Abstract
The development of neutralizing antibodies against blood coagulation factor VIII (FVIII), referred to clinically as “inhibitors”, is the most challenging and deleterious adverse event to occur following intravenous infusions of FVIII to treat hemophilia A. Inhibitors occlude FVIII surfaces that must bind to activated phospholipid membranes, the serine proteinase factor IXa, and other components of the ‘intrinsic tenase complex’ in order to carry out its important role in accelerating blood coagulation. Inhibitors develop in up to one of every three patients, yet remarkably, a substantial majority of severe hemophilia A patients, who circulate no detectable FVIII antigen or activity, acquire immune tolerance to FVIII during initial infusions or else after intensive FVIII therapy to overcome their inhibitor. The design of less immunogenic FVIII proteins through identification and modification (“de-immunization”) of immunodominant T-cell epitopes is an important goal. For patients who develop persistent inhibitors, modification of B-cell epitopes through substitution of surface-exposed amino acid side chains and/or attachment of bulky moieties to interfere with FVIII attachment to antibodies and memory B cells is a promising approach. Both experimental and computational methods are being employed to achieve these goals. Future therapies for hemophilia A, as well as other monogenic deficiency diseases, are likely to involve administration of less immunogenic proteins in conjunction with other novel immunotherapies to promote a regulatory cellular environment promoting durable immune tolerance.
Keywords: Factor VIII, Epitopes, Anti-drug antibodies, Immunogenicity, Antigenicity
1. Introduction
The development of neutralizing anti-drug antibodies (ADAs) in patients has derailed translation to the clinic of several promising protein drugs designed to be administered intravenously, subcutaneously and/or via gene therapy. There is a growing appreciation of the compelling need to avoid and/or manage these deleterious immune responses in order to fulfill the promise of potentially lifesaving therapies, e.g. protein replacement therapies for genetic diseases such as hemophilia A and B (Factor VIII (FVIII) and Factor IX (FIX) deficiency, respectively), Gaucher’s disease (glucocere-brosidase deficiency) and Fabry disease (alpha galactosidase deficiency). In addition to replacement therapies for genetic diseases, significant efforts have gone into the engineering of various proteins to alter or enhance their physiological roles and thereby achieve or improve therapeutic efficacy in patients. For example, addition of disulfide bonds and other amino acid sequence substitutions can increase the structural stability of proteins, while rational sequence modifications can result in stronger or weaker receptor-ligand binding avidities, changes in phosphorylation or glycosylation sites, and alterations of virtually any targeted activity of the therapeutic protein of interest. The scientific literature is replete with well-executed studies demonstrating that rationally improved, sequence-modified proteins exhibit the desired effects in vitro.
Preclinical testing of proposed therapeutic protein drugs includes tests to predict their immunogenicity in humans. A detailed description of regulatory requirements for immunogenicity testing is beyond the scope of this review, however several preclinical methods are consistently employed, notably the use of animal models to test whether the experimental protein drug induces an anti-drug antibody response. The animals are considered surrogates for patients, and the underlying assumption (or hope) is that proteins that do not show immunogenicity in mice, or rats, or dogs, or monkeys will also be immunologically “silent” when administered to patients.
Unfortunately, ADAs have developed in clinical trials and in post-marketing surveillance of several drugs, leading to cancellation of projects close to the end of the translational pipeline, after the expenditure of enormous effort and millions of dollars. In addition to the financial aspects, the clinical risks to patients who develop ADAs can be serious, especially if the ADAs cross-react with endogenous proteins. For example, a PEGylated recombinant thrombopoeitin (TPO) molecule administered to healthy volunteers and patients elicited ADAs in several individuals that bound to their endogenous TPO, resulting in prolonged thrombocytopenia [1]. Another recent example is the development of ADAs in 11% of hemophilia patients receiving a recombinant factor VIIa, vatreptacog alfa, during phase III confirmatory testing [2, 3]. This protein had three amino acid substitutions that altered its conformation and potency as a procoagulant factor. These cases illustrate the fact that even minor sequence or structural changes may provoke neutralizing antibodies, which can develop following CD4 T-effector recognition of “foreign” peptides on antigen presenting cells (APCs). For this to happen, the peptides must be processed by and presented on APCs [4], and this peptide-HLA complex must then be recognized by a T-cell receptor (TCR), resulting in cytokine secretion and hence B-cell maturation to antibody-secreting plasma cells.
The ability to accurately predict which amino acid sequences or modifications are likely to induce HLA-restricted responses leading to antibody production would significantly reduce risks to patients and increase the successful translation of promising drugs to the clinic. Accordingly, epitope prediction algorithms are continually improving [5–8], in tandem with larger data training sets consisting of peptide-MHC binding data, identification of HLA-restricted T-cell epitopes, etc. At present, however, these computer programs significantly over-predict potential epitopes. This is due in part to the tremendous diversity of TCRs, which cannot be properly accounted for in attempts to predict formation of an HLA-peptide-TCR immunological synapse, and to the difficulty in predicting which peptides will be presented on APCs following processing of the protein antigen in the MHC compartment. Thus, although these programs are tremendously useful in generating lists of potential epitopes and also, importantly, in deducing the MHC binding registers of some confirmed peptide epitopes, prediction algorithms still generally fall short of accurately and comprehensively predicting risks of developing clinically significant ADAs.
1.1. Hemophilia A and “inhibitor” antibodies
Hemophilia A (HA) is an X-linked bleeding disorder resulting from lack of or dysfunctional FVIII, a non-enzymatic protein cofactor that accelerates blood coagulation at a critical control point in the clotting cascade. Patients with severe HA have less than 1% normal FVIII activity, and almost all of these individuals have no detectable circulating FVIII antigen. Genetic deficiency diseases such as severe HA present a unique opportunity to study the course of anti-drug immune responses. An interesting aspect of such studies is that the development of ADAs often does not preclude further exposure to the immunogenic protein. This is because the patients do not circulate an endogenous functional protein, and hence there is no risk of cross-reacting antibodies exacerbating their disease. Indeed, clinical Immune Tolerance Induction protocols entail intensive FVIII treatment in an effort to reduce antibody titers. Neutralizing ADAs are the most serious complication of FVIII replacement therapy in HA, affecting up to 1 in 3 patients. Thus studies of immune responses to infused FVIII have the potential to improve outcomes for HA patients, and the scientific knowledge thus gained is also highly applicable to the development of antibodies against other therapeutic proteins.
