Abstract
Autoimmune diseases affect 7.5% of the U.S. population, and are among the leading causes of death and disability. A striking feature of many autoimmune diseases is their increased prevalence in females, but the underlying mechanisms have remained unclear. Using high-resolution global transcriptome analyses we demonstrate a female-biased molecular signature associated with autoimmune disease susceptibility, and linked to extensive sex-dependent, co-expression networks. This signature was independent of biological age and sex-hormone regulation, and regulated by the transcription factor VGLL3, which also had a strong female biased expression. On a genome-wide level, VGLL3-regulated genes had a strong association with multiple autoimmune diseases including lupus, scleroderma and Sjögren’s syndrome and had a prominent transcriptomic overlap with inflammatory processes in cutaneous lupus. These results identify VGLL3-regulated gene network as a novel inflammatory pathway promoting female-biased autoimmunity, they demonstrate the importance of studying immunological processes in females and males separately, and open up new avenues for therapeutic development.
Introduction
Autoimmune diseases are characterized by immune responses to self-antigens that result in tissue damage. It is estimated that autoimmune diseases affect 7.5% of the U.S. population, compared to 2.8% for cancer and 6.9% for heart diseases, and are among the leading causes of death and disability. Currently there is no cure, and commonly used immunosuppressant treatments can lead to devastating side effects such as serious infections and cancer1–3.
Many autoimmune diseases feature increased prevalence in females, ranging from systemic disorders such as systemic lupus erythematosus (SLE) (female-to-male ratio 9:1) to organ-specific diseases such as Grave’s disease (female-to-male 7:1)2,3, whereas infectious disease risks are higher in men4. Overall 78% of the people affected with autoimmune diseases are women2,3. Sex hormones are among the most studied factors for contributions to this sex bias. The role of sex hormones has been best studied in mouse models of SLE where androgen is protective whereas estrogen accelerates disease5. In humans, however, the relationship between sex hormones and autoimmunity appears to be more complicated. For example, when SLE occurs in men, the disease is often more severe, and many autoimmune diseases commonly have their onset before puberty or after menopause5–7. In addition, there is evidence that post-menopausal hormonal therapy does not increase disease activity or the risk of major flares in women with SLE5,8,9.
Skin as the biggest organ in human is the front line of immune protection and is a sensitive indicator of immune dysregulation10. Skin changes are prominently manifested in autoimmune diseases such as SLE. Of the eleven criteria for the diagnosis of SLE, four are cutaneous in nature, namely malar rash (butterfly-shaped rash across cheeks and nose), discoid rash (raised red patches), photosensitivity (skin rash as a result of unusual reaction to sunlight) and mucosal ulcers. Collectively, skin involvement is present in 72–85% of SLE patients11. Systemic sclerosis, an autoimmune disease with a female-to-male prevalence ratio of 11:1, is characterized by skin symptoms including thickening and itching12.
To understand the cause of female-biased susceptibility of autoimmune diseases in humans, we investigated the sexual dimorphisms of human skin. We identified a female-biased molecular signature significantly associated with autoimmune diseases susceptibility. Sex differences extended beyond the signature to genome-wide co-expression networks involving processes such as complement activation and phagocytosis. We further identified VGLL3, one of the sex-biased transcription factors uncovered in our analyses, as a critical regulator of the female-biased inflammatory genes, including BAFF/TNFSF13B and ITGAM, encoding for the integrin molecule αM, which is a therapeutic target13 and a genetic risk factor for SLE14, respectively. On a genome-wide level, VGLL3 targets had a strong association with multiple autoimmune diseases including lupus, systemic sclerosis and Sjögren’s syndrome and had a prominent transcriptomic overlap with inflammatory processes in cutaneous lupus. VGLL3 was also required for the optimal response to interferons (IFN) in monocytes and salivary gland cells. Our results uncovered a sex-hormone independent mechanism predisposing females to autoimmune diseases, and provided a foundation towards development of novel, targeted treatment measures.
Results
Sex differences in human skin
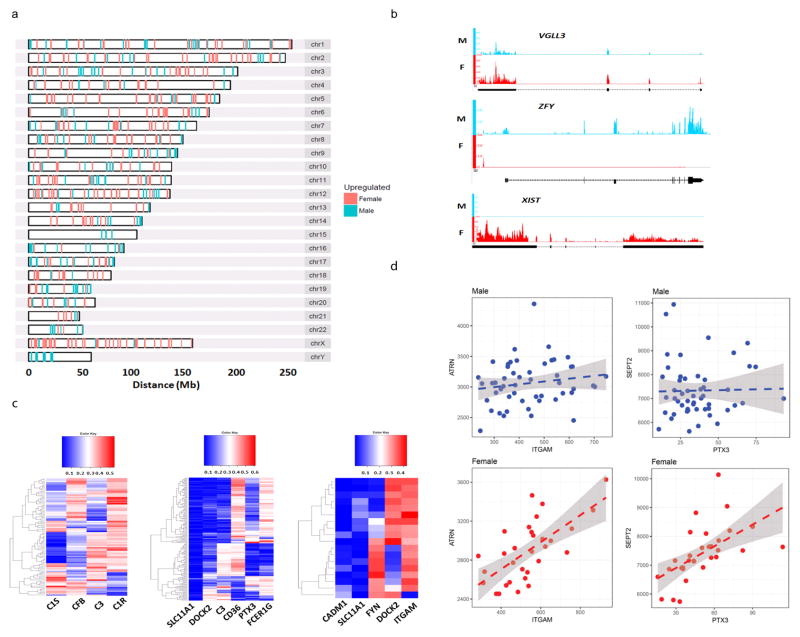
We analyzed 31 female and 51 male skin biopsy samples from healthy donors by whole genome RNA-sequencing (RNA-seq). We identified 661 genes differentially expressed between the two sexes (FDR ≤ 0.1) (Supplementary Table 1). 268 genes were upregulated in males (i.e. male-biased), including 26 genes on the Y chromosome and 6 genes on the X. 393 genes were upregulated in females (i.e. female-biased), including 55 genes on the X chromosome (Fig. 1a). As expected, known sex-biased expression, such as that of XIST and ZFY, was reproduced in our datasets (Fig. 1b). Of the 55 genes that escaped X-inactivation, 7 have orthologues on the Y chromosome, whose expression in males potentially enables dosage compensation (Supplementary Fig. 1a–d). 48 genes did not have Y-linked orthologues, supporting that incomplete X-inactivation may contribute to sexually dimorphic traits (Supplementary Fig. 1e–g).
Figure 1. Identification of sex-biased genes from human skin biopsies.
a, chromosomal locations of female- and male-biased genes. b, raw RNA-Seq reads in female and male skin for XIST, ZFY, and VGLL3. c, sex-biased co-expression correlation for genes in the complement activation, phagocytosis regulation, T cell proliferation functional categories, respectively (left-to-right). d, sex-specific co-expression correlation for the ITGAM-ATRN and PTX3-SEPT2 gene pairs.
Sex differences may extend beyond the 661 differentially expressed genes (DEGs) to their associated networks. To test this, we conducted gene-gene correlation analysis between the sex DEGs against all other genes in male and female, separately (details described in Methods). Indeed, sex-biased co-expression correlations were found from the gene pair level to pathway- and genome-wide levels, involving genes in various immunological processes such as phagocytosis and complement activation (Fig. 1c, d, Supplementary Fig. 2a). In total, we identified 124,521 gene-gene pairs that only showed significant results in female samples (FDR ≤ 0.1) but not in male samples (P-value > 0.5); conversely, 158,303 gene-gene pairs showed only significance in male samples (FDR ≤ 0.1) but not in female samples (P -value > 0.5). We further compared the correlation results with those obtained from published skin microarray datasets15, and obtained high correlation concordance (Supplementary Fig. 2b). This finding indicates the presence of sex-biased, genome-wide networks and suggests that the biologic effect of the sex bias is much greater than what can be anticipated from the initial DEG list.
