Introduction
Deletion of the long arm of chromosome 4 has been reported with variable clinical findings including developmental delay and intellectual disability. We report an individual with a 4q35.2 deletion (1.2 Mb in size) detected by chromosomal microarray analysis, which to our knowledge, is the first individual with a submicroscopic interstitial deletion identifying three genes implicated in his clinical presentation and findings.
Clinical report
The proband was born to nonconsanguineous parents following a 33-week gestation complicated by maternal diabetes, alcohol use during the first trimester, and premature rupture of membranes. His apgar scores were 6 and 6 at 1 and 5 min, respectively, and his birth weight was 3.19 kg (25th centile). Because of respiratory distress, he received mechanical ventilation for 1 day following delivery and phototherapy for jaundice and hyperbilirubinemia until 6 days. A left cephalohematoma and bruising were reported at birth. His hematocrit on day 7 was 34.4. An echocardiogram at 2 days of age showed a large left-to-right patent ductus arteriosus and a small left-to-right atrial shunt. Repeat echocardiogram at 6 days of age continued to show a patent ductus arteriosus, patent foramen ovale, right ventricular hypertrophy, flattened septum, tricuspid regurgitation, and mildly increased cardiac wall thickness. No surgical intervention was recommended. The head MRI scan was interpreted as normal. During early childhood, autism, attention-deficit hyperactivity disorder, obsessive compulsive disorder, and intellectual disability were diagnosed along with posttraumatic stress disorder, self-injury, pica, and aggression. Sleep disturbances were identified by 3 years of age when he would not sleep until about 11:00 pm daily and for only 1–2 h at a time. He currently sleeps for only 4 h per night. The irregular sleep patterns were treated with melatonin but with limited success. His history of sensitivity to red food coloring and easy bruisability was noted throughout life. His developmental milestones were delayed with walking at 17 months and talking at 27 months. The Preschool Language Scale-3 was performed at the age of 32 months and he scored at the first percentile or an age equivalent of 10–12 months for auditory comprehension, expressive communication, and total language score. At the age of 4 years and 6 months, the Vineland Adaptive Behavior Scale showed age equivalent scores ranging from 17 months (socialization) to 32 months (motor skills). The Childhood Autism Rating Scale generated an overall score of 41 with a score greater than 36 consistent with autism. The Autism Diagnostic Observation Schedule-Generic generated an overall score of 15 with the cut-off score for autism being 12 or higher.
The hematogram at the age of 1 year showed a white blood cell count of 7300, red blood cell count of 5.19, hemoglobin of 14.1, hematocrit of 41.6, and platelet count of 217 000. At 4 years and 7 months, the hematogram was normal including a platelet count of 302 000. At 12 years his complete blood count was normal (platelets 198 K/μl), his electrolytes were normal, prothrombin time was normal at 13.7 s, and partial thromboplastin time, activated was normal at 31s.
Results
On physical examination at the age of 12 years and 9 months, he was quiet and cooperative with height 147.5 cm (20th centile), weight 37.6 kg (20th centile), and head circumference 52.5 cm (25th centile). Inner canthal distance was 3.4 cm (85th centile), outer canthal distance was 7.7 cm (10th centile), interpupillary distance was 5.7 cm (75th centile), and palpebral fissure length was 2.5 cm (25th centile). Ear length was 5.7 cm (40th centile). He had flattening of the right occiput compared with the left, which may be related to head positioning as an infant. His hair texture was coarse with an abnormal single posterior hair whorl. He manifested malar hypoplasia, brushfield spots, a short nose with anteverted nares, a smooth and flat philtrum, a thin upper lip, a broad mouth, and prognathism (Fig. 1). He had a high-arched palate. His hand length was 15.7 cm (10th centile) and top finger length was 6.4 cm (10th centile). He had fifth finger clinodactyly, hyperflexible fingers, and shallow nail beds. He had short fifth toes bilaterally, left sided nail hypoplasia of the fifth toe, toe malalignment, and length discrepancy with toes on the right appearing longer than the toes on the left. A large ecchymotic area (6 8 cm) was present on the right lateral knee area not related to recognized trauma or skin abrasion.
Fig. 1.