The most predictive factor for development of anti-FVIII ADAs, referred to clinically as “inhibitors”, is the F8 gene mutation, with multi-exon deletions and early nonsense mutations carrying a high risk, inversion mutations an intermediate risk, and missense mutations the lowest risk [9]. Intensity of FVIII treatment and other environmental factors also contribute to inhibitor risk [10–12], and there is growing interest in delineating the synergistic roles of other genetic factors such as sequence variations in immunoregulatory genes in predisposing some individuals to ADAs [13, 14].
Almost half of severe HA patients have an inversion mutation at intron 22 of this 26-exon, 2332-amino-acid protein, and it has been proposed that low levels of one or more partial FVIII proteins translated from the interrupted F8 mRNA sequence and from a ubiquitously expressed shorter transcript termed F8B [15] are expressed intracellularly [16]. In principle, this could result in central tolerance to FVIII sequences with the exception of those encoded by the inversion site itself (FVIII residues 2124–2125). However, the observation of T-cell responses to FVIII C2 domain sequences, which are encoded by both the F8 and F8B genes, in severe HA patients [17, 18] (and K. Pratt, unpublished data) argues that multiple T-cell epitopes can contribute to inhibitor responses in patients with inversions as well as other F8 gene mutations.
2. T-cell and B-cell epitope mapping
Cytokine secretion and proliferation of human CD4 T cells from HA and acquired HA patients and even from some healthy controls has been demonstrated following stimulation with FVIII peptides corresponding to multiple FVIII domains [18–22]. Definitive identification of several T-cell epitopes has been accomplished through cloning, expansion and characterization of FVIII-specific CD4 T-cell clones and polyclonal lines [22–26]. The use of peptide-loaded HLA-DRB1 tetramers [27, 28] has greatly facilitated the mapping of T-cell epitopes in FVIII and isolation of CD4 T-cell clones and lines [24–26, 29], although the size of the FVIII protein and the available blood volumes from inhibitor patients, who are usually infants, remain a daunting challenge to comprehensive epitope mapping. Nevertheless, further mapping of immunodominant T-cell epitopes in FVIII remains a strong priority, as this knowledge is essential for understanding mechanisms of inhibitor responses, as well as of the acquired tolerance to FVIII that ~2/3 of inhibitor patients are fortunate to eventually achieve. Interestingly, the eradication of clinically significant levels of neutralizing anti- FVIII antibodies does not require deletion of all FVIII-specific T cells, as demonstrated by a recent study in which oligoclonal FVIII-specific T-cell clones and lines were isolated and expanded from a successfully tolerized patient in whom anti-FVIII antibodies were undetectable by ELISA assay [30].
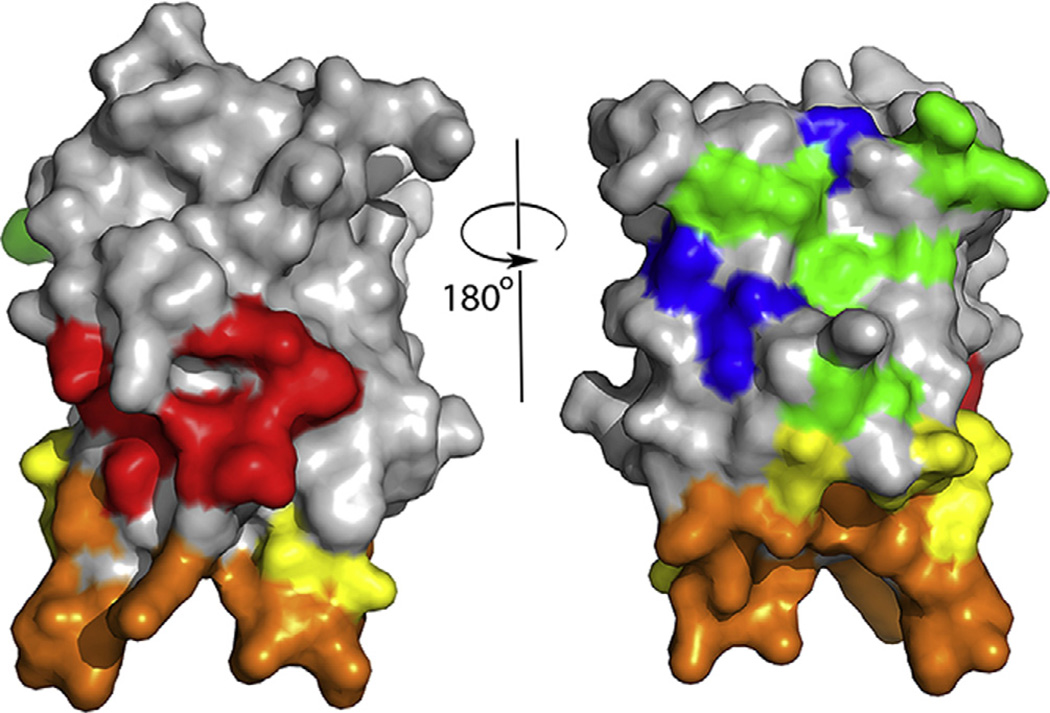
Tremendous progress has been made over the past several years in mapping of B-cell epitopes recognized by neutralizing anti-FVIII antibodies. The FVIII domain specificity and roles of some residues were determined by biochemical experiments, including the elegant use of porcine-FVIII hybrid proteins to map the domain specificity of FVIII antibodies [31–34]. The first definitive picture of a FVIII B-cell epitope was revealed by the crystal structure of the FVIII C2 domain bound to the patient-derived human monoclonal antibody Fab fragment BO2C11 [35]. This antibody blocks FVIII binding to phospholipid membranes and von Willebrand factor [36], and the crystal structure confirmed the participation of specific amino acid side chains in these processes that had been proposed based on the FVIII-C2 domain crystal structure [37] and on mutagenesis studies [38, 39]. More recently, competition ELISA experiments have identified partially overlapping surfaces on the FVIII C2 and A2 domains recognized by neutralizing antibodies [40, 41]. Higher-resolution mapping approaches have included affinity-directed mass spectrometry [42, 43], phage display [44], hydrogen–deuterium exchange mass spectrometry [45], X-ray scattering [46], in silico predictions [47, 48] and crystallographic studies [46, 49]. In addition, comprehensive high-resolution mapping of the minimal B-cell epitopes on the FVIII-C2 domain surface, with minimal epitopes defined as the amino acid side chains that contribute significantly to antigen–antibody binding avidities, has been accomplished using a targeted mutagenesis plus surface plasmon resonance (SPR) strategy to map epitopes recognized by neutralizing anti-FVIII monoclonal antibodies [50, 51]. This approach offers significant advantages for the design of less antigenic FVIII proteins, as it can suggest specific amino acid substitutions to allow the proteins to evade existing antibodies and to lessen the risk of memory B-cell stimulation. Fig. 1 summarizes the results of SPR-based mapping of epitopes recognized by 11 neutralizing anti-FVIII monoclonal antibodies.
