Abstract
The neuropeptide Calcitonin Gene-Related Peptide (CGRP) is a well-characterized neurotransmitter. However, little is known about the role of CGRP in osteogenesis and vascular genesis during the developmental formation of bone. In the present study, we assessed the abundance of CGRP mRNA and the mRNA of osteogenesis and vascular genesis markers in the foetal mouse mandible and leg bone (tibia). We also analysed the expression and localization of CGRP, osteopontin (OPN) and vascular endothelial growth factor (VEGF-A) using in situ hybridization and immunohistochemical localization in the mouse mandible and tibia at embryonic days 12.5 (E12.5), E14.5, E17.5, and postnatal day 1 (P1). CGRP was clearly detected in the mandible relative to the tibia at E14.5. Hybridization using an anti-sense probe for CGRP was not detected in the mandible at P1. Hybridization with an anti-sense probe for OPN was detected at E14.5, later in the mandible and at P1 in Meckel’s cartilage. However, OPN was only detected in the tibia at E17.5 and later. The abundance of CGRP mRNA differed between the mandible and tibia. The level of vasculogenesis markers, such as VEGF-A, was similar to that of CGRP in the mandible. The levels of VEGF-A, cluster of differentiation 31 (CD31) and lymphatic vessel endothelial hyaluronan receptor 1 (LIVE-1) differed from that of OPN in the mandible. In contrast, the levels of VEGF-A, CD31, matrix metalloproteinase-2 (MMP-2), collagen I (Col I), collagen II (Col II) and OPN mRNA differed from E12.5 to P1 (P<0.001) in the tibia. The abundance of mRNA of CGRP and bone matrix markers (Col I, Col II, and OPN) was low at P5 in the tibia. These differences in CGRP and other mRNAs may induce a different manner of ossification between the mandible and tibia. Therefore, a time lag of ossification occurs between the mandible and tibia during foetal development.
Key words: CGRP, mandible, vasculogenesis, osteogenesis, development
Introduction
The neuropeptide Calcitonin Gene-Related Peptide (CGRP) is a neurotransmitter involved in vasculogenesis during muscle differentiation and organ development. CGRP first appears in the mouse at embryonic day 16.5 and localizes around blood vessels and cartilaginous bone matrix in the limbs.1 CGRP is involved in the formation and remodelling of bone, muscle, and connective tissues. During fracture healing and remodelling, CGRP regulates bone formation.2,3 Bone metabolism may be affected by nerves and by CGRP,4 which regulates osteoclasts and osteoblasts5-9 and epidermal stem cells.10 CGRP promotes osteogenesis in vitro11 and plays a specific role in regulating bone resorption during experimental tooth movement.12,13 It also participates in periodontal tissue regeneration, particularly alveolar bone regeneration.14 Transgenic mice expressing CGRP in osteoblasts display increased bone density during bone formation, in contrast to mice lacking CGRP expression. This phenotype indicates that osteopaenia results from decreased bone formation.15,16 A CGRP-deficient mouse model was analysed to understand the physiological role of CGRP in bone remodelling.17 Cranial neural crest cell-derived vascular endothelial growth factor (VEGF-A) regulates the proliferation, vascularization and ossification of cartilage and membranous bone.18-21 Calvarial and mandibular malformations were observed following the deletion of VEGF-A in neural crest cells.19,20 Mice lacking the secreted isoform of VEGF-A showed craniofacial defects including cleft palate, unfused cranial sutures, and shorter jaws.22 OPN is a major non-collagen glycoprotein in the bone matrix that regulates mineralization and bone resorption. OPN is primarily expressed in the lower hypertrophic cell zone at the site of endochondral bone formation of long bones and the cartilage matrix.23-25 Furthermore, Ichikawa et al. observed OPN-positive sensory neurons in the spinal and trigeminal ganglion.26 Moreover, insulin-like growth factor system, perlecan, dentin matrix protein, and matrix extracellular phosphoprotein affected the formation of mandibular condylar cartilage in foetal mice and humans.27-29 However, there are no reports regarding CGRP, VEGF-A, or OPN mRNA levels during mandibular bone formation. Conversely, previous studies have demonstrated a role for Meckel’s cartilage anterior to the intramembranous ossification centre during ossification of the mouse mandible.30 Receptor activator of nuclear factor-kappa B (RANKL), osteoprotegerin (OPG)31 and MMPs32 acts to degrade Meckel’s cartilage during endochondral bone formation in the mandible. Interleukin-beta also alters chondrocyte activity in Meckel’s cartilage during mandible formation.33 However, there are many events that influence Meckel’s cartilage during mandible formation. For example, CGRP in osteoblasts is a marker for increasing bone density.34 However, there are few reports on the relationship between CGRP, OPN and osteocalcin during the formation of bone under cartilage. In general, the expression of and correlation between CGRP and osteogenic markers in the developing mandible during craniofacial formation have remained unclear. Here, we use real-time PCR to quantify CGRP mRNA and the mRNA of other bone and cartilage morphological markers to differentiate between terminal mouse mandible and leg bone (tibia). Moreover, we obtained localization data on these markers using in situ hybridization levels in the two regions.
