Abstract
As several model genomes have been sequenced, the elucidation of protein function is the next challenge toward the understanding of biological processes in health and disease. We have generated a human ORFeome resource and established a functional genomics and proteomics analysis pipeline to address the major topics in the post-genome-sequencing era: the identification of human genes and splice forms, and the determination of protein localization, activity, and interaction. Combined with the understanding of when and where gene products are expressed in normal and diseased conditions, we create information that is essential for understanding the interplay of genes and proteins in the complex biological network. We have implemented bioinformatics tools and databases that are suitable to store, analyze, and integrate the different types of data from high-throughput experiments and to include further annotation that is based on external information. All information is presented in a Web database (http://www.dkfz.de/LIFEdb). It is exploited for the identification of disease-relevant genes and proteins for diagnosis and therapy.
Two types of high-throughput human sequence resources are available. (1) The genome is almost finished (Lander et al. 2001; Venter et al. 2001). (2) A large number of EST and full-length cDNA sequences have been collected, mostly in dedicated large-scale projects (Adams et al. 1992; Nomura et al. 1994; Wiemann et al. 2001; Strausberg et al. 2002; Ota et al. 2004). In combination, these two resources have been instrumental in identifying the genes that are dispersed throughout the genome, and in defining the “transcriptome,” that is, the many mRNA variants that are transcribed and processed from these genes. The variability of the transcriptome mostly derives from the alternative use of promoters, exons, and polyadenylation sites, making it significantly more complex than the genome (Brett et al. 2002). In the “post genome sequencing era,” the identification of novel human genes and transcripts will continue for some more time, as the number of human genes is still unclear but believed to be higher than the presently “known” 23,000 that have LocusLink (http://www.ncbi.nlm.nih.gov/LocusLink/; Wheeler et al. 2004) records. The other major challenge is to unravel the exact biological functions and interactions of all these genes and their products. The level of knowledge for any “known” gene varies considerably, from simply having determined the nucleotide sequence to having identified presumably all biological functions of the gene products, functional RNA or encoded protein, in the cellular context. Key questions that need to be answered to determine the biological activity of a gene product are as follows: (1) When is the gene expressed during growth and development? This is one central question for the identification of disease-relevant genes and can be addressed, for example, by expression profiling of healthy and diseased tissues. (2) In which tissues and cell types is the gene expressed, and where in the cell does the gene product execute its activity? (3) What biological activity does the gene product have, and how does the cell react to elevated or reduced levels, for example, of protein concentration or activity. (4) How is the protein activity regulated in the cell? (5) What is the biological context in which the protein acts, and what are the interaction partners, which determine the possible suite of substrates and the biochemical pathways of which a particular protein is part? In combination, these questions are central toward the identification of potential drug targets. To this end, resources and strategies need to be developed that are suitable to tackle a large number of genes and proteins in parallel, to achieve a high throughput while providing meaningful and significant information. Such strategies are commonly termed “functional genomics” and “proteomics.”
Despite the undisputed importance of functional RNAs, here we focus on genes giving rise to protein products, as we have put our initial focus on this subset of genes. Furthermore, the importance of regulatory elements in 5′- and 3′-UTRs should not be neglected, which determine the stability (Bashirullah et al. 2001), expression level (Hentze et al. 1987), or localization (Dalgleish et al. 2001) of mRNAs. Nevertheless, we focus on the analysis of the ORFeome for its immediate applicability in high-throughput experimentation.
Knowledge of gene sequences and of the deduced protein sequences is of primary importance in the process of determining protein function and disease relation. However, in silico analysis of gene and protein sequences is not sufficient to answer most of the questions raised above. Instead, in vitro and in vivo studies are necessary to be carried out to understand the biological activity and context of a protein. Full-length cDNAs (Wiemann et al. 2001; Strausberg et al. 2002; Ota et al. 2004) are of primary importance as they provide the immediate means to express the encoded proteins in living cells, and to analyze the effects of perturbations of these cellular systems. We have contributed to the identification of the ORFeome by means of generating and sequencing full-length cDNAs on a large scale (Wiemann et al. 2001), and by subcloning the ORFs to generate resources for experimental exploitation (Simpson et al. 2000). In recent years, we have constantly expanded the range of high-throughput experiments to address the biological function and disease relevance of human genes and encoded proteins (Wiemann et al. 2003). Here we describe the modules of our functional genomics and proteomics pipeline (Fig. 1). With help of a concrete example (cDNA DKFZp434P097, accession no. AL136895), we demonstrate how these modules have been integrated to create hypotheses in the prioritization process for a more detailed characterization of proteins.
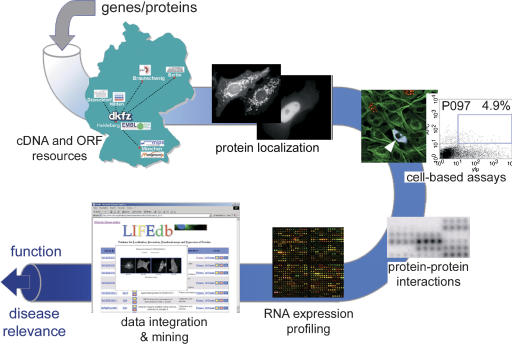
Figure 1.
The functional genomics and proteomics pipeline. Starting with the large-scale production and molecular analysis of cDNAs, a human ORFeome resource is generated. This physical resource is systematically exploited in high-throughput applications of protein localization, cell-based assays, and proteomics applications. Information that is derived from these experiments is integrated with expression profiling data from clinical studies and external information to allow for an efficient mining of data. The output is functionally characterized genes and proteins with their possible disease relations. The results are presented through http://www.dkfz.de/LIFEdb.
RESULTS
Full-Length cDNAs: The German cDNA Consortium
To generate the clone resources for the sequencing efforts of the German cDNA Consortium, we have improved technologies for library construction. Using full-length enrichment and cDNA size fractionation, sublibraries with average insert sizes up to 7 kb were generated. According to BLAST analysis of >12,000 5′-ESTs, the representation of full-length clones is up to 70% also for transcripts >5 kb (Wellenreuther et al. 2004). So far, we have produced 32 libraries; >560,000 clones have been arrayed. The libraries and individual clones are available through the German Resource Center for Genome Research (http://www.rzpd.de).