Fig. 1.
Front and back views (rotated 180°) of the FVIII C2 domain crystal structure [37], with surface-exposed amino acids colored to indicate the 5 partially-overlapping B-cell epitopes recognized by 11 neutralizing monoclonal antibodies. The antibodies and epitopes were originally denoted Types A, AB, B, BC and C on the basis of competition ELISA experiments, and the antibodies inhibited distinct binding interactions and functions of FVIII [40]. Thus the identification of specific amino acids comprising these epitopes also indicates which residues and surfaces interact with phospholipid membranes, von Willebrand factor, and components of the intrinsic tenase complex. Type A: red; Type AB: orange; Type B: yellow; Type BC: green; Type C: blue. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3. Modification of T-cell and B-cell epitopes
Two types of sequence-modified FVIII proteins can be envisioned that could lead to more therapeutic options for HA patients and improved clinical outcomes: (1) less immunogenic proteins having rationally modified T-cell epitopes, and (2) less antigenic FVIII proteins that retain FVIII procoagulant function even in the presence of a pre-existing inhibitory antibody response. Experimental and computational approaches may both be employed in efforts to design more immunotolerant protein therapeutics. Regarding the design of less immunogenic FVIII proteins, if further experimental analysis of patient samples reveals only a limited number of immunodominant, promiscuous T-cell epitopes, it would be sensible to modify these amino acid sequences, e.g. by substitution of one or more “anchor” residues that engage the MHC peptide-binding groove in multiple HLAs. This will require characterizing the HLA restriction of the original T-cell response and evaluating, e.g. by peptide-MHC binding assays and/or computational methods [6, 29, 52–57], the unwanted possibility that the substitution(s) produced a neoepitope that would stimulate new HLA-restricted T-effector responses. It is likely that HLA typing of HA patients will become routine in the not-too-distant future, which would enable immunologists to identify a reasonable fraction of the patients who might be at increased risk of responding to specific immunodominant T-cell epitopes. The cost of manufacturing and obtaining regulatory approval for new FVIII proteins is quite high. Nevertheless, if there turns out to be just a small set of immunogenic “hot spots”, then production of several FVIII proteins, and stratification of patients according to their evidence-based risk of responding to these epitopes, would lead to improved matching of patients to appropriate, minimally immunogenic FVIII products.
Regarding the design of less antigenic FVIII proteins, the prototype for this approach is the use of porcine FVIII (pFVIII) to treat patients with high-titer antibodies against therapeutic human FVIII. Plasma-derived pFVIII was part of the hemophilia physician’s armamentarium two decades ago, and its use saved lives because the neutralizing antibodies against human FVIII generally did not bind pFVIII. However, its use was discontinued due to concerns about cross-species transfer of infectious agents. A recombinant pFVIII has recently been approved and is now available as a potential “bypass” therapeutic that enables many inhibitor patients to achieve hemostasis. Long-term use of this protein replacement therapy is, however, compromised in those patients who proceed to develop neutralizing antibodies against pFVIII. PFVIII has an 83% sequence identity with human FVIII (excluding the B domains of both proteins, which are removed during FVIII activation and are not present in recombinant therapeutic B-domain-deleted proteins) [58]. A large fraction of these sequence variations occur at residues that are not exposed to the protein surface and therefore do not comprise parts of B-cell epitopes. Therefore, by consulting available FVIII crystal structures, it is clearly feasible to design novel, less antigenic FVIII proteins having amino acid substitutions (relative to human FVIII) at specific surface-exposed side chain positions. The resulting increased sequence identity of such proteins with human FVIII would decrease the chances of introducing a neoepitope that would stimulate CD4 T cells to provide help for antibodies against the designed protein. Analyzing sequences of homologous FVIII proteins (orthologs) from other species is another productive approach to designing new therapeutics [59], as evolution has already optimized their stability. FVIII proteins from other species besides pigs may have therapeutic efficacy comparable to that of pFVIII, however the goal of minimizing the potential number of new T-cell epitopes argues for additional, rational modifications of the FVIII sequence to reduce its immunogenicity.
In addition to homology-based approaches, strategic amino acid substitutions in FVIII have been made, and will continue to be made, based on biochemical and biophysical information about FVIII, including its interactions with both antibodies and with components of the membrane-bound intrinsic factor tenase complex. The antigenicity of specific FVIII regions has been modified by rational and alanine-scanning mutagenesis, identifying epitopes and suggesting possible sequence modifications to block antibody binding [31, 32, 38, 39, 60–63].