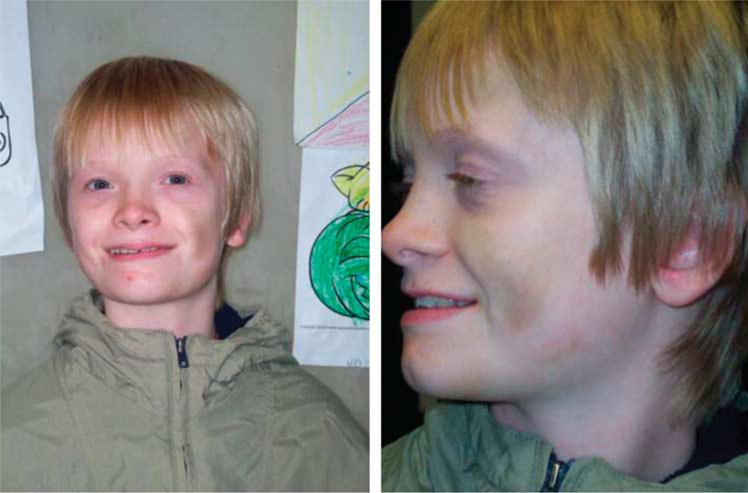
Frontal and profile facial views of the proband at the age of 12 years showing malar hypoplasia, a short nose with anteverted nares, a smooth and flat philtrum, a thin upper lip, prognathism, and a broad mouth. Note the ecchymotic area on the left cheek.
High resolution chromosome and fragile X DNA analyses were normal, but later, chromosomal microarray analysis was carried out clinically (CMDX laboratory, Irvine, California, USA) using the 105 K Oligo HD Scan to rule out microdeletions or microduplications. The microarray study showed a 1.2 Mb deletion (187.47–188.66 Mb from the p-terminus) of the chromosome 4q35.2 band, verified using fluorescent in-situ hybridization analysis (Fig. 2). The participant’s parents were unavailable for study.
Fig. 2.
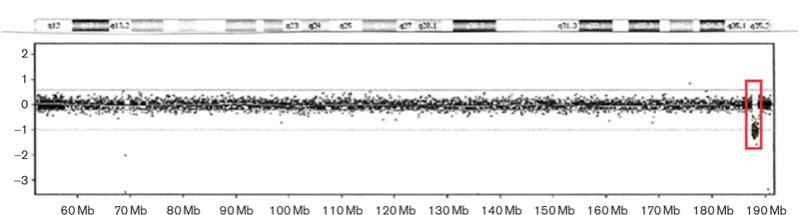
Chromosome microarray hybridization showing the location of the 1.2 Mb deletion of the 4q35.2 region occurring at 187.47–188.66 Mb from the p-terminus. The deleted region is indicated by the inserted box.
Discussion
Interestingly, three genes are known to be located within the chromosome band including the melatonin receptor 1A (MTNR1A), a high affinity melatonin receptor that likely mediates melatonin receptors in the hypothalamic suprachiasmatic nucleus or site of the circadian clock (Slaugenhaupt et al., 1995). Wu et al. (2006), reported that melatonin receptor 1A is expressed in many nuclei of the human hypothalamus, suggesting that melatonin may directly modulate hypothalamic physiological processes through this receptor. A disruption in this gene could lead to irregular and disruptive sleep patterns seen in the proband. The fat tumor suppressor (Drosophila) homolog 1 (FAT1) gene, located in this chromosome region, is involved in nerve cell adhesion and is part of the cadherin gene family, important in many developmental processes (Dunne et al., 1995). Disturbance of the FAT1 gene is associated with both bipolar affective disorders and autism (Abou et al., 2008; Chien et al., 2010). Although the assigned deletion includes both the MTNR1A and FAT1 genes, it is located very near to the 3′ end of the F11 gene involved with blood coagulation and positioned at 187 187 118–187 210 835 bp. The F11 gene participates in blood coagulation as a catalyst in the conversion of factor XI to factor XIa in the presence of calcium ions (Asakai et al., 1987). The F11 gene may be impacted by the deletion process as reported in other deletions of chromosome regions through disruption of cis-regulation of neighboring genes (Bittel et al., 2003, 2005). Loss of function of this gene may contribute to the history of easy bruisability in the proband and the large ecchymotic area observed during the examination.
Several other cases of terminal 4q deletions have been described including a 7-year-old nondysmorphic boy with mild intellectual disability reported by Balikova et al. (2007) due to a 4q35.2 qter deletion (about 1.3 Mb in size) detected with a 32 K Bacterial Artificial Chromosome array (see Table 1 for summary). In addition, array comparative genomic hybridization was used to identify a de-novo terminal deletion of the long arm of chromosome 4, del(4)(q32.3) in a 5-year-old girl with facial and digital dysmorphism, a complex congenital heart defect, a large occipital encephalocele, postnatal growth deficiency, and developmental delay (Quadrelli et al., 2007). In 2008, a 3-year-old boy with dysmorphic facial features, fifth finger clinodactyly, hypospadias, and severe developmental delay was reported to have a de-novo distal deletion of 4q33, detected at about 20 Mb in size by using a 1 M resolution array comparative genomic hybridization and subsequent fluorescent in-situ hybridization analysis (Kitsiou-Tzeli et al., 2008). A 6-year-old boy presenting with short stature, eczema, and developmental delay was reported with an interstitial deletion at 4q33–q35 (Mdzin et al., 2008). More recently, Rossi et al. (2009) reported a 17-year-old girl with a 16.4 Mb deletion of 4q34.1–q35.2 presenting with Pierre–Robin sequence, cardiac abnormalities, and developmental delay. An individual reported by Chien et al. (2010) with hypotonia during infancy and autism was found to have a 4q35.1–35.2 deletion, which included the region deleted in our proband. The above patients possessed distal 4q deletions that overlapped with the proband, but no reports of individuals with an interstitial deletion were identical to the deletion found in the proband.