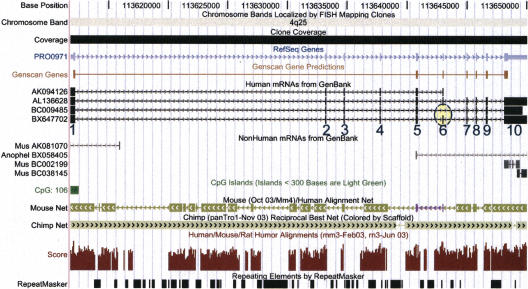
Since 1997, when the German cDNA Consortium was founded, 13,376 cDNAs (43.5 Mb) have been fully sequenced (Wiemann et al. 2001). The Consortium's focus is on the identification of novel genes. Initial annotation is done at MIPS (http://mips.gsf.de/projects/cdna), and functional annotation of the genes is carried out in an international collaboration (Imanishi et al. 2004), in which cDNAs of major full-length cDNA projects worldwide (U.S. Mammalian Gene Collection, Japan FLJ project, German cDNA Consortium) are systematically analyzed in silico. The UCSC genome browser (Kent et al. 2002) screen shot that is shown in Figure 2 impressively demonstrates the power of combining genome (“Clone Coverage”) and full-length cDNA sequences (“Human mRNAs from GenBank”) to immediately visualize the gene structures with exons and introns. Redundancy in cDNA coverage (full-length and EST sequences) is prerequisite to identify splice variants, like the exon-skipping event of exon 6 in the MGC cDNA IMAGE:3623656 (accession no. BC009485), compared with the German cDNA Consortium cDNA DKFZp686P0859 (accession no. BX647702), which is highlighted with a yellow circle. For this gene no full-length cDNA coverage is apparent in species other than human, nor any functional prediction possible by means of computerized protein analysis. The alignment of human, mouse, and rat sequences extends the exonic regions, and reaches similar values within intronic sequences (“Mouse Net” and “Score”). Large-scale cDNA sequencing was prerequisite for the identification of this and of many more genes.
Figure 2.
UCSC genome browser view of the gene locus of PRO0971. The exons (numbered bars) and introns (connecting lines) are immediately apparent when cDNAs are aligned with the genome sequence. Arrow heads in the intron lines indicate the orientation of the gene (left to right), with a CpG island (green bar, “CpG: 106”) supporting the 5′-end of the gene and transcript. Multiple coverage of the gene with individual cDNAs (accession nos. BC009485 from the MGC, AK094126 from the FLJ project, and BX647702 from the German cDNA Consortium) helps to identify putative splice variants. An example of exon skipping in the IMAGE:3623656 cDNA (BC009485) as compared with the DKFZp686P0859 cDNA (BX647702) is highlighted within the yellow circle. The UCSC genome browser is at http://genome.ucsc.edu/cgi-bin/hgGateway.
All sequences generated by the German cDNA Consortium are manually annotated to identify and classify the ORFs. Nevertheless, experimental validation of all predictions is mandatory. For many proteins, experimental analysis creates the first functional information at all when no homology information is available. In this line we process full-ORF clones to contribute to the human ORFeome resource, and exploit them in our functional genomics pipeline.
Building a Human ORFeome Resource
The Gateway technology (Hartley et al. 2000) was established early in the project as a highly reliable and efficient method to systematically clone PCR-amplified ORFs into a range of different expression systems (Simpson et al. 2000). We have amplified and cloned >1000 different ORFs from human cDNAs that had been precharacterized by full-length or EST sequencing. The success rate in obtaining PCR products of the expected size in PCR was independent (>80%-90%) of the ORF size. However, although ORFs up to 2 kb could be cloned with a success rate higher than 90%, longer ORFs had a decreased success rate in cloning. This size bias could be attributed to a reduced efficiency of the Gateway BP reactions with long PCR products, and to the error rate of the PCR enzymes, which becomes more prominent with longer products. The latter resulted in elevated levels of frameshift mutations that rendered the resulting clones useless. Such clones consequently reduced the success rate in the entry clone statistics. Because of the apparent size bias in successful Gateway cloning, we modified several parameters in the amplification and cloning procedure. PCR was then carried out at lower temperatures to favor proofreading and to reduce the number of frame-shift mutations. Products longer than 3 kb have been routinely purified in agarose gels prior to the BP reaction, and the conditions of the recombination reactions have been optimized (ratio of vector/insert, reaction time). These modifications have led to an elevated success rate (80%) in the cloning reactions, which was then also achieved for ORFs longer than 4 kb.
Systematic Subcellular Localization of Proteins
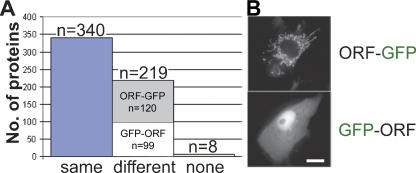
Using the ORFeome resource described above, 650 previously uncharacterized proteins have been localized on the subcellular level. ORFs were expressed both as N-terminal and C-terminal fusions with fluorescent proteins in mammalian cells and analyzed with a fluorescent microscope. The intracellular localization of the GFP fusion protein was influenced by the orientation of the ORF relative to the tag in 39% of the proteins. The statistics shown in Figure 3A were based on the analysis of 567 different proteins that had been selected without bias for subcellular compartments. Signal peptides that are frequently present at the N- or C-terminal ends of proteins are in many cases masked by the color tag that either precedes (GFP-ORF) or follows (ORF-GFP) the protein under investigation. In such cases, one of the fusion proteins frequently localizes to a wrong cellular compartment (Fig. 3B) depending on the orientation of the tag relative to the ORF. The correct localization could always be determined by including other information (e.g., protein similarity). We did not observe a strong tendency for one orientation (ORF-GFP or GFP-ORF) to have a higher rate of correct localizations; hence, we routinely investigated both orientations in downstream experiments. However, we excluded constructs and fusion proteins localizing artificially from further experimentation. Even though these fusion proteins create effects in downstream applications, the data would be artificial and should not be used to characterize and annotate the respective proteins.
Figure 3.
Effect of the orientation of the GFP-tag relative to the ORF. (A) For 340 proteins of 567 tested, both orientations resulted in the correct localization of the fusion proteins (same). Another 219 proteins localized differently in the two orientations. Of these, 120 localizations were correctly localizing with the ORF-GFP construct, and 99 fusion proteins localized correctly in the GFP-ORF orientation. Eight expression constructs did not show any detectable expression (none). (B) An example of a mitochondrial protein (upper image). The fusion protein mislocalized (lower image) when the signal peptide at its N terminus was blocked by the GFP-tag. The cytoplasmic and nuclear staining of the GFP-ORF fusion protein is also the default localization of GFP alone. The bar indicates 10 μm.
We frequently observed the protein localization to be highly dynamic. For example, the protein encoded by cDNA DKFZp434P097 localized to the cytoplasm in ∼80% of the transfected cells, whereas in the other 20% either a nuclear or a mixed cytoplasmic and nuclear staining was observed. The fraction of cells with nuclear staining was dependent on the cell density (low cell density—preferentially cytoplasm, high cell density—more frequently in the nucleus). Therefore, protein localization should be considered a dynamic and context-dependent process, not a static feature.