Using a structure- and energetics-guided approach, our laboratory has carried out a proof-of-principle study modifying specific amino acid residues within an immunodominant B-cell and T-cell epitope in FVIII [50]. SPR experiments measuring the affinities of sequence-modified FVIII-C2 proteins to the patient-derived monoclonal antibody BO2C11 [50, 51] revealed that only six FVIII amino acid side chains contributed significant binding affinity, despite the fact that a high-resolution crystal structure of the C2-BO2C11 complex [35] had identified 15 residues that contacted the antibody surface. The existence of such binding “hot spots” is not an unusual feature of interfaces between protein–protein complexes [64], and SPR is an efficient technique with which to identify critical residues and contacts. Interestingly, the residues in the beta-hairpin turn region that contributes most of the antibody-binding affinity also comprise an immunodominant HLA-DRB1*01:01-restricted T-cell epitope recognized by several HA patients with this allele [24–26], as well as an epitope recognized by T cells from a hemophilia A mouse model [65]. B-domain-deleted (BDD)-FVIII proteins with amino acid substitutions F2196A, F2196K and M2199A were generated, and all showed specific activities and binding to von Willebrand factor that were similar to wild-type BDD-FVIII. BDD-FVIII-F2196K was particularly promising, as this single substitution allowed the protein to avoid neutralization by both BO2C11 and a murine monoclonal antibody targeted against an overlapping site on FVIII [50]. FVIII-C2 proteins with single amino acid substitutions at F2196 were also markedly less stimulatory to T-cell clones isolated from inhibitor subjects with the allele HLA-DRB1*01:01 [66]. These studies support the concept that identification and rational mutagenesis of immunodominant epitopes can produce less immunogenic and antigenic therapeutic FVIII proteins.
Some residues contributing to T-cell and B-cell epitopes are essential for FVIII structural stability and/or procoagulant activity and will thus not be candidates for substitution. Nevertheless, it appears highly feasible to design proteins that are both less antigenic and less immunogenic than currently available FVIII products. It is important to note that individuals with mild HA have between 5% and 10% normal FVIII activity, and many are not even identified as having HA until surgery, e.g. tooth extraction or traumatic injury. From the standpoint of protein design, this is encouraging: a therapeutic protein that was less immunogenic and/or antigenic could provide therapeutic benefit to patients, even if the modifications resulted in a significant (>80%) loss of FVIII specific activity.
4. Many amino acid sequence variations are not immunogenic
It is important to keep one’s perspective regarding the likelihood of minor amino acid sequence variations provoking serious adverse immune responses. This is, appropriately, an enormous concern in the production of therapeutic proteins. However, our immune systems daily encounter many nonself substances, not to mention self-proteins modified by post-translational and environmental processes, that do not require immediate neutralization, phagocytosis, clearance, etc., and that instead elicit a tolerogenic response. Such acquired tolerogenic responses are common, and they can be a barrier to effective vaccine development, immunotherapies for cancer, etc. It is quite apparent that many FVIII sequences, although foreign to the HA patient, do not provoke high-titer neutralizing antibodies, and that most inhibitor responses are transient and followed by acquired tolerance to infused FVIII. Immune Tolerance Induction therapies (via intensive FVIII treatment) succeed in 70% of cases. A primary current challenge is to identify the small set of FVIII sequences that are highly immunogenic and the characteristics of patients who are at increased risk of developing a neutralizing antibody response. An even more important challenge is to discern why most but not all HA patients tolerate infusions of this foreign or partly-foreign protein, and to apply this knowledge to enable more patients to avoid or overcome ADA responses.
Human genome sequencing studies are revealing the tremendous genetic diversity of the human population, including sequence variations in genes related to protein deficiency diseases such as HA. Three non-synonymous single nucleotide polymorphisms (ns-SNPs) encoding single amino acid variants at FVIII residues 484, 1241 and 2238 are more common in African and African American populations than in White populations [67]. Identification of these population variants created a concern that amino acid sequence differences between infused FVIII and a HA patient’s endogenous FVIII protein (if any is translated) could contribute to the increased inhibitor incidence that has been seen consistently in African American patients [68]. However, a study of almost 400 African American and White American subjects [57], as well as studies of smaller cohorts of Black and White South Africans and of American and European HA populations [69–71], showed no statistical association between these ns-SNPs and inhibitors. Furthermore, quantitative peptide-MHC binding assays did not indicate promiscuous binding of these peptides to 11 common HLA-DRB1 proteins, and tetramer-guided epitope mapping experiments also did not reveal CD4 T-cell responses following stimulation with the peptides, whereas tetanus-specific positive controls consistently gave the expected results [57]. Although it is certainly possible that some patients will show T-cell responses to these polymorphic FVIII sequences, their incidence is likely to be quite low, comparable to the infrequent development of inhibitors in mild HA due to FVIII missense mutations. Clearly, other genetic factors must be contributing to the clinically noted race-associated differences in FVIII inhibitor responses. Similar approaches to evaluate HLA-associated risks of other “mismatched” protein therapies, particularly in the case of sequence-modified endogenous proteins, as opposed to genetic deficiency diseases where multiple epitopes may be involved, would be appropriate and indeed are increasingly being pursued by pharmaceutical and biotechnology companies [52].
5. Synergy of epitope modification/masking with novel tolerogenic immunotherapies
It should be noted that additional types of alterations to protein structures, besides amino acid sequence substitutions, may affect immunogenicity and antigenicity. Attachment of the PEG moiety, or alterations in carbohydrate structure, or fusions with other protein sequences, notably the Fc region of immunoglobulins, albumin, etc. may be used to extend the half-life of therapeutic proteins by altering or masking epitopes, and these modifications may also be immunomodulatory. Presentation of antigens together with Fc regions can promote immune tolerance. This has been demonstrated for FVIII by the Scott lab’s engineering of B cells to present Fc proteins fused to the FVIII A2 and C2 domains [72–74]. A FVIII-Fc protein having an extended half-life in the circulation is now in the clinic [75, 76], and a recent animal model study indicates it may have desirable immunomodulatory properties as well (see N. Gupta et al., this issue). FVIII fusion proteins incorporating other protein or non-protein moieties (PEG, albumin, etc.) may also have altered immunogenicity or antigenicity. Pioneering work by Dr. Annie deGroot has identified “Tregitope” sequences in immunoglobulins and other proteins, which bind to MHC and promote regulatory T cells responses [77]. The tantalizing possibility that peptide therapies utilizing Tregitope sequences could decrease hemophilic inhibitor responses has yet to be explored systematically.