Association between female-biased genes and autoimmunity
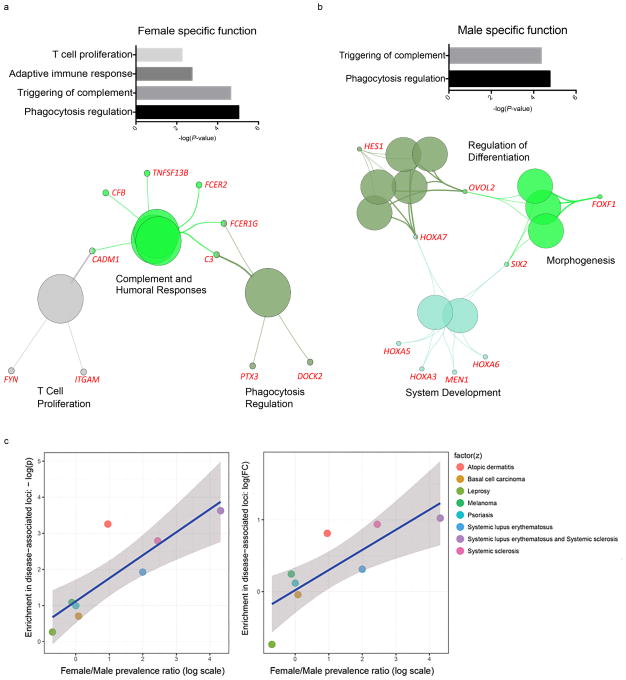
Analyses of biological functions enriched in the DEGs revealed that female-, but not male-, biased genes were enriched for immunological and inflammatory processes (Fig. 2a,b, Supplementary Fig. 3a). In addition, network analysis primarily organized female-biased genes into complement activation pathways that are known to be dysregulated in autoimmune diseases (Fig. 2a).
Figure 2. Female-biased genes are associated with autoimmune processes.
a, functional categories enriched in female-biased genes. b, functional categories enriched in male-biased genes. c, correlation between enrichment in disease-associated loci with female/male disease prevalence ratio for female-biased DEGs.
The sex-specific up-regulation of immune genes in females led us to hypothesize that the female-biased gene signature associates with high susceptibility to autoimmune diseases. We detected significant overlap between female-biased genes and common disease loci of SLE and systemic sclerosis, two female-dominant autoimmune diseases (P < 0.05, Fig. 2c). Among female-biased genes, female-to-male prevalence ratio is significantly correlated with the enrichment of disease-associated loci, measured by P-value (ρ = 0.83; P = 1.5 × 10−2) and observed-to-expected fold change (ρ = 0.88; P = 7.2 × 10−3; Fig. 2c). There was no association between male-biased genes and autoimmune diseases (data not shown). We also implemented a sampling approach to estimate the empirical P-values for the enrichment, and the results are highly concordant with the hypergeometric enrichment analysis (Supplementary Fig. 3b,c; details described in Methods).
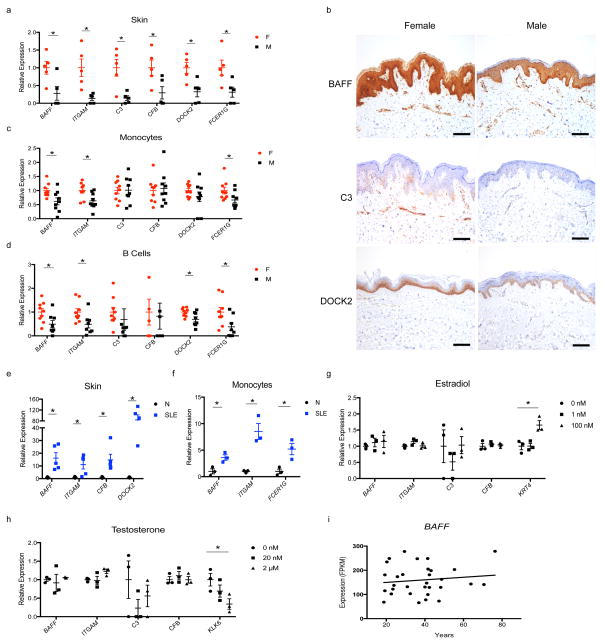
We confirmed the increased expression of the immune genes in female skin (Fig. 3a,b), including BAFF (also known as TNFSF13B), which is frequently elevated in SLE patients and served as the first approved target for biologic therapy for SLE13, and ITGAM, whose variants are associated with SLE susceptibility14. Consistent with the systemic feature of SLE symptoms, the female-biased pattern of risk gene expression was not restricted to skin, but was also detected to variable degrees in monocytes, B cells, and T cells (Fig. 3c,d, Supplementary Fig. 3d). We further observed an elevation in their expression in skin and monocytes from SLE patients compared to sex-matched healthy controls (Fig. 3e,f), supporting their involvement in SLE pathogenesis. Collectively, these results suggest that the female-biased inflammatory genes associate with high susceptibility to autoimmune processes.
Figure 3. Female-biased immune gene expression is dependent on SLE disease states but not sex hormone levels.
a, qRT-PCR of female-biased immune genes in whole skin of healthy humans (n=5 each sex). b, immunohistochemistry images of female-biased immune genes in skin of healthy humans. Scale bar, 50 μm. c, qRT-PCR of female-biased immune genes in monocytes of healthy humans (n=9 each sex). d, qRT-PCR of female-biased immune genes in B cells of healthy humans (n=9 female, 8 male). e, qRT-PCR of female-biased immune genes in skin of SLE patients (SLE) and healthy subjects (N) (n=5 each). f, qRT-PCR of female-biased immune genes in monocytes of SLE patients (SLE) and healthy subjects (N) (n=3 each). g, gene expression by RNA-Seq in primary human keratinocytes with or without estradiol treatment (n=3 independent experiments). h, gene expression by RNA-Seq in primary human keratinocytes with or without testosterone treatment (n=3 independent experiments). i, scatter plot of BAFF expression from RNA-Seq of human skin biopsies versus age at biopsy. Female, F. Male, M. Mean ± s.e.m, * P<0.05, Student’s t-test.
Molecular mechanism for female-biased risk gene expression
To search for the molecular mechanism underlying sex-biased risk gene expression, we examined the effects of physiological or 100-fold concentrated estradiol or testosterone on gene expression by RNA-Seq in primary human keratinocytes. The sex hormones did not alter the expressions of the female-biased immune genes (Fig. 3g,h). More broadly, none of the 661 sex-DEGs were significantly regulated by estradiol or testosterone in the given settings (data not shown). To address the possibility that keratinocytes lose their responsiveness to sex hormones upon ex vivo culture, we turned to our transcriptomics data from skin and reasoned that the gene expression should decrease with age if they were regulated by sex hormones. We observed no correlation between expression and biological age for the genes investigated (Fig. 3i, Supplementary Fig. 3e–i). Overall, we found no compelling evidence supporting the direct regulation of female-biased risk genes by sex hormones.
