution comparisons of samples become ever more feasible. This has created a growing need for a streamlined, user-friendly, yet versatile pipeline to obtain and view chromatin contact maps.
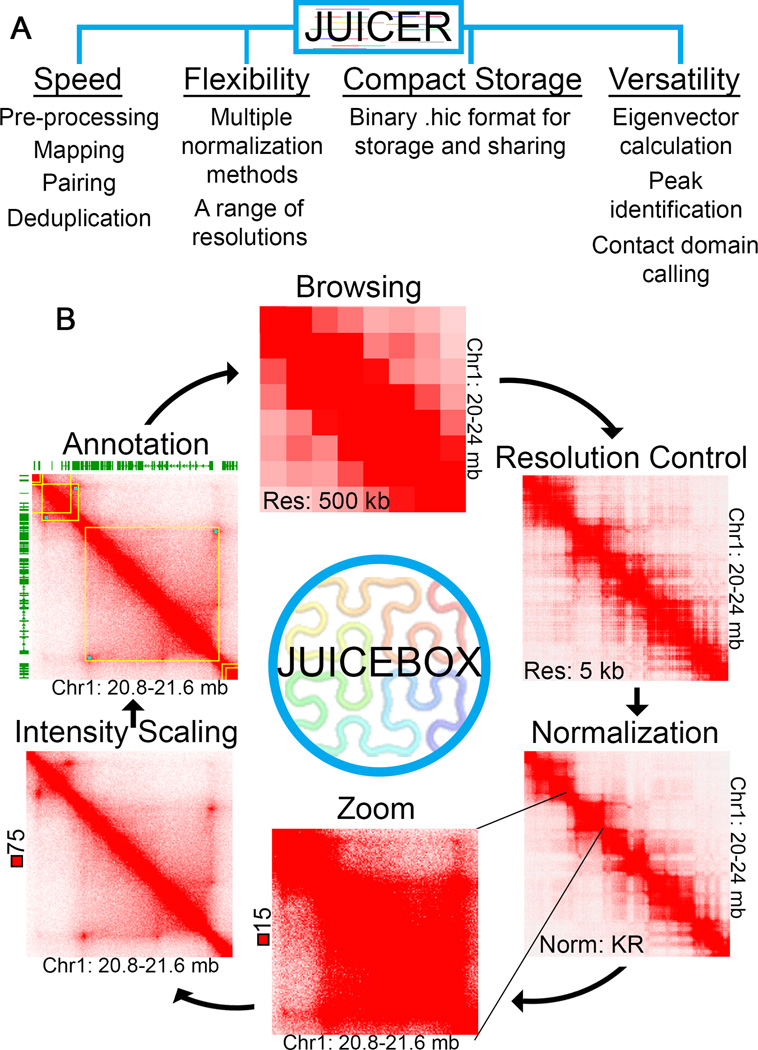
Quality pipelines for Hi-C data analysis must perform numerous tasks in a computationally efficient manner, including: processing ligation-joined reads, performing alignment, pairing reads, removing duplicates, and performing normalizations. Also needed is the ability to store contact matrices within a compact, yet easily retrievable, format. Juicer automates these steps, incorporating parallel processing to reduce the amount of time it takes to go from raw reads to useable contact coordinates (Figure 1A). These contacts are then stored within a compact binary format, a storage scheme so efficient that it will likely become the most common format for storing and sharing Hi-C data.
Figure 1. Functionality of Juicer and Juicebox.
(A) Juicer streamlines Hi-C processing while maintaining flexibility. All standard Hi-C processing steps are performed by Juicer, as are the creation of contact maps using several different normalization methods and at many different resolutions. Processed Hi-C contact maps are stored in a compact binary format, which can be used in conjunction with Juicebox as well as with incorporated tools for feature identification. (B) Juicebox allows dynamic browsing of Hi-C data. Through Juicebox, Hi-C contacts are viewed as browsable maps, providing users with control over the resolution, normalization method, zoom, and intensity scale. Juicebox also supports the overlay of 2D and 1D annotation tracks. Hi-C data from GM12878 cells processed by Juicer and Juicebox is shown for one region at different resolutions, normalizations, zoom, and intensity scales.
One enticing aspect of Juicer is the automatic normalization of Hi-C data at many different resolutions. While several normalization techniques have been proposed (Hu et al., 2012; Imakaev et al., 2012; Rao et al., 2014; Yaffe and Tanay, 2011), it remains uncertain which is most representative of the actual chromatin interactome. Juicer performs three different normalization techniques and stores each along with unnormalized contacts in its compact format. This allows for the comparison of normalization methods and provides flexibility for downstream analysis.