These and other creative immunotherapeutic approaches to promote tolerance could certainly be combined with infusions of less immunogenic FVIII proteins to maximize the probability of improving patient outcomes. It is well known that Immune Tolerance Induction through intensive FVIII treatment succeeds more often when the initial inhibitor titer is fairly low. The design of less immunogenic and antigenic FVIII proteins can be expected to eliminate some but not all neutralizing ADAs. Even if antibody titers are only reduced, this could provide a therapeutic window that would improve the success rate of Immune Tolerance Induction or other tolerance-promoting therapies. The research process itself is synergistic. For example, our recent characterization of hemophilia-patient-derived T-cell clones and lines enabled Kim et al. to employ state-of-the-art methods to express a FVIII-specific TCR on CD4 T cells and then expand them under conditions to produce regulatory T cells that home to FVIII molecules, where they become activated and exert their immunosuppressive effect [78]. As our understanding of anti-FVIII immune responses and acquired tolerance to FVIII improves, we expect this will lead to the development and refinement of new therapies that will impact not only inhibitor patients, but also individuals with other immune and autoimmune disorders.
References
- 1.Li J, Yang C, Xia Y, Bertino A, Glaspy J, et al. Thrombocytopenia caused by the development of antibodies to thrombopoietin. Blood. 2001;98:3241–3248. doi: 10.1182/blood.v98.12.3241. [DOI] [PubMed] [Google Scholar]
- 2.Mahlangu JN, Weldingh KN, Lentz SR, Kaicker S, Karim FA, et al. Changes in the amino acid sequence of the rFVIIa analog, vatreptacog alfa, are associated with clinical immunogenicity. J. Thromb. Haemost. doi: 10.1111/jth.13141. in press. [DOI] [PubMed] [Google Scholar]
- 3.Lentz SR, Ehrenforth S, Karim FA, Matsushita T, Weldingh KN, et al. Recombinant factor VIIa analog in the management of hemophilia with inhibitors: results from a multicenter, randomized, controlled trial of vatreptacog alfa. J. Thromb. Haemost. 2014;12:1244–1253. doi: 10.1111/jth.12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Haren SD, Wroblewska A, Herczenik E, Kaijen PH, Ruminska A, et al. Limited promiscuity of HLA-DRB1 presented peptides derived of blood coagulation factor VIII. PLoS One. 2013;8:e80239. doi: 10.1371/journal.pone.0080239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nielsen M, Lund O, Buus S, Lundegaard C. MHC class II epitope predictive algorithms. Immunology. 2010;130:319–328. doi: 10.1111/j.1365-2567.2010.03268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim Y, Ponomarenko J, Zhu Z, Tamang D, Wang P, et al. Immune epitope database analysis resource. Nucleic Acids Res. 2012;40:W525–W530. doi: 10.1093/nar/gks438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh H, Raghava GP. ProPred: prediction of HLA-DR binding sites. Bioinformatics. 2001;17:1236–1237. doi: 10.1093/bioinformatics/17.12.1236. [DOI] [PubMed] [Google Scholar]
- 8.Schafer JR, Jesdale BM, George JA, Kouttab NM, De Groot AS. Prediction of well-conserved HIV-1 ligands using a matrix-based algorithm, EpiMatrix. Vaccine. 1998;16:1880–1884. doi: 10.1016/s0264-410x(98)00173-x. [DOI] [PubMed] [Google Scholar]
- 9.Gouw SC, van den Berg HM, Oldenburg J, Astermark J, de Groot PG, et al. F8 gene mutation type and inhibitor development in patients with severe hemophilia A: systematic review and meta-analysis. Blood. 2012;119:2922–2934. doi: 10.1182/blood-2011-09-379453. [DOI] [PubMed] [Google Scholar]
- 10.Gouw SC, van den Berg HM. The multifactorial etiology of inhibitor development in hemophilia: genetics and environment. Semin. Thromb. Hemost. 2009;35:723–734. doi: 10.1055/s-0029-1245105. [DOI] [PubMed] [Google Scholar]
- 11.Gouw SC, van den Berg HM, Fischer K, Auerswald G, Carcao M, et al. Intensity of factor VIII treatment and inhibitor development in children with severe hemophilia A: the RODIN study. Blood. 2013;121:4046–4055. doi: 10.1182/blood-2012-09-457036. [DOI] [PubMed] [Google Scholar]
- 12.Astermark J, Altisent C, Batorova A, Diniz MJ, Gringeri A, et al. Non-genetic risk factors and the development of inhibitors in haemophilia: a comprehensive review and consensus report. Haemophilia. 2010;16:747–766. doi: 10.1111/j.1365-2516.2010.02231.x. [DOI] [PubMed] [Google Scholar]
- 13.Astermark J, Donfield SM, Gomperts ED, Schwarz J, Menius ED, et al. The polygenic nature of inhibitors in hemophilia A: results from the Hemophilia Inhibitor Genetics Study (HIGS) Combined Cohort. Blood. 2013;121:1446–1454. doi: 10.1182/blood-2012-06-434803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bardi E, Astermark J. Genetic risk factors for inhibitors in haemophilia A. Eur. J. Haematol. 2015;94(Suppl. 77):7–10. doi: 10.1111/ejh.12495. [DOI] [PubMed] [Google Scholar]
- 15.Levinson B, Kenwrick S, Gamel P, Fisher K, Gitschier J. Evidence for a third transcript from the human factor VIII gene. Genomics. 1992;14:585–589. doi: 10.1016/s0888-7543(05)80155-7. [DOI] [PubMed] [Google Scholar]
- 16.Pandey GS, Yanover C, Miller-Jenkins LM, Garfield S, Cole SA, et al. Endogenous factor VIII synthesis from the intron 22-inverted F8 locus may modulate the immunogenicity of replacement therapy for hemophilia A. Nat. Med. 2013;19:1318–1324. doi: 10.1038/nm.3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pratt KP. Inhibitory antibodies in hemophilia A. Curr. Opin. Hematol. 2012;19:399–405. doi: 10.1097/MOH.0b013e328356ed37. [DOI] [PubMed] [Google Scholar]