High-Throughput Cellular Assays to Unravel Protein Function
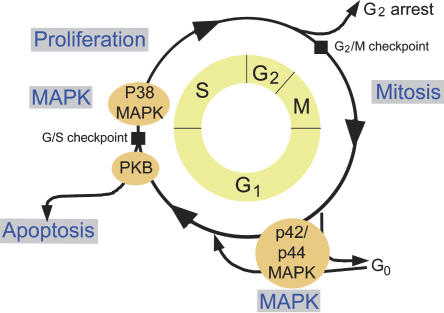
We focused on the development of assays that significantly contribute to the validation of novel proteins as targets for diagnostics and therapy. To this end, we have established a range of assays that investigate the activity of proteins during different points of the cell cycle (Fig. 4). Proteins were selected for analysis based on novelty, and on the differential expression of the corresponding genes in tumor tissues. The assays are based on the effect of protein overexpression that is monitored after transfection and expression in NIH3T3 cells, and which are carried out in 96-well format. Phosphorylated histone H3 indicates cells that have entered into mitosis (M). The apoptosis assay identifies proteins that affect the activation of caspase-3. A proliferation assay measures the effect of the overexpressed protein on BrdU incorporation during the DNA synthesis (S) phase. The MAP kinase assay detects changes in phosphorylation of p42/p44 (ERK1/2) after transfection in the first growth phase (G1). A P38/MAPK and PKB assays are in development.
Figure 4.
Established assays (gray boxes) to address processes of the cell cycle (yellow circle). G1, S, G2, and M are the phases of the cell cycle (G, growth; S, DNA synthesis; M, mitosis).
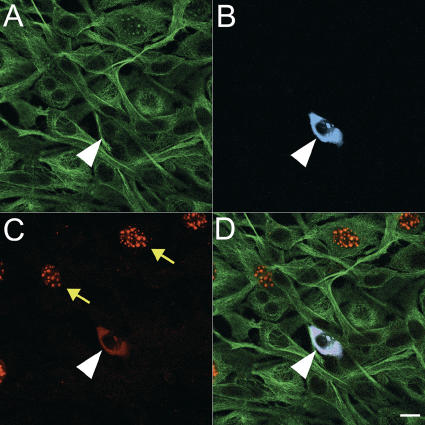
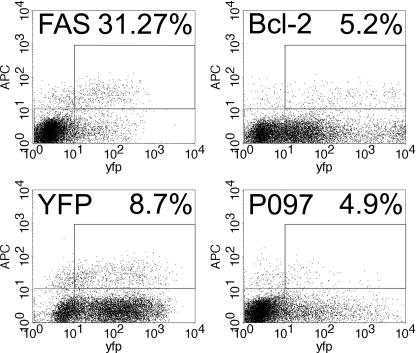
The effect of challenging the protein encoded by cDNA DKFZp434P097 in the mitosis assay is shown in Figure 5. The overlay (D) shows the cytoplasmic colocalization of the recombinant protein with the phosphorylated histone H3 protein. Overexpression of the recombinant protein resulted in a change of the subcellular localization of the phosphorylated histone H3 protein from the nucleus (arrows in Fig. 5C) to the cytoplasm. The same protein, when analyzed in the apoptosis assay, turned out to be a strong inhibitor of caspase-3 activation (Fig. 6). The observed effect was even stronger than that seen with Bcl-2 (Hockenbery et al. 1990), which is a positive control for inhibition.
Figure 5.
Effect of protein overexpression during mitosis. The protein encoded by cDNA DKFZp434P097 was overexpressed as a CFP fusion protein in NIH-3T3 cells (B), antitubulin staining (A). Phosphorylated histone H3 in mitotic cells was detected with a specific antibody (C), which shows a punctuate staining pattern in nuclei of cells in prophase (yellow arrows). The overlay (D) shows colocalization of the DKZp434P097 and the phosphohistone H3 proteins in the cytoplasm. The white arrowhead in A-D marks a cell expressing the DKFZp434P097 protein. Bar, 10 μm.
Figure 6.
Identification of apoptosis modulators. Shown are plots of the fluorescence intensity in the YFP channel (expression of the recombinant proteins) against the level of activated caspase-3 (measured with an APC-labeled antibody). NIH3T3 cells were transfected with ORFs that were C- or N-terminally tagged with YFP. After 24 h, the cells were stained with an antibody directed against the active form of caspase-3 and measured by FACS. For every protein, the percentage of transfected cells (YFP > 10e1) that were positive for activated caspase-3 (APC > 10e1) is given as compared with the transfected cells (YFP > 10e1) that were negative in active caspase-3 (APC < 10e1). APC is fluorescence of the secondary antibody labeled with allophycocyanine. FAS (Chinnaiyan et al. 1995) is the receptor for the cytokine ligand known as FASL. Activated Fas results in the formation of Death-inducing signaling complex, which ultimately leads to cell death (activator control). Bcl-2 (Hockenbery et al. 1990) is an integral protein of the inner mitochondrial membrane that blocks apoptotic death (inhibitor control). YFP is the YFP protein. P097 is the DKFZp434P097 protein. All proteins were expressed as fusion proteins with YFP.
In the proliferation assay, the DKFZp434P097 protein did not have any significant effect on the passage of cells through S phase. Also, the p42/p44 MAPK assay did not result in a detectable effect of that protein. Further experiments are needed to determine the cause of the observed effects. The apparent colocalization of the protein with phosphohistone H3 in the mitosis assay also requires validation. Immediate questions arise as to the way the protein is regulated and if its expression level is altered, for example, in tumor cells.
Two general aspects should be noted. First, the fraction of transfected cells will always be well below 100% when plasmid constructs are used. Second, even when the cells had been synchronized prior to transfection, not all cells would simultaneously be in M phase. As a consequence, a considerable number of cells need to be analyzed to draw significant conclusions. The latter point is extended in the section on data analysis.
Protein Arrays to Determine Protein Interactions
We further exploit the ORF resource to express and purify the encoded proteins. The Gateway cloned ORFs are fused to various tags (GST, His6, MBP, and NusA) to enhance solubility of the fusion proteins and to allow for automated purification. Soluble fusion proteins are spotted on glass slides and incubated, for example, with a suite of protein kinases to identify potential substrates of these kinases. Twenty different proteins encoded by novel cDNAs were selected to prove the experimental setup. The protein encoded by cDNA DKFZp434P097 was an in vitro substrate of P42 MAPK and of CDK2/Cyclin E, as shown in Figure 7. The corresponding evolutionarily conserved kinase motifs were found in a sequence search (http://scansite.mit.edu; Obenauer et al. 2003). The DKFZp434P097 protein has two consensus motifs for proline-dependent serine/threonine kinases (S583 and T650) in the C-terminal domain of the coding region, and two potential motifs for cyclin-dependent kinases (S77 and T650). Both kinases belong to kinase families that regulate mitotic events. The physiological sequence of phosphorylation events to activate the protein remains to be established as well as the in vivo testing of phosphorylation.