In addition to the creation of contact maps, Juicer includes tools to identify several different chromatin features, a process that has been a difficult yet important part of Hi-C data analysis. Algorithms to identify compartments, domains, and chromatin loops are included in Juicer, yet these may be more useable for human data than for other organisms. For example, eigenvector analysis for identifying compartments is done at 500 kb resolution or lower, which permits identification of the compartments seen in humans at coarse resolution (Lieberman-Aiden et al., 2009). However, eigenvector at higher resolutions may be necessary for organisms with smaller genomes or for exploring compartmentalization at finer resolutions.
Hi-C contact maps also display domains formed from the intra-association of genomic regions to the exclusion of others. Domains found in various organisms include compartment (A/B) domains, topologically associating domains (TADs), chromosomally interacting domains (CIDs), globules, and small contact domains. The similarities or differences between these domain types have not been well-defined (Rowley and Corces, 2016). TADs are the most frequently studied type of contact domain, and have generally been identified utilizing a contact directionality index and hidden markov model (Dixon et al., 2012). This type of feature is currently not supported by Juicer, thus TAD calling still requires external analysis. Instead, Juicer identifies smaller contact domains through Arrowhead transformation (Rao et al. 2014). Additionally, through the HiCCUPS algorithm, Juicer includes identification of loops (i.e. peaks or bright spots in the 2-D contact plot) which correspond to interactions between CTCF binding sites in humans and mice (Rao et al., 2014). It should be noted that contact domains identified by Juicer often correspond to CTCF loops (Rao et al. 2014). However, the presence or nature of loops in other organisms has not been examined, thus the use of this feature identification tools in non-mammalian organisms should be done with careful consideration.
The output of analyses carried out with Juicer can be visualized using Juicebox, which is a genome browser specific to the two-dimensional nature of Hi-C contacts (Figure 1B). The ability to quickly visualize Hi-C data at different resolutions, intensities, and genomic loci is essential for quality analysis. Until now, contact heatmaps have been static for score intensity and/or resolution and have generally required production for each locus of interest. Juicebox takes the compact Juicer output and produces a browseable heatmap with adjustable zoom, intensity, and resolution. The ability to dynamically control intensity, resolution, and zoom makes the discovery of fine-point features much more feasible. Juicebox also allows loading of custom 2-D features and 1-D tracks, which enhances discovery of overlapping features. Additionally, one can switch between different normalization schemes as well as view contacts in relation to the expected decay. This flexibility in how contact maps are browsed contributes to our ability to find meaningful trends in the data.
In addition, Juicebox allows sample comparison side by side or as a ratio to each other. However, caution should be used when comparing samples by means of a Hi-C ratio. Due to an inherent ratio bias, bins with low contact counts will display a high degree of difference thereby biasing for interactions in relation to their distance from the contact map diagonal. In lieu of ratios, some have used subtraction contact maps for comparison between samples (Li et al., 2015), but this method encounters the opposite bias, and it is not clear which method represents a more accurate depiction of biologically relevant interaction changes. Thus any sample comparison map must be considered in light of the inherent diagonal bias. Despite this, Juicebox allows confirmation of sample differences by allowing one to view samples side by side.
Overall, Juicer and Juicebox represent a significant advance in the accessibility of chromatin contact data. Contact maps can be easily produced and visualized dynamically. Many aspects of these tools are useful for chromatin contact data from any organism or condition; however, the included tools for feature identification were optimized for human data and their use in other systems must be approached cautiously. Regardless, these tools give researchers the flexibility to tailor analysis to not only the specific dataset, but to the questions being posed. The fast and dynamic functionality of Juicer and Juicebox makes them likely to become the standard pipeline and genome browser for chromatin 3D conformation data. We suggest that the hic format for processed Hi-C data should become the standard in the field, allowing for the creation of a repository for easy access and sharing of 3D conformation data by the scientific community.
References
- Dixon, et al. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand, et al. Cell Syst. 2016a;3:99–101. doi: 10.1016/j.cels.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand, et al. Cell Syst. 2016b;3:95–98. doi: 10.1016/j.cels.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, et al. Bioinformatics. 2012;28:3131–3133. doi: 10.1093/bioinformatics/bts570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imakaev, et al. Nat. Methods. 2012;9:999–1003. doi: 10.1038/nmeth.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, et al. Mol. Cell. 2015;58:216–231. doi: 10.1016/j.molcel.2015.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden, et al. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, et al. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley, Corces Curr. Opin. Cell Biol. 2016;40:8–14. doi: 10.1016/j.ceb.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe, Tanay Nat. Genet. 2011;43:1059–1065. doi: 10.1038/ng.947. [DOI] [PubMed] [Google Scholar]