- 18.Reding MT, Okita DK, Diethelm-Okita BM, Anderson TA, Conti-Fine BM. Human CD4+ T-cell epitope repertoire on the C2 domain of coagulation factor VIII. J. Thromb. Haemost. 2003;1:1777–1784. doi: 10.1046/j.1538-7836.2003.00251.x. [DOI] [PubMed] [Google Scholar]
- 19.Reding MT, Wu H, Krampf M, Okita DK, Diethelm-Okita BM, et al. Sensitization of CD4+ T cells to coagulation factor VIII: response in congenital and acquired hemophilia patients and in healthy subjects. Thromb. Haemost. 2000;84:643–652. [PubMed] [Google Scholar]
- 20.Reding MT, Okita DK, Diethelm-Okita BM, Anderson TA, Conti-Fine BM. Epitope repertoire of human CD4(+) T cells on the A3 domain of coagulation factor VIII. J. Thromb. Haemost. 2004;2:1385–1394. doi: 10.1111/j.1538-7836.2004.00850.x. [DOI] [PubMed] [Google Scholar]
- 21.Hu GL, Okita DK, Conti-Fine BM. T cell recognition of the A2 domain of coagulation factor VIII in hemophilia patients and healthy subjects. J. Thromb. Haemost. 2004;2:1908–1917. doi: 10.1111/j.1538-7836.2004.00918.x. [DOI] [PubMed] [Google Scholar]
- 22.Jones TD, Phillips WJ, Smith BJ, Bamford CA, Nayee PD, et al. Identification and removal of a promiscuous CD4+ T cell epitope from the C1 domain of factor VIII. J. Thromb. Haemost. 2005;3:991–1000. doi: 10.1111/j.1538-7836.2005.01309.x. [DOI] [PubMed] [Google Scholar]
- 23.Jacquemin M, Vantomme V, Buhot C, Lavend’homme R, Burny W, et al. CD4 + T-cell clones specific for wild-type factor VIII: a molecular mechanism responsible for a higher incidence of inhibitor formation in mild/moderate hemophilia A. Blood. 2003;101:1351–1358. doi: 10.1182/blood-2002-05-1369. [DOI] [PubMed] [Google Scholar]
- 24.James EA, Kwok WW, Ettinger RA, Thompson AR, Pratt KP. T-cell responses over time in a mild hemophilia A inhibitor subject: epitope identification and transient immunogenicity of the corresponding self-peptide. J. Thromb. Haemost. 2007;5:2399–2407. doi: 10.1111/j.1538-7836.2007.02762.x. [DOI] [PubMed] [Google Scholar]
- 25.Ettinger RA, James EA, Kwok WW, Thompson AR, Pratt KP. Lineages of human T-cell clones, including T helper 17/T helper 1 cells, isolated at different stages of anti-factor VIII immune responses. Blood. 2009;114:1423–1428. doi: 10.1182/blood-2009-01-200725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ettinger RA, James EA, Kwok WW, Thompson AR, Pratt KP. HLA-DR-restricted T-cell responses to factor VIII epitopes in a mild haemophilia A family with missense substitution A2201P. Haemophilia. 2010;16:44–55. doi: 10.1111/j.1365-2516.2008.01905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novak EJ, Liu AW, Gebe JA, Falk BA, Nepom GT, et al. Tetramer-guided epitope mapping: rapid identification and characterization of immunodominant CD4+ T cell epitopes from complex antigens. J. Immunol. 2001;166:6665–6670. doi: 10.4049/jimmunol.166.11.6665. [DOI] [PubMed] [Google Scholar]
- 28.Nepom GT, Buckner JH, Novak EJ, Reichstetter S, Reijonen H, et al. HLA class II tetramers: tools for direct analysis of antigen-specific CD4+ T cells. Arthritis Rheum. 2002;46:5–12. doi: 10.1002/1529-0131(200201)46:1<5::AID-ART10063>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 29.James EA, van Haren SD, Ettinger RA, Fijnvandraat K, Liberman JA, et al. T-cell responses in two unrelated hemophilia A inhibitor subjects include an epitope at the factor VIII R593C missense site. J. Thromb. Haemost. 2011;9:689–699. doi: 10.1111/j.1538-7836.2011.04202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pautard B, D’Oiron R, Li Thiao Te V, Lavend’homme R, Saint-Remy JM, et al. Successful immune tolerance induction by FVIII in hemophilia A patients with inhibitor may occur without deletion of FVIII-specific T cells. J. Thromb. Haemost. 2011;9:1163–1170. doi: 10.1111/j.1538-7836.2011.04267.x. [DOI] [PubMed] [Google Scholar]
- 31.Healey JF, Lubin IM, Nakai H, Saenko EL, Hoyer LW, et al. Residues 484–508 contain a major determinant of the inhibitory epitope in the A2 domain of human factor VIII. J. Biol. Chem. 1995;270:14505–14509. doi: 10.1074/jbc.270.24.14505. [DOI] [PubMed] [Google Scholar]
- 32.Lubin IM, Healey JF, Barrow RT, Scandella D, Lollar P. Analysis of the human factor VIII A2 inhibitor epitope by alanine scanning mutagenesis. J. Biol. Chem. 1997;272:30191–30195. doi: 10.1074/jbc.272.48.30191. [DOI] [PubMed] [Google Scholar]
- 33.Lollar P, Healey JF, Barrow RT, Parker ET. Factor VIII inhibitors. Adv. Exp. Med. Biol. 2001;489:65–73. doi: 10.1007/978-1-4615-1277-6_6. [DOI] [PubMed] [Google Scholar]
- 34.Lollar P. Mapping factor VIII inhibitor epitopes using hybrid human/porcine factor VIII molecules. Haematologica. 2000;85:26–28. (discussion 28-30) [PubMed] [Google Scholar]
- 35.Spiegel PC, Jr, Jacquemin M, Saint-Remy JM, Stoddard BL, Pratt KP. Structure of a factor VIII C2 domain-immunoglobulin G4kappa Fab complex: identification of an inhibitory antibody epitope on the surface of factor VIII. Blood. 2001;98:13–19. doi: 10.1182/blood.v98.1.13. [DOI] [PubMed] [Google Scholar]
- 36.Jacquemin MG, Desqueper BG, Benhida A, Vander Elst L, Hoylaerts MF, et al. Mechanism and kinetics of factor VIII inactivation: study with an IgG4 monoclonal antibody derived from a hemophilia A patient with inhibitor. Blood. 1998;92:496–506. [PubMed] [Google Scholar]