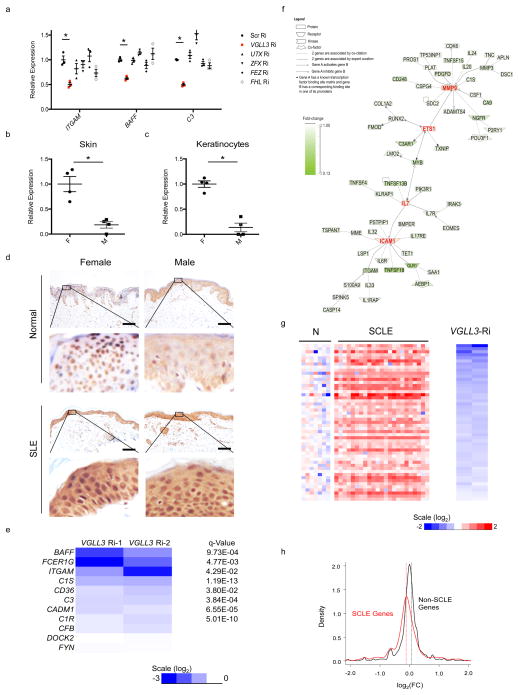
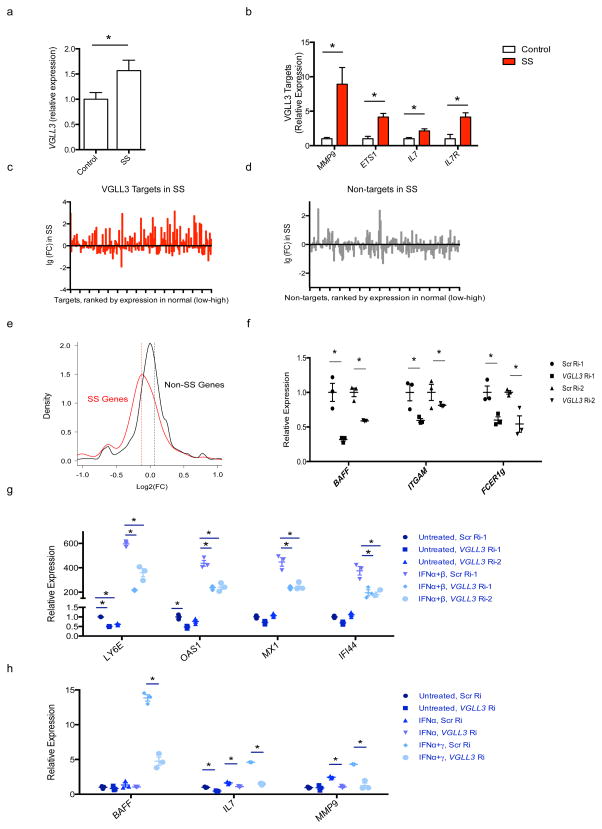
Another potential mechanism could be that the risk genes are regulated by sex-biased transcription factors. We identified 8 putative female-biased transcription factors based on their annotated function from the top 100 genes that are most significantly female-biased (ranked by FDR, Supplementary Tables 1,2). 6 of the 8 genes were expressed in keratinocytes based on transcriptomic analyses of 3 different female primary keratinocytes (Supplementary Table 2). We were able to achieve efficient knockdown for 5 of the 6 genes by RNAi (Supplementary Fig. 4a–e; Supplementary Table 2). We found that RNAi of VGLL3, but not UTX, ZFX, FEZ or FHL, decreased ITGAM and BAFF mRNA abundance (Fig. 4a, Supplementary Fig. 4a–e). VGLL3 knockdown did not affect the expression of UTX or ZFX, suggesting its effect on these SLE-associated genes is specific (Supplementary Fig. 4f).
Figure 4. VGLL3 regulates genes associated with autoimmune diseases.
a, qRT-PCR of ITGAM, BAFF, and C3 upon RNAi of scrambled siRNA (Scr), VGLL3, UTX, ZFX, FEZ, FHL in primary human keratinocytes (n=3 independent experiments). b, qRT-PCR of VGLL3 in normal female (F) and male (M) skin (n=4 each). c, qRT-PCR of VGLL3 in primary human keratinocytes (n=4 each). d, immunohistochemistry of VGLL3 in healthy (Normal) and SLE patient (SLE) skin. Scale bar, 50 μm. e, log2(FC) and q-Value of the 10 female-biased immune transcripts upon VGLL3 RNAi in primary human keratinocytes from RNA-Seq experiments. f, literature-based network analysis of VGLL3-regulated autoimmune disease genes. g, log2(FC) levels of VGLL3 targets in skin of healthy (N) subjects and SCLE patients (SCLE) as well as upon VGLL3 RNAi. h, density plot of log2(FC) levels upon VGLL3 knockdown for SCLE and non-SCLE genes. P = 2.53 × 10−8. Mann-Whitney-Wilcoxon test. (a–c) Mean ± s.e.m, *, P<0.05, Student’s t-test.
VGLL3 is a homologue of the vestigial gene of Drosophila, a cofactor of the TEF-1 (transcription enhancer factor-1) homologue Scalloped16, and it exhibits sex-dependent dominance in salmon17. The elevated expression of VGLL3 (FDR = 7.2 × 10−4) in females was confirmed in skin and in keratinocytes (Fig. 4b,c). In healthy skin, consistent with its transcriptional functions, VGLL3 exhibited a nuclear localization that is more distinct in females than in males (Fig. 4d). By contrast in lesional SLE skin, VGLL3 was concentrated in the nucleus in both sexes, indicating disease-dependent regulation (Fig. 4d).
RNA-Seq of female primary human keratinocytes showed that in addition to BAFF and ITGAM, VGLL3 knockdown down-regulated 7 out of 10 female-biased immune genes that were expressed in proliferating or post-confluent keratinocytes (Fig. 4e). At the q<0.05, |log2FC|>0.5 threshold, there were a total of 208 genes decreased by VGLL3 RNAi in keratinocytes (Supplementary Table 3). To test if genetic variants that affect the functions or expression of VGLL3 would also affect the expression of VGLL3 targets, we conducted eQTL analysis surrounding the ±1 Mb region of VGLL3 (details described in Methods). The strongest cis-eQTL signal was found at chr3:87902673 (P = 4 × 10−5) (Supplementary Fig. 5a). Furthermore, we identified nine targets of VGLL3 that were significantly associated with the VGLL3-associated cis-eQTLs, indicating trans-eQTL effect (Supplementary Fig. 5b). We observed strong enrichment for VGLL3 targets among the female-biased genes (P = 7.7 × 10−7), but not male-biased genes. All of the top 10 pathways enriched in VGLL3 targets were immune-related (Supplementary Fig. 5c), and the top three diseases enriched included autoimmune diseases (97 genes/47%, P = 3.63 × 10−12). Network analysis of the 97 genes revealed additional nodes of autoimmune pathogenesis (Fig. 4f). MMP-9 is elevated in patients of SLE, systemic sclerosis, multiple sclerosis (MS), Sjögren’s syndrome (SS), polymyositis, and rheumatoid arthritis (RA)18. ETS1 variants confer susceptibility to SLE, RA, and ankylosing spondylitis from genome-wide association studies19. Excess interleukin 7 is present in patients of Sjögren’s syndrome, MS and RA, promotes autoimmunity in lupus mice, and predicts clinical response to IFN-β in MS20–22. The adhesion molecule ICAM-1 is upregulated in MS patient brains, murine lupus nephritis and arthritis, and has been advocated for controlling autoimmune diseases23–25. Collectively these lines of evidence support a role of VGLL3 in promoting the expression of multiple autoimmune disease genes.