Figure 7.
In vitro phosphorylation of arrayed proteins. Purified proteins were arrayed in quadruplicate on glass slides, and incubated with different protein kinases in the presence of [γ-33P]ATP. Rb is the retinoblastoma protein (Lee et al. 1987), which served as positive control. GFP-GST is purified fusion protein of GFP with a GST-tag, which should not be phosphorylated by the kinases. The protein from cDNA DKFZp434P097 was expressed as a fusion protein with the C terminus of GST. (A) The array was incubated with CDK2/cyclin E kinase. (B) The array was incubated with p42 MAPK kinase.
DNA Microarrays to Probe for Disease Relevance
Initially, we focused on the construction of disease-specific microarrays. However, in our recent analyses we use global cDNA clone or oligonucleotide collections, now allowing for the examination of expression changes of almost every gene. We have identified gene expression patterns for kidney tumors (Boer et al. 2001; H. Sültmann, A. v. Heydebreck, W. Huber, R. Kuner, A. Buneß, M. Vogt, B. Gunawan, M. Vingron, L. Füzesi, and A. Poustka, in prep.), as well as brain and breast tumors (J. Schneider, H. Sültmann, M. Asslaber, F. Ploner, H. Samonigg, K. Zatloukal, A. Poustka, unpubl.). The genes belonging to these signatures are funneled into the functional genomics pipeline to add disease relevance to the high-throughput analysis of genes and proteins.
The transcript corresponding to cDNA DKFZp434P097 did not show significant differential expression in the tumors we have analyzed so far (a study of estrogen receptor positive vs. negative breast tumors). However, the data of Huang et al. (2003) identified the gene of cDNA DKFZp434P097 to be up-regulated in patients who suffered from tumor recurrence after surgery, as compared with patients without tumor recurrence (p = 0.017, Wilcoxon rank sum test). Note that this gene was only one in a long list of several hundred differentially expressed genes from that analysis, and that expression profiling alone would not have provided enough reason to follow up, especially on this gene and protein. Only the combination of expression profiling data with the results from functional assays provides focus and leads to biologically relevant discovery.
Data Storage, Analysis, and Integration
Databases
Functional genomics experiments generate diverse types of data (e.g., sequence, numerical, text, and picture) in large amounts that need to be stored, analyzed, and delivered to the community. Experiments include a series of complex steps that require an uninterrupted tracking of samples with tight quality control. We have developed databases that enable the scientists and technicians at the bench to handle all relevant data, to identify bottlenecks, and to optimize their work. For example, the success rate in the ORF cloning process was identified to require optimization especially for ORFs >3 kb in size. Following the labwork, the data need to be actively managed, processed, and made accessible.
Data Analysis and Statistics
The development of computational tools for quality control, visualization, and post hoc calibration of the data was a prerequisite for obtaining satisfactory sensitivity and specificity in our microarray studies (von Heydebreck et al. 2001; Huber et al. 2002, 2003; Huber and Gentleman 2004) and high-throughput assays.
For example, for the proliferation assay, we needed to adjust for varying levels of background signal in each image frame, and variable amounts of spectral overlap between the CFP and DAPI spectra. The nature of the assay did not allow us to define a fixed threshold between “non-expressing” and “expressing” cells. We needed to detect the specific effects of just very slight overexpression, and to avoid the trivial findings that would be obtained from gross overexpression, which tends to shut down the cells' ability to enter S phase nonspecifically and irrespective of the protein's primary function. Local regression models are statistical tools that allow for this type of analysis, but they are computationally intensive and are usually applied with manual tweaking. To analyze the data from thousands of wells, we developed a fully automated procedure that comprises the statistical model, objective choice of parameters, quality control, and documentation of all steps of the analysis.
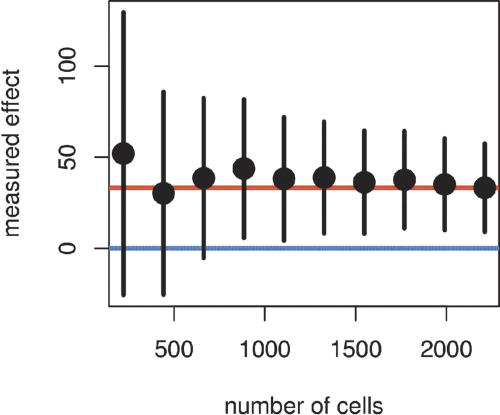
Statistically significant and biologically useful conclusions cannot be drawn from the anecdotal investigation of one or a few cells. Conclusions need to be based on several observations to account for the biological and experimental noise from the assay. Assays where the proteins are expected to have gradual effects on the readout (e.g., cell proliferation, apoptosis; see also Fig. 6) require larger numbers of cells to be analyzed. This was also outlined in a recent RNAi screen for proteins relevant in TRAIL-induced apoptosis (Aza-Blanc et al. 2003). These authors noted the need for the numbers of cells examined to be in the order of 4000. In our own experiments, we investigated at least 2000-4000 cells for every ORF-tag combination to allow for statistically relevant conclusions. We achieved these and higher numbers by combining the data from two or more wells from one or several 96-well microtiter plates. Although proteins that cause extreme effects could also be detected from analyzing a smaller number of cells, the rate of false negatives and false positives of the overall experiment would be unacceptably high. In Figure 8 we show a statistical power analysis that is based on real data. Approximately, the width of the confidence range for an effect estimated from the observation of n cells is proportional to 1/n1/2. With numbers <1000 cells, the results fluctuate strongly around the true effect (red line), and would result in rates of false negatives and false positives of >5%.
Figure 8.
Statistical power analysis for the number of cells. The plot shows means (dots) and 95% confidence intervals (vertical bars) of the measured effect on the proliferation rate of transfection with cyclin A (a positive control in the assay) as a function of the number of cells analyzed. The effect was measured by a robust local regression of the anti-BrdU intensity on the intensity from the YFP-tag (arbitrary fluorescence units). The dependence on the number of cells was simulated by random sampling from the full data set with 2211 cells. The red line represents the approximate true effect, and the blue line no effect. In this example, we would have detected cyclin A as an activator of cell proliferation with 95% probability only for cell numbers ≧1000. Conversely, we would have assigned an activating effect to a protein that is in fact neutral with <5% probability. To reliably detect modifiers of cell proliferation that are subtler, or to achieve probabilities better than 95%, cell numbers must be even higher.