- 37.Pratt KP, Shen BW, Takeshima K, Davie EW, Fujikawa K, et al. Structure of the C2 domain of human factor VIII at 1.5 A resolution. Nature. 1999;402:439–442. doi: 10.1038/46601. [DOI] [PubMed] [Google Scholar]
- 38.Barrow RT, Healey JF, Jacquemin MG, Saint-Remy JM, Lollar P. Antigenicity of putative phospholipid membrane-binding residues in factor VIII. Blood. 2001;97:169–174. doi: 10.1182/blood.v97.1.169. [DOI] [PubMed] [Google Scholar]
- 39.Gilbert GE, Kaufman RJ, Arena AA, Miao H, Pipe SW. Four hydrophobic amino acids of the factor VIII C2 domain are constituents of both the membrane-binding and von Willebrand factor-binding motifs. J. Biol. Chem. 2002;277:6374–6381. doi: 10.1074/jbc.M104732200. [DOI] [PubMed] [Google Scholar]
- 40.Meeks SL, Healey JF, Parker ET, Barrow RT, Lollar P. Antihuman factor VIII C2 domain antibodies in hemophilia A mice recognize a functionally complex continuous spectrum of epitopes dominated by inhibitors of factor VIII activation. Blood. 2007;110:4234–4242. doi: 10.1182/blood-2007-06-096842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Markovitz RC, Healey JF, Parker ET, Meeks SL, Lollar P. The diversity of the immune response to the A2 domain of human factor VIII. Blood. 2013;121:2785–2795. doi: 10.1182/blood-2012-09-456582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ansong C, Miles SM, Fay PJ. Epitope mapping factor VIII A2 domain by affinity-directed mass spectrometry: residues 497-510 and 584-593 comprise a discontinuous epitope for the monoclonal antibody R8B12. J. Thromb. Haemost. 2006;4:842–847. doi: 10.1111/j.1538-7836.2006.01831.x. [DOI] [PubMed] [Google Scholar]
- 43.Griffiths AE, Wang W, Hagen FK, Fay PJ. Use of affinity-directed liquid chromatography-mass spectrometry to map the epitopes of a factor VIII inhibitor antibody fraction. J. Thromb. Haemost. 2011;9:1534–1540. doi: 10.1111/j.1538-7836.2011.04397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kahle J, Orlowski A, Stichel D, Becker-Peters K, Kabiri A, et al. Epitope mapping via selection of anti-FVIII antibody-specific phage-presented peptide ligands that mimic the antibody binding sites. Thromb. Haemost. 2015;113(2):396–405. doi: 10.1160/TH14-01-0101. [DOI] [PubMed] [Google Scholar]
- 45.Sevy AM, Healey JF, Deng W, Spiegel PC, Meeks SL, et al. Epitope mapping of inhibitory antibodies targeting the C2 domain of coagulation factor VIII by hydrogen-deuterium exchange mass spectrometry. J. Thromb. Haemost. 2013;11:2128–2136. doi: 10.1111/jth.12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walter JD, Werther RA, Polozova MS, Pohlman J, Healey JF, et al. Characterization and solution structure of the factor VIII C2 domain in a ternary complex with classical and non-classical inhibitor antibodies. J. Biol. Chem. 2013;288:9905–9914. doi: 10.1074/jbc.M112.424564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lebreton A, Moreau V, Lapalud P, Cayzac C, Andre S, et al. Discontinuous epitopes on the C2 domain of coagulation Factor VIII mapped by computer-designed synthetic peptides. Br. J. Haematol. 2011;155:487–497. doi: 10.1111/j.1365-2141.2011.08878.x. [DOI] [PubMed] [Google Scholar]
- 48.Lebreton A, Simon N, Moreau V, Demolombe V, Cayzac C, et al. Computer-predicted peptides that mimic discontinuous epitopes on the A2 domain of factor VIII. Haemophilia. 2015;21:e193–e201. doi: 10.1111/hae.12575. [DOI] [PubMed] [Google Scholar]
- 49.Walter JD, Werther RA, Brison CM, Cragerud RK, Healey JF, et al. Structure of the factor VIII C2 domain in a ternary complex with 2 inhibitor antibodies reveals classical and nonclassical epitopes. Blood. 2013;122:4270–4278. doi: 10.1182/blood-2013-08-519124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin JC, Ettinger RA, Schuman JT, Zhang AH, Wamiq-Adhami M, et al. Six amino acid residues in a 1200 A2 interface mediate binding of factor VIII to an IgG4kappa inhibitory antibody. PLoS One. 2015;10:e0116577. doi: 10.1371/journal.pone.0116577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nguyen PC, Lewis KB, Ettinger RA, Schuman JT, Lin JC, et al. High-resolution mapping of epitopes on the C2 domain of factor VIII by analysis of point mutants using surface plasmon resonance. Blood. 2014;123:2732–2739. doi: 10.1182/blood-2013-09-527275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perry LC, Jones TD, Baker MP. New approaches to prediction of immune responses to therapeutic proteins during preclinical development. Drugs R D. 2008;9:385–396. doi: 10.2165/0126839-200809060-00004. [DOI] [PubMed] [Google Scholar]
- 53.Bryson CJ, Jones TD, Baker MP. Prediction of immunogenicity of therapeutic proteins: validity of computational tools. BioDrugs. 2010;24:1–8. doi: 10.2165/11318560-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 54.Brinks V, Weinbuch D, Baker M, Dean Y, Stas P, et al. Preclinical models used for immunogenicity prediction of therapeutic proteins. Pharm. Res. 2013;30:1719–1728. doi: 10.1007/s11095-013-1062-z. [DOI] [PubMed] [Google Scholar]