VGLL3 targets in autoimmune diseases
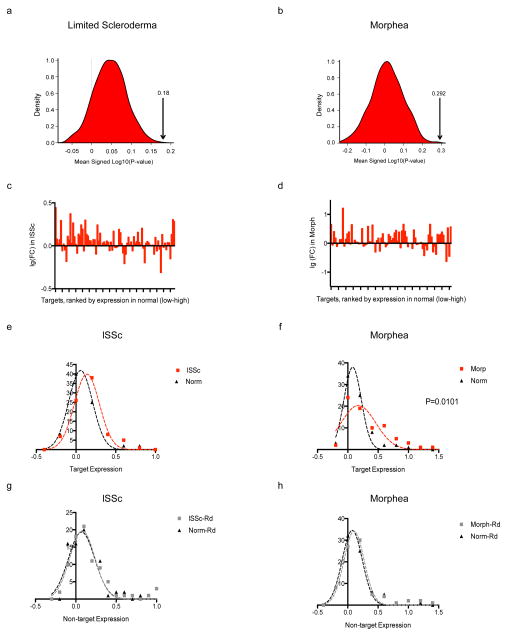
If VGLL3 contributes to higher susceptibility to autoimmune diseases in females, one prediction is that VGLL3 targets are associated more tightly with such female-biased diseases compared to diseases that do not exhibit significant sex differences. To test this, we performed transcriptomic analyses on skin biopsies from subacute cutaneous lupus erythematosus (SCLE), morphea, and systemic sclerosis patients. SCLE is a female-biased, lupus-specific eruption that features prominent skin involvement. We first attempted to identify the subset of VGLL3 targets upregulated in SCLE, using less stringent criteria to be inclusive at the identification stage. Crossing of VGLL3 targets (genes decreased by VGLL3 RNAi at the q<0.05, FC < 0.8 threshold) with gene sets dysregulated in SCLE (genes upregulated in SCLE at the q<0.05, FC ≥1.5 threshold) revealed a significant overlap (P = 2.9 × 10−5), specifically 51 SCLE-increased genes that were positively regulated by VGLL3 (Fig. 4g; Supplementary Table 4). The overlap included bona-fide type I IFN (IFN-α and IFN-β) response genes LY6E, OAS1, MX1, and IFI44 (refs. 26,27), consistent with the central role of type I IFN in the pathogenesis of SLE28,29. Similarly, we found that upon VGLL3 knockdown, SCLE-increased genes (Supplementary Table 5; Supplementary Fig. 6a) exhibited significant down-regulation genome-wide compared to non-SCLE genes (P = 2.53 × 10−8) (Fig. 4h). By contrast, genes increased in plaque psoriasis (Supplementary Table 5), a chronic inflammatory skin condition that has no sex bias30, showed minimal correlation with VGLL3 regulation (Supplementary Fig. 6b,c). Likewise, SCLE-increased genes showed minimal correlation with regulation by Fez, whose knockdown was unable to reduce the expression of female-biased autoimmune genes (Fig. 4a), or Fyn which is not known to exhibit transcription factor activities (Supplementary Fig. 6d,e), suggesting that the regulation of SCLE-increased genes is specific to VGLL3. Intriguingly, sex differences in the expression of VGLL3 and the targets of VGLL3 including BAFF and ITGAM were diminished when comparing male and female SCLE patients, consistent with VGLL3 being a sex-biased risk factor prior to disease manifestation and a general regulator brought to comparable functional levels in the two sexes as autoimmune conditions arise (Supplementary Fig. 6f). Consistent with that scenario is the similar nuclear localization of VGLL3 in involved skin of SCLE in both males and females (Fig. 4d). Similarly, VGLL3-regulated genes were expressed significantly higher in lesional skin of morphea (female: male 4:131, P = 3 × 10−4), and limited scleroderma, a subtype of systemic sclerosis (female: male 4:132, P = 1.4 × 10−2) (Fig. 5a,b, Supplementary Fig. 6g). Gene-by-gene analysis of top VGLL3 targets that are expressed in scleroderma and morphea33 showed that a majority of targets were increased in these diseases states compared with normal skin (Fig. 5c–h).
Figure 5. VGLL3 targets are involved in multiple autoimmune conditions.
Density plot of the null distribution for the mean signed log10 P-value as compared to the mean of VGLL3 targets indicated by the arrows in limited scleroderma (a) and morphea (b). a,b, P < 0.001. c, d, Expression changes of VGLL3 targets in limited scleroderma (lSSc) (c) and morphea (d). e, f, Expression of VGLL3 targets in diseased (lSSc in e or morphea in f) or normal skin. P = 0.0412 (e); 0.0101 (f). g, h, Expression of non-targets in diseased (lSSc in g or morphea in h) or normal skin. P = 0.8686 (g); 0.6449 (h). Mann-Whitney U test.
To address the role of VGLL3 in a sex-biased autoimmune condition not primarily located in skin we extended our analyses to Sjögren’s syndrome, an autoimmune condition characterized by inflammation of salivary and lacrimal glands with reported female-to-male ratio as high as 20:1 (ref.34). Examination of published dataset35 showed VGLL3 mRNA expression was increased in parotid tissue of primary Sjögren’s syndrome patients compared to healthy control subjects (Fig. 6a). Concurrently, the VGLL3 ‘node’ targets MMP9, ETS1, IL7 and IL7R were also elevated in SS patients (Fig. 6b). Notably, the IL-7 axis has been shown to be pivotal in the pathogenesis of Sjögren’s syndrome, with both IL-7 and its receptor being overexpressed36,37. IL-7 enhances Th1 response and T cell-dependent monocyte and B cell activation and promotes IFN-γ–CXCR3 ligand-mediated lymphocyte infiltration of target organs37,38. Furthermore, IL-7 has been shown to be a successful therapeutic target in this syndrome38. Gene-by-gene view showed that VGLL3 target genes had increased expression in inflamed parotid tissue compared to normal (Fig. 6c), a trend not observed with non-targets (Fig. 6d). Collectively, VGLL3 target expression was increased in Sjögren’s syndrome, compared with non-targets (Supplementary Fig. 6h), and Sjögren’s syndrome-upregulated genes were significantly decreased upon VGLL3 knockdown (P < 2.2 × 10−16) (Fig. 6e). Consistent with SCLE, VGLL3 was mainly localized in the cell nucleus in affected Sjögren’s syndrome tissue (data not shown). Collectively, VGLL3 target expression was increased in all four autoimmune diseases, compared with non-diseased tissue, which was not observed for non-targets. Therefore, VGLL3-regulated genes are involved in multiple female-biased autoimmune diseases.