Data Integration
We have implemented tools for the automated analysis of sequences (Del Val et al. 2004). Custom-made tasks aid in the annotation process of cDNAs and proteins, and to retrieve and integrate information from remote databases and servers. These tasks are run on a weekly basis, to constantly provide up-to-date information. The output of the annotation tasks is directly inserted into a local SQL database, from where it can be retrieved and integrated with the experimental results. Data from external sources such as the NCBI are regularly imported into the database, and we map commonly used biological identifiers (e.g., RefSeq IDs, gene names) to in-house data. To jointly visualize, analyze, and interpret data from multiple experiments, we use a federated data management architecture (MS .NET). We developed the LIFEdb Web interface (http://www.dkfz.de/LIFEdb) to assemble and disseminate the experimental information and annotation to the community (Bannasch et al. 2004).
DISCUSSION
The Functional Genomics and Proteomics Pipeline
We have described a systematic pipeline, which begins with the large-scale production and sequence analysis of cDNAs, and aims at the identification and full-length cloning of novel human genes and splice variants. We exploit the cDNA resource by subcloning the ORFs into different vector systems to allow for mammalian and bacterial expression of the recombinant proteins. The mammalian expression system is used to produce fusion proteins with GFP and variants (CFP, YFP) in order to determine the subcellular localization, and to identify the relevant clone resources for high-throughput cellular assays. In these assays, we investigate central cellular pathways with high disease relevance. In combination we create information on a large number of novel proteins. This information is used to generate hypotheses, to define prioritized candidates and more detailed experiments for further investigation toward protein function and potential roles in diseases. We have demonstrated the power of our approach with the protein encoded by cDNA DKFZp434P097 as an example.
This cDNA was cloned and sequenced within the German cDNA Consortium. Both the sequence (accession no. AL136895) and the clone (via http://www.rzpd.de) are publicly available. The protein has a dynamic subcellular localization in the cytoplasm and/or in the nucleus. In the mitosis assay, the protein colocalizes in the cytoplasm with the phosphorylated histone H3. None of the cells that overexpressed the protein was found in mitosis, not even those in which the protein was located in the nucleus. Phosphorylation of DNA-binding histone 3 occurs in mitosis and correlates with DNA condensation. Such DNA condensation was not seen in the overexpressing cells, whereas nontransfected cells show clear DNA condensation (Fig. 5C,D). We hypothesize that histone 3 is not properly transported in the nucleus when DKFZp434P097 is overexpressed and consequently DNA condensation is abolished. The overexpression of this protein does not affect the passage through S phase, nor does it affect the p42/p44 MAPK signaling pathway. However, it strongly inhibits apoptosis. The recombinant protein is an in vitro substrate of several protein kinases that are relevant in cell cycle control. Interestingly, several potential phosphorylation sites are located in a presumed C-terminal activation domain (Kim et al. 2003), which might imply protein localization and/or activity to be regulated by phosphorylation. The transcript of DKFZp434P097 does not appear to be differentially expressed between estrogen receptor negative and positive breast cancers, but we have found it to be positively associated with tumor recurrence in breast cancer from the data of Huang et al. (2003).
The protein encoded by cDNA DKFZp434P097 contains a predicted Winged DNA-binding domain (http://www.ebi.ac.uk/interprol; Mulder et al. 2003) and is highly similar (89% identical) to a mouse coiled-coil transcriptional coactivator (Kim et al. 2003). The mouse protein was recently described to be involved in estrogen-receptor-mediated induction of gene expression. Based on all this information, we hypothesize that this protein resides in the cytoplasm until activation by external stimuli (e.g., via phosphorylation). Once activated, the protein would translocate into the nucleus, where it could bind transcriptional coactivators to induce gene expression. Although overexpression does not induce cell proliferation, the increased expression in recurrent breast tumors might go well in line with the repressed apoptotic rate we found in overexpressing cells. In combination, we have systematically generated data and information that can be used to build a hypothesis. This, on one hand, prioritizes the protein for further characterization and, on the other hand, opens paths to follow in this analysis. For example, the phosphorylation status of the protein could be investigated in vivo, the localization and the effects in cell-based assays could be studied in response to different external stimuli, and a possible direct protein interaction with phosphorylated histone H3 could be investigated.
Applying the described high-throughput approaches, and other automated cell-based assays that are focused on protein secretion, Golgi integrity, and calcium signaling, we have identified several candidate proteins and are currently investigating their biomedical impact. The integration of large-scale genomics resources with diverse high-throughput experiments offers great power toward the systematical generation of functional and disease-relevant information.
Challenges
Several critical issues remain, however, that need to be tackled within a foreseeable future to put this approach forward:
Comprehensiveness of the human ORFeome resource. The one gene-one protein paradigm has proven to be wrong. Instead, many if not most genes are transcribed and processed into several mRNA variants. Alternative uses of promoters and exons generate a diversity of mRNAs that has not yet been completely unraveled. The effect of cSNPs causing amino acid exchanges is just beginning to be investigated for a role in disease processes. Appropriate clone resources will be required to determine the possible effects of the individual variants.
The quality control of ORF resources and high-throughput experimentation must be tight, as amino acid substitutions that may derive from the original cDNA clone or from PCR amplification of ORFs in the subcloning process may influence the biological activity of the expressed protein. Quantitative measurements that rely on cell cultures, complex biochemical reactions, and sensitive optical and electronic equipment are susceptible to many sources of experimental variability. For example, microscope lamps deteriorate, or cells' behavior can be affected by cell density and their well position in a microtiter plate.
A single high-throughput assay by itself rarely provides clear-cut yes/no answers. Rather, biologically coherent “stories” emerge through the integration of multiple, independent experiments, and prior biological knowledge. Thus, the range of assayed conditions needs to grow constantly, as the number of biological processes is huge and the conclusions drawn from experimentation strictly depend on the relevance of the applied assay. For example, a hypothesis that could be based on the expression profiling data of breast cancer recurrences for the DKFZp434P097 gene might be that the gene, or rather the protein, could stimulate tumor development and/or progression. However, the results of the mitosis assay, where overexpression of the protein disturbs the “normal” M phase, and from the proliferation assay, where overexpression did not result in a significant effect, need deeper analysis. Only the combination of multiple experimental and computational approaches is suited to define candidates that are primary targets for disease-relevant research.