Materials and Methods
Sample preparation
All laboratory animals were procured from the Nippon Medical Science Animal Resource Laboratory and were bred at the Animal Testing Centre of the Department of Dentistry, Nippon Dental University. Male mice (Cler Japan, Inc., Tokyo, Japan) were maintained on a solid pellet diet (MF; Oriental Yeast Inc., Tokyo, Japan). The mice were used on embryonic days 12.5 (E12.5), E14.5, E17.5, and E18.5 and postnatal days 0 (P0), P1, and P5. The animals were sacrificed using an overdose of pentobarbital, and the right mandible and tibia were subsequently removed. Fresh samples were isolated and prepared from the right mandible and tibia from four groups. Tissue from each stage (n=4) was used for light microscopy studies and mRNA analyses using real-time RT-PCR.
Analysis of mRNA using real-time RT-PCR
Isolation of total RNA
The mandible and tibia were removed from each mouse (E12.5, E14.5, E17.5, E18.5, P0, P1 and P5) by immediately scraping after sacrifice and then storing at - 80°C. The mandible and tibia were cut into small pieces, and 20-50 mg of total RNA was isolated from the samples using an RNeasy Mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. Contaminating DNA was removed using RNase-free DNase (DNA-free; Ambion, Austin, TX, USA), and the total RNA was quantified using a spectrophotometer (Biowave S2100; Cambridge, UK) at an absorbance of 260 nm. The samples were stored at -80°C until further use. Total RNA was converted to cDNA using 0.4 µM random hexamers (N808-0127; Applied Biosystems, Carlsbad, CA, USA) in a mixture containing dNTPs at 1 mM each, 20 units of RNase inhibitor (2311A; TaKaRa, Tokyo, Japan), 5 units of AMV reverse transcriptase XL (2620A; TaKaRa), 25 mM Tris-HCl (pH 8.3), 50 mM KCl, 2 mM DTT, and 5 mM MgCl2. The following thermal PCR incubation conditions were used: 30°C for 10 min, 42°C for 30 min, 90°C for 5 min, and 5°C for 5 min. We obtained separate connective tissue samples with mesenchymal cells from Meckel’s cartilage using a microscope (LEICA MZ16, Leica Microsystems, Bensheim, Germany) to separate the mandible bone-like connective tissue at E12.5.
Quantitative real-time RT-PCR
Quantitative real-time RT-PCR was performed using the Applied Biosystems 7300 Fast Real-Time PCR System (Applied Biosystems,ì) according to the manufacturer’s instructions. Each amplification mixture (50 µL) contained 100 ng of cDNA, 900 nM forward primer, 900 nM reverse primer, 250 nM fluorogenic probe, and 25 µL of Universal PCR Master Mix (Applied Biosystems). The PCR cycling parameters were 50°C for 2 min and 95°C for 10 min, followed by 50 cycles of 95°C for 15 s and 60°C for 1 min. The following sequences were amplified: embryonic CGRP (Mm00801463_g1; Applied Biosystems), VEGF-A (vascular endothelial growth factor A, Mm00437304_m1, Applied Biosystems, CA, USA), PECAM (CD31), (Mm01242584_m1; Applied Biosystems), anti-collagen I (Col I) (At02200180_g1; Applied Biosystems), anti-collagen II (Col II) (Mm 01309565_m1; Applied Biosystems), OPN (Mm00436767_m1; Applied Biosystems), matrix metalloproteinase 2 (MMP-2) (Mm00439498_m1; Applied Biosystems), and lymphatic vessel endothelial hyaluronan receptor 1 (LYVE-1) (Mm004 75056_m1; Applied Biosystems). TaqMan Rodent GAPDH Control Reagents with the VIC Probe were also used (Applied Biosystems). Levels of the amplified mouse cDNAs were normalized to those of GAPDH (rodent GAPDH primers and probes were obtained from “Assays-On-Demand”, Applied Biosystems). The threshold cycle (Ct), defined as the cycle at which amplification of the PCR product enters the exponential phase, was determined for each gene by plotting the fluorescence level versus cycle number on a logarithmic scale. The relative expression levels of the genes of interest (Col I, Col II, VEGF-A, CD31, LIVE-1, MMP-2, OPN, CGRP) were estimated by calculating the ΔCt value, defined as the difference in the Ct values of the targets and the reference gene (GAPDH), according to the manufacturer’s recommendations. The ΔCt was inversely proportional to the level of each mRNA transcript present in the muscle samples from the mice. A higher Ct value corresponded to a lower mRNA level. The levels of amplified mouse cDNAs were expressed as the quantity divided by the value of the same gene at E12.5 on the Y-axis of each graph.