- 55.Moise L, Song C, Martin WD, Tassone R, De Groot AS, et al. Effect of HLA DR epitope de-immunization of Factor VIII in vitro and in vivo. Clin. Immunol. 2012;142:320–331. doi: 10.1016/j.clim.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paul S, Kolla RV, Sidney J, Weiskopf D, Fleri W, et al. Evaluating the immunogenicity of protein drugs by applying in vitro MHC binding data and the immune epitope database and analysis resource. Clin. Dev. Immunol. 2013;2013:467852. doi: 10.1155/2013/467852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gunasekera D, Ettinger RA, Nakaya Fletcher S, James EA, Liu M, et al. Factor VIII gene variants and inhibitor risk in African American hemophilia A patients. Blood. 2015;126:895–904. doi: 10.1182/blood-2014-09-599365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Doering CB, Healey JF, Parker ET, Barrow RT, Lollar P. High level expression of recombinant porcine coagulation factor VIII. J. Biol. Chem. 2002;277:38345–38349. doi: 10.1074/jbc.M206959200. [DOI] [PubMed] [Google Scholar]
- 59.Zakas PM, Vanijcharoenkarn K, Markovitz RC, Meeks SL, Doering CB. Expanding the ortholog approach for hemophilia treatment complicated by factor VIII inhibitors. J. Thromb. Haemost. 2014;13:72–81. doi: 10.1111/jth.12755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lubin IM, Healey JF, Scandella D, Runge MS, Lollar P. Elimination of a major inhibitor epitope in factor VIII. J. Biol. Chem. 1994;269:8639–8641. [PubMed] [Google Scholar]
- 61.Lollar P. Characterization of factor VIII B-cell inhibitory epitopes. Thromb. Haemost. 1999;82:505–508. [PubMed] [Google Scholar]
- 62.Lollar P. Pathogenic antibodies to coagulation factors. Part one: factor VIII and factor IX. J. Thromb. Haemost. 2004;2:1082–1095. doi: 10.1111/j.1538-7836.2004.00802.x. [DOI] [PubMed] [Google Scholar]
- 63.Scandella D, Gilbert GE, Shima M, Nakai H, Eagleson C, et al. Some factor VIII inhibitor antibodies recognize a common epitope corresponding to C2 domain amino acids 2248 through 2312, which overlap a phospholipid-binding site. Blood. 1995;86:1811–1819. [PubMed] [Google Scholar]
- 64.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 65.Pratt KP, Qian J, Ellaban E, Okita DK, Diethelm-Okita BM, et al. Immunodominant T-cell epitopes in the factor VIII C2 domain are located within an inhibitory antibody binding site. Thromb. Haemost. 2004;92:522–528. doi: 10.1160/TH03-12-0755. [DOI] [PubMed] [Google Scholar]
- 66.Ettinger RA, James EA, Puranik K, Thompson AR, Matthews DC, Pratt KP. Factor VIII proteins having a rationally modified, immunodominant T-cell epitope demonstrate normal procoagulant activity, bind to VWF with high affinity, and are markedly less stimulatory to FVIII-specific human T cells. Blood. 2013;122(21):574. [Google Scholar]
- 67.Viel KR, Machiah DK, Warren DM, Khachidze M, Buil A, et al. A sequence variation scan of the coagulation factor VIII (FVIII) structural gene and associations with plasma FVIII activity levels. Blood. 2007;109:3713–3724. doi: 10.1182/blood-2006-06-026104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Viel KR, Ameri A, Abshire TC, Iyer RV, Watts RG, et al. Inhibitors of factor VIII in black patients with hemophilia. Engl. J. Med. N. 2009;360:1618–1627. doi: 10.1056/NEJMoa075760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lochan A, Macaulay S, Chen WC, Mahlangu JN, Krause A. Genetic factors influencing inhibitor development in a cohort of South African haemophilia A patients. Haemophilia. 2014;20:687–692. doi: 10.1111/hae.12436. [DOI] [PubMed] [Google Scholar]
- 70.Miller CH, Benson J, Ellingsen D, Driggers J, Payne A, et al. F8 and F9 mutations in US haemophilia patients: correlation with history of inhibitor and race/ethnicity. Haemophilia. 2012;18:375–382. doi: 10.1111/j.1365-2516.2011.02700.x. [DOI] [PubMed] [Google Scholar]
- 71.Schwarz J, Astermark J, Menius ED, Carrington M, Donfield SM, et al. F8 haplotype and inhibitor risk: results from the Hemophilia Inhibitor Genetics Study (HIGS) Combined Cohort. Haemophilia. 2013;19:113–118. doi: 10.1111/hae.12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lei TC, Scott DW. Induction of tolerance to factor VIII inhibitors by gene therapy with immunodominant A2 and C2 domains presented by B cells as Ig fusion proteins. Blood. 2005;105:4865–4870. doi: 10.1182/blood-2004-11-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lei TC, Su Y, Scott DW. Tolerance induction via a B-cell delivered gene therapy-based protocol: optimization and role of the Ig scaffold. Cell. Immunol. 2005;235:12–20. doi: 10.1016/j.cellimm.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 74.Skupsky J, Saltis M, Song C, Rossi R, Nelson D, et al. Gene therapy for tolerance and vice versa: a case for hemophilia. Curr. Opin. Mol. Ther. 2010;12:509–518. [PubMed] [Google Scholar]
- 75.Mahlangu J, Powell JS, Ragni MV, Chowdary P, Josephson NC, et al. Phase 3 study of recombinant factor VIII Fc fusion protein in severe hemophilia A. Blood. 2014;123:317–325. doi: 10.1182/blood-2013-10-529974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shapiro AD, Ragni MV, Kulkarni R, Oldenberg J, Srivastava A, et al. Recombinant factor VIII Fc fusion protein: extended-interval dosing maintains low bleeding rates and correlates with von Willebrand factor levels. J. Thromb. Haemost. 2014;12:1788–1800. doi: 10.1111/jth.12723. [DOI] [PubMed] [Google Scholar]
- 77.De Groot AS, Moise L, McMurry JA, Wambre E, Van Overtvelt L, et al. Activation of natural regulatory T cells by IgG Fc-derived peptide “Tregitopes”. Blood. 2008;112:3303–3311. doi: 10.1182/blood-2008-02-138073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim YC, Zhang AH, Su Y, Rieder SA, Rossi RJ, et al. Engineered antigen-specific human regulatory T cells: immunosuppression of FVIII-specific T- and B-cell responses. Blood. 2015;125:1107–1115. doi: 10.1182/blood-2014-04-566786. [DOI] [PMC free article] [PubMed] [Google Scholar]