Careful optimization and consideration of the sensitivity and specificity of the assays are critical. To detect the response of the perturbation of a cellular system through over- or under-expression of the protein, true effects need to be distinguished from experimental and biological noise. However, this cannot be accomplished by simply increasing the effect, for example, by using a larger perturbation: in most cases, nonphysiological artifacts would be measured in consequence. In fact, the perturbation, and also its effect, should be kept small to remain as close as possible at the physiological condition. This requires minimization of the noise through clever experimental design, improved data acquisition systems. Experimental design issues include the choice of the assay system (e.g., is the cell type used suitable to really answer the biomedical question? Is the cell type compatible with the assay system?), the number of cells that are analyzed, the choice of controls, and the stability and robustness of the data acquisition system. The use of sharp statistical tools needs to provide discrimination between signal and noise as well as a thorough understanding of the unavoidable noise-induced uncertainty.
Standardized data formats and standardized, machine-readable descriptions of experimental and analysis procedures are urgently needed to allow for the exchange of data and the comparison and integration of functional genomics' results. The experience of the microarray community (Brazma et al. 2001) and nascent efforts from the proteomics community (Hermjakob et al. 2004) provide some orientation. We are using XML for the exchange of data with other databases, and actively contribute to the definition of a commonly accepted and used standard for high-throughput functional genomics.
METHODS
cDNA Libraries and Sequencing
First-strand cDNA was generated with the SMART cDNA library construction kit (Clontech). After second-strand synthesis, cDNA was size-fractionated by preparative agarose gel electrophoresis. Three to six size fractions from each cDNA were separately amplified and cloned into plasmid vectors. Every sublibrary was quality controlled by plasmid preparation and restriction digest of clone pools of 5000 to 10,000 clones (R. Wellenreuther, I. Schupp, The German cDNA Consortium, A. Poustka, and S. Wiemann, in prep.). cDNA sequencing was done as described (Wiemann et al. 2001). In brief, clones were arrayed and replicated, and the DNA was extracted. 5′-ESTs were generated and analyzed by BLAST (Altschul et al. 1997). According to BLAST results of 5′-ESTs, candidate clones were selected for further sequencing. Of these, 3′-ESTs were generated, and after BLAST analysis, clones were selected for full-length sequencing by primer walking. Complete sequenced cDNAs were computationally analyzed and annotated at the Munich Institute for Protein Sequences (Mewes et al. 2002).
Subcloning of ORFs—Gateway
To allow for a systematic ORF cloning, we use the Gateway technology (Invitrogen), which is based on cloning by site-specific recombination (Hartley et al. 2000; Simpson et al. 2000). ORFs are amplified in 96-well format by two-step PCR using the “expand high fidelity PCR system” (Roche). In the first PCR, primer pairs with gene-specific sequence and short Gateway overhangs of 9 and 11 bp for forward and reverse primers, respectively, were used. The gene-specific sequences of the 5′-primers are fixed to the initiator start codon, and 3′-primers are fixed to the end of coding region but leaving out the stop codon, thus allowing N- and C-terminal translation fusions to be generated. The gene-specific parts of primers were designed with the PRIDE program (Haas et al. 1998) and purchased from commercial vendors. The Gateway recombination sites were completed in the second PCR. PCR products were cloned without prior purification in a BP reaction into the Gateway donor vector pDONR201. For each ORF, eight entry clones were picked into 96-well plates and analyzed for the presence of the ORF by colony PCR. Positive entry clones were completely sequence-verified to exclude frameshift mutations. The ORF cloning process is managed with help of the LIFEdb tracking database (Bannasch et al. 2004).
Subcellular Localization
For subcellular localization as well as for cell-based assays, the ORFs were subcloned in 96-well format into ECFP and EYFP expression vectors (Clontech), that had been made Gateway-compatible (Simpson et al. 2000). These vectors generated N-terminal fusions with the YFP and C-terminal fusions with CFP. Expression constructs were isolated in 96-well format using a robotic workstation (Biorobot 9600 QIAGEN) and Qiawell Ultra plasmid preps (QIAGEN). Transfection of Vero cells (ATCC CCL81), and subsequent image acquisition and analysis were described by Simpson et al. (2000).
Cell-Based Assays
Plasmid DNA was prepared in a 96-well format using a Multi-Probe IIex robot (Perkin Elmer) and a Millipore Montage Plasmid Miniprep96 Kit. DNA masterplates were generated with standardized DNA concentrations. NIH3T3 cells (ATCC CRL-1658) were transfected in an array of 12 chamber slides using a MultiProbe IIex robot encased by a perspex housing. Fixing and staining were also performed with the pipetting robot. Images (12 bit, 1280 × 1024 pixels) were taken with a fully automated high-content screening microscope (Liebel et al. 2003) and analyzed with an automated segmentation and analysis algorithm in Lab-view (National Instruments). Data acquisition of apoptosis and MAPkinase assays was with an FACS Calibur in a 96-well format. Irrespective of the data acquisition system, the final output was tables of fluorescence intensities taken in different channels for the individual cells. On average, 20,000 cells were analyzed for every condition. The data analysis for the assays was performed in three steps: image segmentation and quantification, within-well analysis, and across-well analysis.
Protein Arrays
Constructs encoding fusion proteins were transformed into BL21-SI cells (Invitrogen). Protein expression was induced with IPTG in 25-mL cultures and continued for 18 h at 20°C. Lysis was performed by freeze/thaw cycles in the presence of lysozyme, followed by sonication. Cell debris was removed by centrifugation, and soluble fusion proteins were purified with affinity columns (e.g., GST fusion proteins with help of Pharmacia spin columns, NusA and MBP fusion proteins were purified with Swell Gel [Pierce] pellets). Purity and yield were assessed by SDS-PAGE. Affinity-purified proteins were spotted on Hydrogel (Perkin Elmer) glass slides with a Perkin Elmer BioChip arrayer. After blocking of unspecific protein binding (0.5% BSA [w/v], 0.2% NP-40 in BSA), arrays were incubated with different protein kinases (CDK2/Cyclin E, ProQinase; ERK2, Calbiochem) for 1 h in the presence of [γ-33P]ATP using the conditions recommended by the suppliers. After washing, the slides were air-dried and exposed to an imager plate. Images were taken using a Typhoon imager (Amersham). Primary data analysis was done with Genepix software (Axon Instruments). Statistical methods and visualizations for data analysis and quality control were based on the R/Bioconductor platform (http://www.bioconductor.org). Spots with signal intensity of twice the background signal were counted as positive.
RNA Expression Profiling
Tumor-specific microarrays for kidney (H. Sültmann, A. v. Heydebreck, W. Huber, R. Kuner, A. Buneß, M. Vogt, B. Gunawan, M. Vingron, L. Füzesi, and A. Poustka, in prep.), brain, and breast (J. Schneider, H. Sültmann, M. Asslaber, F. Ploner, H. Samonigg, K. Zatloukal, A. Poustka, unpubl.) cancer were designed by screening whole-genome clone collections with the tumor samples of interest and selecting those clones for which high expression and differential expression was evident. In addition, clones selected from literature surveys were added. Global microarrays were constructed using the 36,000 cDNA clones of the UniGene RZPD 3.1 collection (German Resource Center for Genome Research). Microarrays were processed, hybridized with more tumor samples, and analyzed as described (H. Sültmann, A. v. Heydebreck, W. Huber, R. Kuner, A. Buneß, M. Vogt, B. Gunawan, M. Vingron, L. Füzesi, and A. Poustka, in prep.).