Table 1.
Sumamry of clinical, developmental and behavioral features
| Quadrelli et al., 2007 | Balikova et al., 2007 | Kitsiou-Tzeli et al., 2008 | Mdzin et al., 2008 | Rossi et al., 2009 | Chien et al., 2010 | Present Case | |
|---|---|---|---|---|---|---|---|
| Features | |||||||
| Age/Sex | 5 years/F | 7 years/M | 3 years/M | 6 years/M | 17 years/F | 8 years/M | 12 years/M |
| Chromosome 4q proximal breakpoint | q32.3 | q35.2 | q33 | q33 | q34.1 | q35.1 | q35.2 |
| Chromosome 4q distal breakpoint | qter | qter | qter | q35 | q35.2 | q35.2 | q35.2 |
| Deletion size (Mb) | 25.7 | 1.3 | 18.9–22.9 | NA | 16.4 | 6.8 | 1.2 |
| FTT/growth retardation/short stature | Yes | No | No | Short stature only | Short stature only | No | No |
| Head circumference | Microcephaly | Normal | Normal | Normal | Normal | Normal | Normal |
| Abnormal skull | Large fontanelle | No | No | No | No | No | No |
| Ear anomaly | Over-folded | No | Protruding, lowset; tags | No | No | No | No |
| Hypertelorism | Yes | No | Yes | No | Yes | No | No |
| Broad nasal bridge | No | No | Yes | Yes | Yes | No | Yes |
| Thin upper lip | No | No | Yes | No | Yes | No | Yes |
| Microstomia | Yes | No | No | Yes | No | No | No |
| Chin anomaly | Micrognathia | No | Microretro gnathia | No | Micrognathia | No | Prognathia |
| Cardiovascular defect | PDA; VSD; coarctation of aorta | No | AS; dysplastic; pulmonic valve | No | Ebstien anomaly; ASD | No | PDA |
| Genital anomalies | Abnormal labia minor; prominent clitoris | No | Hypospadias | No | No | No | No |
| Digital anomalies | Overtapering fingers; fifth clinodactyly; malaligned toes | No | Fifth finger clinodactyly; tapering fingers | Absent IP joint on 5th finger; hypoplastic nails | Fifth finger clinodactyly | No | Fifth finger clinodactyly; hyperflexible; toe malalignment |
| Vision and hearing problems | No | No | No | No | Myopia | No | No |
| Developmental delay/intellectual disability | Mild | Mild | Severe | Moderate | Mild | Motor and speech delay | Severe |
| Neurological/behavioral problems | Occipital encephalocele; Arnold–Chiari malformation | No | Normal MRI; EEG; ultrasound; hypotonia; feeding difficulties | No | ADHD; depression; bouts of anger | Hypotonia | ADHD; aggression |
| Autism | No | No | No | No | No | Yes | Yes |
| Other | Facial hemangioma; short, upward slanting palpebral fissures | U-shaped cleft palate; long philtrum; short nose with anteverted nares; low posterior hair line; inguinal hernia | Eczema on feet; epicanthal folds; long philtrum; widely spaced nipples; anteverted nares | Nystagmus; short nose; Pierre–Robin sequence; primary amenorrhea | Sleep disorder; easy bruising |
ADHD, attention deficit hyperactivity disorder; AS, aortic supravalvular membrane; F, female; FTT, failure to thrive; IP joint, interphalangeal joint; M, male; NA, not available; PDA, patent ductus arteriosus; VSD, ventral septal defect.
One could speculate that the findings in the proband may fit the definition of a contiguous gene deletion syndrome as facial, cardiac, digital, and genital anomalies are often seen including Pierre–Robin sequence, cognitive, neurological, and behavioral problems. Fragile sites are reported in this chromosome region which may contribute to the deletion breakpoints (Sutherland and Hecht, 1985; Kahkonen 1988; Butler, 1998). The authors encourage the reporting of other individuals with deletions involving the long arm of 4q to better define the clinical presentation and the use of chromosomal microarray studies for genotype–phenotype correlations.
Footnotes
Conflicts of interest
There are no conflicts of interest.