Figure 6. VGLL3 regulation of genes altered in Sjögren’s syndrome.
a, VGLL3 mRNA levels as fold change in control versus Sjögren’s syndrome (SS) parotid tissue. b, mRNA levels of MMP9, ETS1, IL7 and IL7R as fold change in control versus SS parotid tissue. c,d, Expression changes of VGLL3 targets (c) and randomly selected non-targets (d) in SS. e, density plot of log2(FC) levels upon VGLL3 knockdown for SS (genes increased at the FC 1.5, q 0.05 threshold in SS) and non-SS genes. P < 2.2 × 10−6. Mann-Whitney-Wilcoxon test. f, qRT-PCR of BAFF, ITGAM, FCER1g upon VGLL3 knockdown by RNAi (n=3 independent experiments) in monocytes. g, qRT-PCR of LY6E, OAS1, MX1 and IFI44 upon VGLL3 knockdown (n=3 independent experiments), with or without IFNα+IFNβ treatment in monocytes. h, mRNA levels of BAFF, IL7 and MMP9 by fold change in cultured salivary gland cells with scrambled RNAi (Scr Ri) or VGLL3 RNAi (Vgll3 Ri). Cells were untreated or treated with IFN-α or IFN-α+IFN-γ. (a,b) Mean± s.e.m. *, q<0.05 , n=24 for SS and n=25 for control. (f, g, h) Mean ± s.e.m, *, P<0.05, Student’s t-test.
To test if VGLL3 regulates pro-autoimmune genes in cell types other than keratinocytes, we studied the response of the female-biased, SLE-induced genes BAFF, ITGAM, and FCER1G to VGLL3 disruption in monocytes. Monocyte alterations are hallmarks of SLE, including increased production of BAFF that is involved in B cell differentiation and T cell activation28. We observed that VGLL3 was required for the optimal expression of BAFF and FCER1G in female monocytes (Fig. 6f), indicating VGLL3 participates in the promotion of female-biased autoimmune gene expression in monocytes. We further examined a potential role of VGLL3 in regulating type I IFN responses in both monocytes and cultured salivary gland cells, given the central role of type I IFN in the pathogenesis of SLE28 and SS39, and our observation of IFN-response genes among the VGLL3 target gene set. Focusing on the LY6E, OAS1, MX1, and IFI44 that are bona fide IFN-I response genes in peripheral blood mononuclear cells26,27, we validated their induction by IFN-α and IFN-β in monocytes, and found that their maximal induction required VGLL3 expression (Fig. 6g). Similar to what we observed in monocytes, VGLL3 was required for the induction of pro-inflammatory genes BAFF, IL7 and MMP9 by IFN-α with or without co-stimulation by IFN-γ in salivary gland cells (Fig. 6h), consistent with published findings that cytokine induction can be dependent on both type I IFN and IFN-γ in these cells40. This observation indicates VGLL3 may promote inflammation events via supporting type I IFN responses.
Discussion
There is a critical need to understand the biological differences between men and women, how they influence different disease manifestations, and response to therapy41,42. Autoimmune diseases represent one of the most prominent examples of sexually dimorphic human diseases, with a striking predominance in females. Our data demonstrate that even in healthy individuals there are widespread sex-dependent differences in the activities of multiple immunological pathways. The sex-biased genes identified in this study overlap with genetic risk variants previously identified for autoimmune diseases including SLE and systemic sclerosis, and their expression is increased in sites of involvement. This finding strongly suggests that they contribute to not only increased disease susceptibility but possibly also heightened disease activity. In this context it is of note that female sex is the strongest risk factor for development of autoimmunity and dwarfs those of identified autoimmune genetic risk variants. Thus, these results provide novel insights into how sex contributes to autoimmune disease etiology. Furthermore, they open up possibilities for the identification of high-risk populations using biomarkers based on these risk genes.
In contrast to the enrichment of pro-autoimmune factor in female-biased genes, male-biased genes were specifically associated with transcription factors, some of which have been implicated in anti-inflammatory processes. For instance, skin grafts engineered to produce HOXA3 conferred reduced inflammatory responses43. HOXA5 is induced by the anti-inflammatory agent colchicine44, and Six2 is repressed in chronic inflammation45. Reduction in Foxf1 abundance is associated with pulmonary inflammation, and Foxf1 loss enhanced CXCL12 production and inflammation46. Hes1 suppresses Toll-like receptor-induced CXCL1 expression47. Further studies will be required to examine the potential roles of these and other male-biased genes in protection from autoimmunity.
Importantly, our results suggest that sex differences in immune regulation extend beyond the DEGs identified in this study to extensive genome-wide co-expression networks. For example, in females, the SLE risk factor ITGAM is positively correlated with ARTN, the monocyte counterpart of the T/B/NK cell dipeptidyl peptidase IV (CD26), whose inhibitors are promising drugs for various autoimmune diseases48, whereas this is not seen in males. Similarly, in females there is a positive correlation between PTX3, which regulates clearance of apoptotic cells49, and SEPT2, a GTP-binding, cytoskeleton-interacting protein and a putative autoantigen in systemic sclerosis50, but not in males. Therefore, the ITGAM–ARTN and PTX3–SEPT2 genes may be regulated and function in a common pathway in females, but not males. The presence of such differentially regulated gene sets on a genome-wide level reveals additional layers of sexually dimorphic immune regulation beyond mRNA levels.
VGLL3 is a putative transcription factor16 and had a prominent female biased expression in our data. We identify VGLL3 as a novel inflammatory pathway in autoimmunity, and critical regulator of female-biased inflammatory processes. Intriguingly, it has recently been shown that VGLL3 in salmon exhibits sex-dependent dominance, promoting later maturation in females17. The finding that VGLL3 promotes the expression of several existing autoimmune disease drug targets and inflammatory genes, including BAFF (SLE), MMP9 (SLE, systemic sclerosis, multiple sclerosis, Sjögren’s syndrome, polymyositis), IL-7 (SLE, Sjögren’s syndrome, multiple sclerosis, rheumatoid arthritis), ICAM-1 (SLE, multiple sclerosis, rheumatoid arthritis), and influences IFN I responses in immune and non-immune cell populations, positions it at the intersection of multiple autoimmune pathways for potential therapeutic targeting. Intriguingly, we demonstrate that in males affected by cutaneous lupus, expression of VGLL3 becomes similar to what is seen in females, and this is associated with VGLL3 translocation in actively inflamed tissue to the nucleus, consistent with its role as a transcription factor, indicating that in affected males this same pathway becomes activated. This makes it an attractive therapeutic target as it is present in diseased tissue of both females and males, and reduction of functional VGLL3 down to what it is observed in healthy males would have to be considered to be unlikely to cause serious side effects. Further studies are required to address the regulation of VGLL3, and the mechanisms involved in its activation with disease onset in males.
In summary, our results identify transcriptomic differences between females and males that are associated with extensive genome-wide co-expression gene networks influencing various immunological processes including various autoimmune processes. Furthermore, these results identify VGLL3-regulated gene network as a novel inflammatory pathway promoting female-biased autoimmunity and they demonstrate the importance of studying immunological processes in females and males separately. As many of these diseases are inadequately controlled with existing treatments, identifying a unifying molecular basis underlying multiple autoimmune diseases may have far-reaching implications for the development of novel therapeutics.
Online methods
Normal and SCLE skin Biopsies
All subjects provided informed consent for normal and SCLE skin biopsies. Cases of SCLE biopsies were identified from the University of Michigan Pathology Database under IRB #HUM72843. Patients who met both clinical and histologic criteria for SCLE were included. Fresh skin samples were acquired according to IRB #HUM66116. All patient recruitment and samples were treated according to the Declaration of Helsinki.