Databases, Annotation, and Analysis
The project-specific LIMS applications were first developed as prototypes in Microsoft (MS) Access and were then ported to an MS SQL Server. The latter allowed for remote access using the MS .NET structure either by custom-made clients or by a Web browser via the MS Internet Information Server (see http://www.dkfz.de/LIFEdb). The database structure is described in Bannasch et al. (2004), automated annotation tasks in Del Val et al. (2004). The assay analysis software was designed as an R package (Ihaka and Gentleman 1996) for the Bioconductor environment (http://www.bioconductor.org).
Acknowledgments
Sequence analysis of full-length cDNAs is carried out in the German cDNA Consortium by Rolf Wambutt and Dagmar Heubner (AGOWA GmbH); Karl Köhrer and Andreas Beyer (BMFZ); Regina Albert, Petra Moosmayer, and Ingo Schupp (DKFZ); Wilhelm Ansorge and Sabine Glassl (EMBL); Michael Böcher and Helmut Blöcker (GBF); Birgit Ottenwälder and Brigitte Obermaier (Medigenomix); Clara Amid and Werner Mewes (Mips-GSF); and André Bahr and Jürgen Lauber (QIAGEN). We appreciate the contribution made by Meher Majety (MAPKinase assay); Christian Schmitt (automation); Detlev Bannasch and Heiko Rosenfelder (databases); Jeremy Simpson (subcellular localization); and Urban Liebel (high-content screening microscopy). We thank Patricia McCabe for critical reading of the manuscript. This work is supported by grants 01GR0101 (DKFZ) of the National Genome Research Network, 01KW9987 and 01KW0012 (DKFZ), and 01KW0013 (EMBL) within the German Genome Project, all from the Bundesministerium für Bildung und Forschung (BMBF).
Footnotes
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2576704.
References
- Adams, M.D., Dubnick, M., Kerlavage, A.R., Moreno, R., Kelley, J.M., Utterback, T.R., Nagle, J.W., Fields, C., and Venter, J.C. 1992. Sequence identification of 2,375 human brain genes. Nature 355: 632-634. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aza-Blanc, P., Cooper, C.L., Wagner, K., Batalov, S., Deveraux, Q.L., and Cooke, M.P. 2003. Identification of modulators of TRAIL-induced apoptosis via RNAi-based phenotypic screening. Mol. Cell 12: 627-637. [DOI] [PubMed] [Google Scholar]
- Bannasch, D., Mehrle, A., Glatting, K.-H., Pepperkok, R., Poustka, A., and Wiemann, S. 2004. LIFEdb: A database for functional genomics experiments integrating information from external sources, and serving as a sample tracking system. Nucleic Acids Res. 32: D505-D508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashirullah, A., Cooperstock, R.L., and Lipshitz, H.D. 2001. Spatial and temporal control of RNA stability. Proc. Natl. Acad. Sci. 98: 7025-7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer, J.M., Huber, W.K., Sultmann, H., Wilmer, F., von Heydebreck, A., Haas, S., Korn, B., Gunawan, B., Vente, A., Fuzesi, L., et al. 2001. Identification and classification of differentially expressed genes in renal cell carcinoma by expression profiling on a global human 31,500-element cDNA array. Genome Res. 11: 1861-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazma, A., Hingamp, P., Quackenbush, J., Sherlock, G., Spellman, P., Stoeckert, C., Aach, J., Ansorge, W., Ball, C.A., Causton, H.C., et al. 2001. Minimum information about a microarray experiment (MIAME)—Toward standards for microarray data. Nat. Genet. 29: 365-371. [DOI] [PubMed] [Google Scholar]
- Brett, D., Pospisil, H., Valcarcel, J., Reich, J., and Bork, P. 2002. Alternative splicing and genome complexity. Nat. Genet. 30: 29-30. [DOI] [PubMed] [Google Scholar]
- Chinnaiyan, A.M., O'Rourke, K., Tewari, M., and Dixit, V.M. 1995. FADD, a novel death domain-containing protein, interacts with the death domain of Fas and initiates apoptosis. Cell 81: 505-512. [DOI] [PubMed] [Google Scholar]
- Dalgleish, G., Veyrune, J.L., Blanchard, J.M., and Hesketh, J. 2001. mRNA localization by a 145-nucleotide region of the c-fos 3′-untranslated region. Links to translation but not stability. J. Biol. Chem. 276: 13593-13599. [DOI] [PubMed] [Google Scholar]
- Del Val, C., Mehrle, A., Falkenhahn, M., Seiler, M., Glatting, K.-H., Poustka, A., Suhai, S., and Wiemann, S. 2004. High-throughput protein analysis integrating bioinformatics and experimental assays Nucl. Acids Res. 32: 742-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas, S., Vingron, M., Poustka, A., and Wiemann, S. 1998. Primer design for large scale sequencing. Nucleic Acids Res. 26: 3006-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley, J.L., Temple, G.F., and Brasch, M.A. 2000. DNA cloning using in vitro site-specific recombination. Genome Res. 10: 1788-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze, M.W., Rouault, T.A., Caughman, S.W., Dancis, A., Harford, J.B., and Klausner, R.D. 1987. A cis-acting element is necessary and sufficient for translational regulation of human ferritin expression in response to iron. Proc. Natl. Acad. Sci. 84: 6730-6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermjakob, H., Montecchi-Palazzi, L., Bader, G., Wojcik, J., Salwinski, L., Ceol, A., Moore, S., Orchard, S., Sarkans, U., von Mering, C., et al. 2004. The HUPO PSI's molecular interaction format—A community standard for the representation of protein interaction data. Nat. Biotechnol. 22: 177-183. [DOI] [PubMed] [Google Scholar]
- Hockenbery, D., Nunez, G., Milliman, C., Schreiber, R.D., and Korsmeyer, S.J. 1990. Bcl-2 is an inner mitochondrial membrane protein that blocks programmed cell death. Nature 348: 334-336. [DOI] [PubMed] [Google Scholar]
- Huang, E., Cheng, S.H., Dressman, H., Pittman, J., Tsou, M.H., Horng, C.F., Bild, A., Iversen, E.S., Liao, M., Chen, C.M., et al. 2003. Gene expression predictors of breast cancer outcomes. Lancet 361: 1590-1596. [DOI] [PubMed] [Google Scholar]
- Huber, W. and Gentleman, R. 2004. matchprobes: A Bioconductor package for the sequence-matching of microarray probe elements. Bioinformatics 2004 Feb 26 [Epub ahead of print]. [DOI] [PubMed]