References
- Abou JR, Becker T, Georgi A, Feulner T, Schumacher J, Stromaier J, et al. Genetic variation of the FAT gene at 4q35 is associated with bipolar affective disorder. Mol Psych. 2008;13:277–284. doi: 10.1038/sj.mp.4002111. [DOI] [PubMed] [Google Scholar]
- Asakai R, Davie EW, Chung DW. Organization of the gene for human factor XI. Biochem. 1987;26:7221–7228. doi: 10.1021/bi00397a004. [DOI] [PubMed] [Google Scholar]
- Balikova I, Menten B, de Ravel T, Le Caignec C, Thienpont B, Urbina M, et al. Subtelomeric imbalances in phenotypically normal individuals. Hum Mutat. 2007;28:958–967. doi: 10.1002/humu.20537. [DOI] [PubMed] [Google Scholar]
- Bittel DC, Kibiryeva N, Talebizadeh Z, Butler MG. Microarray analysis of gene/transcript expression in Prader–Willi syndrome: deletion versus UPD. J Med Genet. 2003;40:568–574. doi: 10.1136/jmg.40.8.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittel DC, Kibiryeva N, Talebizadeh Z, Driscoll DJ, Butler MG. Microarray analysis of gene/transcript expression in Angelman syndrome: deletion versus UPD. Genomics. 2005;85:85–91. doi: 10.1016/j.ygeno.2004.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MG. Frequency and distribution of chromosome fragile sites or lesions in males with mental retardation: a descriptive study. J Tennessee Acad Sci. 1998;73:87–99. [PMC free article] [PubMed] [Google Scholar]
- Chien WH, Gau SSF, Wu YY, Huang US, Fang JS, Chen YJ, et al. Identification and molecular characterization of two novel chromosomal deletions associated with autism. Clin Genet. 2010;78:449–456. doi: 10.1111/j.1399-0004.2010.01395.x. [DOI] [PubMed] [Google Scholar]
- Dunne J, Hanby AM, Poulsom R, Jones TA, Sheer D, Chin WG, et al. Molecular cloning and tissue expression of FAT, the human homologue of the Drosophila fat gene that is located on chromosome 4q34–q35 and encodes a putative adhesion molecule. Genomics. 1995;30:207–223. doi: 10.1006/geno.1995.9884. [DOI] [PubMed] [Google Scholar]
- Kahkonen M. Population cytogenetics of folate-sensitive fragile sites. I. Common sites. Hum Genet. 1988;80:344–348. doi: 10.1007/BF00273649. [DOI] [PubMed] [Google Scholar]
- Kitsiou-Tzeli S, Sismani C, Koumbaris G, Ioannides M, Kanavakis E, Kolialexi A, et al. Distal del(4) (q33) syndrome: detailed clinical presentation and molecular description with array-CGH. Eur J Med Genet. 2008;51:61–67. doi: 10.1016/j.ejmg.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Mdzin R, Ko C, Abdul Latif Z, Zakaria Z. Interstitial deletion of the distal long arm of chromosome 4, del (4)(q33–35), in association with paternal balanced translocation. Singapore Med J. 2008;49:336–339. [PubMed] [Google Scholar]
- Quadrelli R, Strehle EM, Vaglio A, Larrandaburu M, Mechoso B, Quadrelli A, et al. A girl with del(4)(q33) and occipital encephalocele: clinical description and molecular genetic characterization of a rare patient. Genet Test. 2007;11:4–10. doi: 10.1089/gte.2006.9995. [DOI] [PubMed] [Google Scholar]
- Rossi MR, DiMaio MS, Xiang B, Lu K, Kaymakcalan H, Seashore M, et al. Clinical and genomic characterization of distal duplications of chromosome 4q: study of two cases and review of the literature. Am J Med Genet Part A. 2009;149A:2788–2794. doi: 10.1002/ajmg.a.33088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaugenhaupt SA, Roca AL, Liebert CB, Altherr MR, Gusella JF, Reppert SM. Mapping of the gene for the Mel1a-melatonin receptor to human chromosome 4 (MTNR1A) and mouse chromosome 8 (Mtnr1a) Genom. 1995;27:355–357. doi: 10.1006/geno.1995.1056. [DOI] [PubMed] [Google Scholar]
- Sutherland GR, Hecht F. Fragile sites on human chromosomes. New York, New York: Oxford University Press; 1985. [Google Scholar]
- Wu YH, Zhou JN, Balesar R, Unmehopa U, Bao A, Jockers R, et al. Distribution of MT1 melatonin receptor immunoreactivity in the human hypothalamus and pituitary gland: colocalization of MT1 with vasopressin, oxytocin, and corticotropin-releasing hormone. J Comp Neurol. 2006;499:897–910. doi: 10.1002/cne.21152. [DOI] [PubMed] [Google Scholar]