Identification of sex-biased genes
RNA-Seq data of 82 normal skin samples (GSE63980) were utilized to identify genes that are differentially expressed between the two sexes in the skin. The sex effect for expression of each gene was modeled by linear regression, with age of patient at biopsy as covariate. Specifically, we utilized the RNA-seq data of normal skin samples from our large cohort that studied the transcriptomes of psoriasis (GSE63980). We obtained the sex and age of biopsy for 82 patients in the cohort, and we used the pipeline and gene model described previously for RNA-seq analysis, including read mapping, assembly, and quantification of gene expression51. We performed analysis for genes only expressed in at least 20% of the normal samples. DESeq was used for expression normalization. Rank-based inverse normalization was then applied on each gene’s normalized expression values. Linear regression was used to model the sex effect for expression each gene (i.e. differential expression analysis), and P-values were computed using Wald-test. We used the age of patient during biopsy as covariate. False discovery rate (FDR) was used to control the multiple testing.
Gene-gene co-expression analysis and functional characterization
We conducted gene-gene correlation analysis between sex DEGs versus all genes in male and female, separately. Gene-gene co-expression was measured by Spearman rank correlation coefficient (ρ); P-values were computed using algorithm AS89, and we computed False Discovery Rate for multiple testing comparison. In total, there are 434,900 gene pairs with significant correlation (FDR ≤ 0.1) in both male and female correlation analyses; we also identified 124,521 gene-gene pairs that only show significant in female samples (FDR <=0.1) but not in male samples (P-value > 0.5); vice versa, 158,303 gene-gene pairs show only significant in male samples (FDR <=0.1) but not in female samples (P-value > 0.5). To assess whether the difference in gene-gene correlation between sexes is significant, we devised a permutation approach. Specifically, we first permuted the sex labels and calculated the difference in gene-gene correlations between the pseudo male and pseudo female samples. The permutation was performed 10,000 times to construct the null distribution for the difference in correlation patterns. The significance was then estimated by comparing the observed gene-gene sex correlation difference to the null distribution for the same gene-gene pair. Functional enrichment analysis was performed using hypergeometric distribution, with biological annotations retrieved from the Gene Ontology, KEGG, and Biocarta. Enriched functions were identified using FDR ≤ 0.1 criteria. Spearman correlation (i.e. ρ) was used to study the difference (ρdiff = ρmale – ρfemale) in co-expression networks between the two sexes, and to study and compare the co-expression networks of the most notable enriched biological functions/pathways between two sexes.
Disease association screening
We retrieved disease associated genetic signals available from the NHGRI catalog52 and the Immunobase (https://www.immunobase.org/), and processed the data to consolidate the disease names and maintain only signals which achieve genome-wide association (P ≤ 5 × 10−8). Combining results from the two sources remained 9,599 variant-to-trait associations. We further merged genetic variants from same locus by using ±500 kb interval as criteria selecting the most significant marker in each locus, and focused on complex skin traits with at least 5 associated loci. Enrichment of sex-biased genes was assessed by hypergeometric test, using all skin-expressing genes from the RNA-seq cohort as background. The female-to-male prevalence ratios for skin-associated traits were retrieved from previous literature data12,30,53,54.
In addition to the hypergeometric enrichment approach to compute the significance between female-bias genes versus common disease loci, we also devised a sampling strategy. For each of the complex skin disease we investigated, we randomly selected the same number of loci from the NHGRI disease catalog, and enumerated the randomly selected loci which comprised of female-bias genes. We repeated the process 10,000 times and constructed the null distribution for the expected number of overlap. We then estimated the significance by comparing the observed overlap against the null distribution. This robust approach present empirical P-values for the eight diseases which are highly concord with what we observed using the enrichment approach, replicating the findings that SLE/Systemic sclerosis and the atopic dermatitis loci are enriched with female-bias genes (Supplementary Fig. 5a). Supplementary Fig. 5b shows the null distribution for the expected overlap for random loci, and red lines illustrates the observed overlap results from the SLE/SS (top) and AD (bottom), respectively.
eQTL analysis
To examine the variants that affect the functions or expression of VGLL3 would also affect the expression of the VGLL3 targets, we first identified the nonsynonymous or splice site variates for VGLL3 using the phase 3 1000 Genomes. Among the nine nonsynonymous and one splice variants identified from 1000 Genome project, all of them are rare variants or variants with low minor allele frequencies, thus we are not able to conduct eQTL analysis on these variants as they are not well-imputed in our genetic cohorts. We then turned into markers that can influence the expression of VGLL3 by conducting a cis-eQTL analysis surrounding ±1 Mb region of VGLL3. The results are shown in Supplementary Fig. 8 and illustrated the strongest cis-eQTL signal at chr3:87902673 (P = 4 × 10−5). We then investigated if this VGLL3 cis-eQTL would influence the expression of VGLL3 target genes (208 genes decreased by VGLL3 knockdown in keratinocytes at the q<0.05, |log2FC|>0.5 threshold; as in Supplementary Table 3), and interestingly, we identified nine VGLL3 targets that are also significantly associated with the cis-eQTLs, indicating trans-eQTL effect (Supplementary Fig. 8).
Cell culture, stimulation, RNAi, and gene expression analyses
Normal human keratinocytes (NHKs) were established from healthy adults as previously described55. Informed consent was obtained from all subjects. and grown in medium 154 CF (ThermoFisher Scientific). NHKs were used at passage 1 or 2. For sex hormone stimulation, estradiol (Sigma E2578) or testosterone (Sigma T1500) was applied to passage 1 cells for 24 h. Monocytes were isolated as indicated below and maintained in RPMI (ATCC) with 10% FBS (ThermoFisher Scientific). A253 salivary gland cells were obtained from ATCC and cultured in McCoy’s 5a medium (ATCC) with 10% FBS (ThermoFisher Scientific). siRNA was introduced by electroporation using Lonza 4D-nucleofector following manufacturer’s instructions. For IFN stimulation, IFN-α (R&D systems) was used at 1000 U/ml, IFN-β (R&D systems) at 1000 U/ml and IFN-γ (R&D systems) at 2000 U/ml. Interferons were applied to cells that had been electroporated with the indicated siRNA for 12 h and RNA was collected by the Qiagen RNeasy plus kit. qRT-PCR was performed on a 7900HT Fast Real-time PCR system (Applied Biosystems) with TaqMan Universal PCR Master Mix (ThermoFisher Scientific). RNA-Seq libraries were prepared using the Illumina Truseq RNA library prep kits and sequenced on the Illumina HiSeq platform. Differential expression analyses were performed using EdgeR, and functional enrichment and literature-based network analyses were performed with the Genomatix software. To study if genes increased in SCLE are regulated by VGLL3 on a genome-wide level, we first defined SCLE-increased genes as genes upregulated in SCLE compared to normal at the FC ≥ 2, q<0.01 threshold (Supplementary Table 5), and defined non-SCLE genes as the rest of genes. We then performed density plots of the log2(FC) levels upon VGLL3 knockdown for SCLE and non-SCLE genes, respectively. Mean expression changes for the two groups of genes were calculated and Mann-Whitney-Wilcoxon test was used for significance.
PBMC isolation
Healthy and SLE patient PBMCs were obtained as part of this study and informed consent was obtained from all subjects. PBMCs were isolated from whole blood using the Ficoll method. Monocyte, T cells and B cells were isolated from PBMC using MACS negative selection kits.
Immunohistochemistry
Formalin fixed, paraffin-embedded specimens on slides were heated for 30 min at 55 °C, rehydrated, and epitope retrieved with Tris-EDTA, pH 9. Slides were blocked, incubated with primary antibody overnight at 4 °C, washed, incubated with secondary antibody, developed with DAB and counterstained using hematoxylin. Images presented are representative of three experiments.