- Huber, W., Von Heydebreck, A., Sultmann, H., Poustka, A., and Vingron, M. 2002. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18 Suppl. 1: S96-S104. [DOI] [PubMed] [Google Scholar]
- ____. 2003. Parameter estimation for the calibration and variance stabilization of microarray data. Stat. Appl. Genet. Mol. Biol. 2: Article 3. [DOI] [PubMed]
- Ihaka, R. and Gentleman, R. 1996. R: A language for data analysis and graphics. J. Comp. Graph. Stat. 5: 299-314. [Google Scholar]
- Imanishi, T., Itoh, T., Suzuki, Y., O'Donovan, C., Fukuchi, S., Koyanagi, K.O., Barrero, R.A., Tamura, T., Yamaguchi-Kabata, Y., Tanino, M., et al. 2004. Integrative annotation of 21,037 human genes validated by full-length cDNA clones. PLoS Biol. 2: E162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, W.J., Sugnet, C.W., Furey, T.S., Roskin, K.M., Pringle, T.H., Zahler, A.M., and Haussler, D. 2002. The human genome browser at UCSC. Genome Res. 12: 996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.H., Li, H., and Stallcup, M.R. 2003. CoCoA, a nuclear receptor coactivator which acts through an N-terminal activation domain of p160 coactivators. Mol. Cell 12: 1537-1549. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Lee, W.H., Bookstein, R., Hong, F., Young, L.J., Shew, J.Y., and Lee, E.Y. 1987. Human retinoblastoma susceptibility gene: Cloning, identification, and sequence. Science 235: 1394-1399. [DOI] [PubMed] [Google Scholar]
- Liebel, U., Starkuviene, V., Erfle, H., Simpson, J.C., Poustka, A., Wiemann, S., and Pepperkok, R. 2003. A microscope-based screening platform for large-scale functional protein analysis in intact cells. FEBS Lett. 554: 394-398. [DOI] [PubMed] [Google Scholar]
- Mewes, H.W., Frishman, D., Guldener, U., Mannhaupt, G., Mayer, K., Mokrejs, M., Morgenstern, B., Munsterkotter, M., Rudd, S., and Weil, B. 2002. MIPS: A database for genomes and protein sequences. Nucleic Acids Res. 30: 31-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder, N.J., Apweiler, R., Attwood, T.K., Bairoch, A., Barrell, D., Bateman, A., Binns, D., Biswas, M., Bradley, P., Bork, P., et al. 2003. The InterPro Database, 2003 brings increased coverage and new features. Nucl. Acids Res. 31: 315-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura, N., Miyajima, N., Sazuka, T., Tanaka, A., Kawarabayasi, Y., Sato, S., Nagase, T., Seki, N., Ishikawa, K., and Tabata, S. 1994. Prediction of the coding sequences of unidentified human genes. I. The coding sequences of 40 new genes (KIAA0001-KIAA0040) deduced by analysis of randomly sampled cDNA clones from human immature myeloid cell line KG-1. DNA Res. 1: 47-56. [DOI] [PubMed] [Google Scholar]
- Obenauer, J.C., Cantley, L.C., and Yaffe, M.B. 2003. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 31: 3635-3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota, T., Suzuki, Y., Nishikawa, T., Otsuki, T., Sugiyama, T., Irie, R., Wakamatsu, A., Hayashi, K., Sato, H., Nagai, K., et al. 2004. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 36: 40-45. [DOI] [PubMed] [Google Scholar]
- Simpson, J.C., Wellenreuther, R., Poustka, A., Pepperkok, R., and Wiemann, S. 2000. Systematic subcellular localization of novel proteins identified by large scale cDNA sequencing. EMBO Rep. 1: 287-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausberg, R.L., Feingold, E.A., Grouse, L.H., Derge, J.G., Klausner, R.D., Collins, F.S., Wagner, L., Shenmen, C.M., Schuler, G.D., Altschul, S.F., et al. 2002. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl. Acad. Sci. 99: 16899-16903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter, J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304-1351. [DOI] [PubMed] [Google Scholar]
- von Heydebreck, A., Huber, W., Poustka, A., and Vingron, M. 2001. Identifying splits with clear separation: A new class discovery method for gene expression data. Bioinformatics 17 Suppl. 1: S107-S114. [DOI] [PubMed] [Google Scholar]
- Wellenreuther, R., Schupp, I., The German cDNA Consortium, Poustka, A., and Wiemann, S. 2004. SMART amplification combined with cDNA size fractionation in order to obtain large full-length clones. BMC Genomics 5: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler, D.L., Church, D.M., Edgar, R., Federhen, S., Helmberg, W., Madden, T.L., Pontius, J.U., Schuler, G.D., Schriml, L.M., Sequeira, E., et al. 2004. Database resources of the National Center for Biotechnology Information: Update. Nucleic Acids Res. 32: D35-D40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemann, S., Weil, B., Wellenreuther, R., Gassenhuber, J., Glassl, S., Ansorge, W., Bocher, M., Blocker, H., Bauersachs, S., Blum, H., et al. 2001. Toward a catalog of human genes and proteins: Sequencing and analysis of 500 novel complete protein coding human cDNAs. Genome Res. 11: 422-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemann, S., Bechtel, S., Bannasch, D., Pepperkok, R., Poustka, A., and German cDNA Network. 2003. The German cDNA network: cDNAs, functional genomics and proteomics. J. Struct. Funct. Genomics 4: 87-96. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://genome.ucsc.edu/cgi-bin/hgGateway; UCSC Genome Browser GoldenPath.
- http://mips.gsf.de/projects/cdna; database with annotation of the cDNAs sequenced by the German cDNA Consortium.
- http://www.dkfz.de/LIFEdb; database with subcellular localizations and protein annotation (the address is case-sensitive).
- http://www.ebi.ac.uk/interpro/; IntroPro database of protein families, domains, and functional sites.
- http://www.ncbi.nlm.nih.gov/LocusLink/; LocusLink database with curated sequence and descriptive information on genetic loci.
- http://www.rzpd.de;not-for-profit service center for genomics and proteomics research.
- http://scansite.mit.edu; Scansite searches for motifs within proteins that are likely to be phosphorylated by specific protein kinases or bind to domains such as SH2 domains, 14-3-3 domains, or PDZ domains.
- http://www.bioconductor.org; Bioconductor is an open source and open development software project for the analysis and comprehension of genomic data.