Statistics
Statistical tests used are described as in Methods and in Figure Legends. Student’s t-tests are two-tailed. The exact p-Values, when not specified in the figures, are as follows:
Figure 3a (left-to-right): 0.026,0.020,0.036,0.012,0.024,0.033
Figure 3c (left-to-right): 0.020,0.014,0.043
Figure 3d (left-to-right): 0.027,0.025,0.026,0.025
Figure 3e (left-to-right): 0.024,0.044,0.036,0.011
Figure 3f (left-to-right): 0.021,0.037,0.043
Figure 3g: 0.041
Figure 3h: 0.043
Figure 4a (left-to-right): 0.011,0.001
Figure 4b: 0.001
Figure 4c: 0.010
Figure 6a: 0.049
Figure 6b (left-to-right): 0.011, 0.0003, 0.0085, 0.004
Figure 6f (left-to-right): 0.032, 0.020, 0.045, 0.048
Figure 6g (left-to-right): 0.00003, 0.004, 0.0004, 0.010, 0.0065, 0.007, 0.007, 0.004, 0.021,0.015,0.016,0.014
Figure 6h (left-to-right): 0.022, 0.0004, 0.002,0.022,0.0003,0.0005,0.013
Supplementary Material
Acknowledgments
We thank A. A. Dlugosz for critical discussions and reading of the manuscript; S. Stoll, Y. Xu, T. Quan, Y. Li, L. Wolterink, and L. Reingold for technical help; and A. Libs for help with biopsy samples and files. The work was in part supported by NIH awards K08-AR060802 (J.E.G.) and R01-AR069071 (J.E.G.), NIH awards numbers R03-AR066337 (J.M.K.) and K08-AR063668 (J.M.K.), the A. Alfred Taubman Medical Research Institute Kenneth and Frances Eisenberg Emerging Scholar Award (J.E.G.), Doris Duke Charitable Foundation Grant #2013106 (J.E.G.), and Pfizer Aspire Award (J.E.G.). We apologize to those researchers whose work was not cited or discussed due to the limitation of the space.
Footnotes
Accession Codes
RNA-Seq GSE63980
Author Contributions Y.L., J.E.G., J.T.E., J.M.K. and J.J.V. designed the study and wrote the manuscript. Y.L., X.X., M.A.B, P.W.H., P.E.S, M.K.S, R.P.N, and C.C.B. collected and analyzed data. L.C.T. and W.R.S. analyzed data. All authors reviewed and commented on the manuscript.
Competing Financial Interests. The authors declare no competing financial interests.
Data availability
The data that support the findings of this study are available from the corresponding author upon request.
References
- 1.AARDA. The Cost Burden of Autoimmune Disease: The Latest Front in the War on Healthcare Spending. 2011. [Google Scholar]
- 2.Fish EN. The X-files in immunity: sex-based differences predispose immune responses. Nat Rev Immunol. 2008;8:737–744. doi: 10.1038/nri2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whitacre CC. Sex differences in autoimmune disease. Nat Immunol. 2001;2:777–780. doi: 10.1038/ni0901-777. [DOI] [PubMed] [Google Scholar]
- 4.Klein SL. The effects of hormones on sex differences in infection: from genes to behavior. Neurosci Biobehav R. 2000;24:627–638. doi: 10.1016/S0149-7634(00)00027-0. [DOI] [PubMed] [Google Scholar]
- 5.Holroyd CR, Edwards CJ. The effects of hormone replacement therapy on autoimmune disease: rheumatoid arthritis and systemic lupus erythematosus. Climacteric. 2009;12:378–386. doi: 10.1080/13697130903025449. [DOI] [PubMed] [Google Scholar]
- 6.Seligman VA, Lum RF, Olson JL, Li H, Criswell LA. Demographic differences in the development of lupus nephritis: a retrospective analysis. Am J Med. 2002;112:726–729. doi: 10.1016/s0002-9343(02)01118-x. [DOI] [PubMed] [Google Scholar]
- 7.Mackay IR. Science, medicine, and the future: Tolerance and autoimmunity. BMJ. 2000;321:93–96. doi: 10.1136/bmj.321.7253.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanchez-Guerrero J, et al. Menopause hormonal therapy in women with systemic lupus erythematosus. Arthritis Rheum. 2007;56:3070–3079. doi: 10.1002/art.22855. [DOI] [PubMed] [Google Scholar]
- 9.Mok CC, et al. Safety of hormonal replacement therapy in postmenopausal patients with systemic lupus erythematosus. Scand J Rheumatol. 1998;27:342–346. doi: 10.1080/03009749850154357. [DOI] [PubMed] [Google Scholar]
- 10.Nestle FO, Di Meglio P, Qin JZ, Nickoloff BJ. Skin immune sentinels in health and disease. Nat Rev Immunol. 2009;9:679–691. doi: 10.1038/nri2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cervera R, et al. Systemic lupus erythematosus: clinical and immunologic patterns of disease expression in a cohort of 1,000 patients. The European Working Party on Systemic Lupus Erythematosus. Medicine (Baltimore) 1993;72:113–124. [PubMed] [Google Scholar]
- 12.Cooper GS, Stroehla BC. The epidemiology of autoimmune diseases. Autoimmun Rev. 2003;2:119–125. doi: 10.1016/s1568-9972(03)00006-5. [DOI] [PubMed] [Google Scholar]
- 13.Vincent FB, Morand EF, Schneider P, Mackay F. The BAFF/APRIL system in SLE pathogenesis. Nat Rev Rheumatol. 2014;10:365–373. doi: 10.1038/nrrheum.2014.33. [DOI] [PubMed] [Google Scholar]
- 14.International Consortium for Systemic Lupus Erythematosus, G. et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grundberg E, et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nature Genetics. 2012;44:1084–+. doi: 10.1038/ng.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maeda T, Chapman DL, Stewart AF. Mammalian vestigial-like 2, a cofactor of TEF-1 and MEF2 transcription factors that promotes skeletal muscle differentiation. J Biol Chem. 2002;277:48889–48898. doi: 10.1074/jbc.M206858200. [DOI] [PubMed] [Google Scholar]
- 17.Barson NJ, et al. Sex-dependent dominance at a single locus maintains variation in age at maturity in salmon. Nature. 2015;528:405–+. doi: 10.1038/nature16062. [DOI] [PubMed] [Google Scholar]
- 18.Ram M, Sherer Y, Shoenfeld Y. Matrix metalloproteinase-9 and autoimmune diseases. J Clin Immunol. 2006;26:299–307. doi: 10.1007/s10875-006-9022-6. [DOI] [PubMed] [Google Scholar]
- 19.Yang W, et al. Genome-wide association study in Asian populations identifies variants in ETS1 and WDFY4 associated with systemic lupus erythematosus. PLoS Genet. 2010;6:e1000841. doi: 10.1371/journal.pgen.1000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee LF, et al. IL-7 Promotes T(H)1 Development and Serum IL-7 Predicts Clinical Response to Interferon-beta in Multiple Sclerosis. Sci Transl Med. 2011;3 doi: 10.1126/scitranslmed.3002400. ARTN93ra68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pickens SR, et al. Characterization of interleukin-7 and interleukin-7 receptor in the pathogenesis of rheumatoid arthritis. Arthritis Rheum. 2011;63:2884–2893. doi: 10.1002/art.30493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bikker A, et al. Increased Expression of Interleukin-7 in Labial Salivary Glands of Patients With Primary Sjogren’s Syndrome Correlates With Increased Inflammation. Arthritis Rheum-Us. 2010;62:969–977. doi: 10.1002/art.27318. [DOI] [PubMed] [Google Scholar]
- 23.Wuthrich RP, Jevnikar AM, Takei F, Glimcher LH, Kelley VE. Intercellular-Adhesion Molecule-1 (Icam-1) Expression Is Upregulated in Autoimmune Murine Lupus Nephritis. American Journal of Pathology. 1990;136:441–450. [PMC free article] [PubMed] [Google Scholar]
- 24.Bo L, et al. Distribution of immunoglobulin superfamily members ICAM-1, −2, −3, and the beta 2 integrin LFA-1 in multiple sclerosis lesions. J Neuropathol Exp Neurol. 1996;55:1060–1072. [PubMed] [Google Scholar]
- 25.Anderson ME, Siahaan TJ. Targeting ICAM-1/LFA-1 interaction for controlling autoimmune diseases: designing peptide and small molecule inhibitors. Peptides. 2003;24:487–501. doi: 10.1016/S0196-9781(03)00083-4. [DOI] [PubMed] [Google Scholar]
- 26.Baechler EC, et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci U S A. 2003;100:2610–2615. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Obermoser G, Pascual V. The interferon-alpha signature of systemic lupus erythematosus. Lupus. 2010;19:1012–1019. doi: 10.1177/0961203310371161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Banchereau J, Pascual V. Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity. 2006;25:383–392. doi: 10.1016/j.immuni.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 29.Ohl K, Tenbrock K. Inflammatory cytokines in systemic lupus erythematosus. J Biomed Biotechnol. 2011;2011:432595. doi: 10.1155/2011/432595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gudjonsson JE, Elder JT. Psoriasis: epidemiology. Clin Dermatol. 2007;25:535–546. doi: 10.1016/j.clindermatol.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 31.Leitenberger JJ, et al. Distinct Autoimmune Syndromes in Morphea A Review of 245 Adult and Pediatric Cases. Arch Dermatol. 2009;145:545–550. doi: 10.1001/archdermatol.2009.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mayes MD, et al. Prevalence, incidence, survival, and disease characteristics of systemic sclerosis in a large US population. Arthritis Rheum-Us. 2003;48:2246–2255. doi: 10.1002/art.11073. [DOI] [PubMed] [Google Scholar]
- 33.Milano A, et al. Molecular Subsets in the Gene Expression Signatures of Scleroderma Skin. Plos One. 2008;3 doi: 10.1371/journal.pone.0002696. ARTNe2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patel R, Shahane A. The epidemiology of Sjogren’s syndrome. Clin Epidemiol. 2014;6:247–255. doi: 10.2147/CLEP.S47399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horvath S, et al. Systems analysis of primary Sjogren’s syndrome pathogenesis in salivary glands identifies shared pathways in human and a mouse model. Arthritis Res Ther. 2012;14:R238. doi: 10.1186/ar4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hillen MR, et al. High soluble IL-7 receptor expression in Sjogren’s syndrome identifies patients with increased immunopathology and dryness. Ann Rheum Dis. 2016 doi: 10.1136/annrheumdis-2016-209236. [DOI] [PubMed] [Google Scholar]
- 37.Bikker A, et al. Il-7-Activated T Cells and Monocytes Drive B Cell Activation in Patients with Primary Sjogren’s Syndrome. Annals of the Rheumatic Diseases. 2011;70:A63–A63. doi: 10.1136/ard.2010.149005.14. [DOI] [Google Scholar]
- 38.Jin JO, Kawai T, Cha S, Yu Q. Interleukin-7 Enhances the Th1 Response to Promote the Development of Sjogren’s Syndrome-like Autoimmune Exocrinopathy in Mice. Arthritis Rheum-Us. 2013;65:2132–2142. doi: 10.1002/art.38007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yao YH, Liu Z, Jallal B, Shen N, Ronnblom L. Type I interferons in Sjogren’s syndrome. Autoimmunity Reviews. 2013;12:558–566. doi: 10.1016/j.autrev.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 40.Jin JO, Shinohara Y, Yu Q. Innate Immune Signaling Induces Interleukin-7 Production from Salivary Gland Cells and Accelerates the Development of Primary Sjogren’s Syndrome in a Mouse Model. Plos One. 2013;8 doi: 10.1371/journal.pone.0077605. ARTNe77605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim AM, Tingen CM, Woodruff TK. Sex bias in trials and treatment must end. Nature. 2010;465:688–689. doi: 10.1038/465688a. [DOI] [PubMed] [Google Scholar]
- 42.Woodruff TK. Sex, equity, and science. Proc Natl Acad Sci U S A. 2014;111:5063–5064. doi: 10.1073/pnas.1404203111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kachgal S, Mace KA, Boudreau NJ. The dual roles of homeobox genes in vascularization and wound healing. Cell Adhes Migr. 2012;6:457–470. doi: 10.4161/cam.22164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ben-Chetrit E, Bergmann S, Sood R. Mechanism of the anti-inflammatory effect of colchicine in rheumatic diseases: a possible new outlook through microarray analysis. Rheumatology. 2006;45:274–282. doi: 10.1093/rheumatology/kei140. [DOI] [PubMed] [Google Scholar]
- 45.Umazume T, et al. Lacrimal Gland Inflammation Deregulates Extracellular Matrix Remodeling and Alters Molecular Signature of Epithelial Stem/Progenitor Cells. Invest Ophth Vis Sci. 2015;56:8392–8402. doi: 10.1167/iovs.15-17477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalin TV, et al. Pulmonary mastocytosis and enhanced lung inflammation in mice heterozygous null for the Foxf1 gene. Am J Resp Cell Mol. 2008;39:390–399. doi: 10.1165/rcmb.2008-0044OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yingli Shang KAaXH. Transcription repressor Hes1 is a selective regulator of TLR-induced CXCL1 expression and neutrophil responses. The Journal of Immunology. 2014;192 [Google Scholar]
- 48.Steinbrecher A, et al. Targeting dipeptidyl peptidase IV (CD26) suppresses autoimmune encephalomyelitis and up-regulates TGF-beta 1 secretion in vivo. J Immunol. 2001;166:2041–2048. doi: 10.4049/jimmunol.166.3.2041. [DOI] [PubMed] [Google Scholar]
- 49.Rovere P, et al. The long pentraxin PTX3 binds to apoptotic cells and regulates their clearance by antigen-presenting dendritic cells. Blood. 2000;96:4300–4306. [PubMed] [Google Scholar]
- 50.Hall PA, Russell SE. The pathobiology of the septin gene family. J Pathol. 2004;204:489–505. doi: 10.1002/path.1654. [DOI] [PubMed] [Google Scholar]
- 51.Tsoi LC, et al. Analysis of long non-coding RNAs highlights tissue-specific expression patterns and epigenetic profiles in normal and psoriatic skin. Genome Biol. 2015;16:24. doi: 10.1186/s13059-014-0570-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Welter D, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Research. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carson CG. Risk factors for developing atopic dermatitis. Dan Med J. 2013;60 [PubMed] [Google Scholar]
- 54.Scrivener Y, Grosshans E, Cribier B. Variations of basal cell carcinomas according to gender, age, location and histopathological subtype. Brit J Dermatol. 2002;147:41–47. doi: 10.1046/j.1365-2133.2002.04804.x. [DOI] [PubMed] [Google Scholar]
- 55.Elder JT, et al. Retinoic acid receptor gene expression in human skin. J Invest Dermatol. 1991;96:425–433. doi: 10.1111/1523-1747.ep12469889. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.