In situ hybridization
DNA fragments of 272 bp corresponding to murine CGRP (GenBank accession number NM_001033954.3), OPN (GenBank accession number NM_001204201.1) and VEGF-A (GenBank accession number NM_00950.4) were sub-cloned into the pGEMT-Easy vector (Promega, Madison, WI, USA) and used for the generation of sense or anti-sense RNA probes. Paraffin embedded intestinal sections (6 µm) from E12.5, E14.5 E17.5 and P1 mice were obtained from Genostaff Co., Ltd., Tokyo, Japan. The E12.5, E14.5, E17.5 and P1 mice were dissected, fixed with Tissue Fixative (Genostaff Co., Ltd), embedded in paraffin using the manufacturer’s proprietary procedures, and cut into 6-µm sections. The tissue sections were then de-paraffinized with xylene and rehydrated through an ethanol series and PBS. The sections were fixed 10% NBF (10% formalin in PBS) for 15 min at RT and then washed with PBS. The sections were treated with 4 µg/ml proteinase K in PBS for 10 min at 37°C, washed with PBS, re-fixed with 10% NBF for 15 min at RT, washed again with PBS, and placed in 0.2 N HCl for 10 min at RT. After washing with PBS, the sections were placed in 1X G-WASH (Genostaff Co., Ltd.), equal to 1X SSC. Hybridization was performed with probes at concentrations of 300 ng/ml in G-Hybo (Genostaff Co., Ltd) for 16 h at 60°C. After hybridization, the sections were washed with 1X G-WASH for 10 min at 60°C, followed by 50% formamide in 1X G-WASH for 10 min at 60°C. Next, the sections were washed twice with 1X G-WASH for 10 min at 60°C, twice with 0.1X G-WASH for 10 min at 60°C, and twice with TBST (0.1% Tween-20 in TBS) at RT. After treatment with 1X G-Block (Genostaff Co., Ltd.) for 15 min at RT, the sections were incubated with anti-DIG AP conjugate (Roche Diagnostics, USA) diluted 1:2000 with 50X G-Block (Genostaff Co., Ltd.) in TBST for 1h at RT. The sections were washed twice with TBST and then incubated in 100 mM NaCl, 50 mM MgCl2, 0.1% Tween-20, and 100 mM Tris-HCl, pH 9.5. Colorimetric reactions were performed with NBT/BCIP solution (Sigma-Aldrich, St. Louis, MO, USA) overnight and then washed with PBS. The sections were counterstained with Kernechtrot stain solution (Muto Pure Chemicals Co., Ltd., Tokyo, Japan), and mounted with G-Mount (Genostaff Co., Ltd.). The stained sections were evaluated using microscopy (DM-2500; Leica Microsystems).
Paraffin-embedded sections for CGRP and CD31
CGRP and CD31 paraffin-embedded blocks for immunohistochemistry were obtained from E14.5 and E17.5 mice from Genostaff Co., Ltd. Mouse embryos were fixed with Tissue Fixative (Genostaff Co., Ltd.), embedded in paraffin using Genostaff’s proprietary procedures and cut into 6-µm sections. The tissue sections used for staining with antibody against CGRP were deparaffinized with xylene and rehydrated through a series of ethanol solutions in PBS. Endogenous peroxidases were blocked with 0.3% H2O2 in methanol for 30 min, followed by incubation with G-Block (Genostaff Co., Ltd.) and the use of an avidin/biotin blocking kit (Vector SP-2001, Vector Laboratories, Burlingame, CA, USA). The sections were then incubated at 4°C overnight with rabbit polyclonal antibodies against CGRP (1 µg/mL; ENZO BML-CA1134, Cosmo Bio, Tokyo, Japan) or normal rabbit Ig (Dako X0936, Agilent Technologies, Santa Clara, CA, USA) as a negative control. The sections were washed two times with TBST for 10 min, followed by TBS for 10 min and subsequent incubation with biotin-conjugated goat anti-rabbit Ig (1:600; Dako E0432, Agilent Technologies) at room temperature for 30 min. The tissue sections to be incubated with antibodies against CD31 were deparaffinized with xylene and rehydrated through a series of ethanol solutions in PBS. Antigen retrieval was performed by microwave treatment with citrate buffer, pH 6.0. Endogenous peroxidases were blocked with 0.3% H2O2 in methanol for 30 min, followed by incubation with G-Block (Genostaff Co., Ltd.) and use of the avidin/biotin blocking kit (Vector SP-2001, Vector Laboratories). The sections were subsequently incubated at 4°C overnight with rabbit polyclonal antibody against CD31 (1 µg/mL; E11110 Spring Bioscience, Pleasanton, CA, USA) or with normal rabbit Ig as a negative control. The sections were washed two times with TBST for 10 min, followed by TBS for 10 min and subsequently incubated with biotin-conjugated goat anti-rabbit Ig (1:600; Dako E0432, Agilent Technologies) at room temperature for 30 min. After washing with TBST and TBS, the sections were incubated with peroxidase-conjugated streptavidin; Nichirei, Tokyo, Japan) at room temperature for 5 min. Peroxidase activity was visualized using diaminobenzidine. The sections were counterstained with Mayer’s Haematoxylin (MUTO Pure Chemicals Co., Ltd., Tokyo, Japan), dehydrated, and subsequently mounted with Malinol (MUTO Pure Chemicals Co., Ltd.). The stained sections were evaluated using microscopy (DM-2500; Leica Microsystems).
Statistical analysis
Differences in the quantitative real-time RT-PCR data among the groups were assessed using two-way analysis of variance (ANOVA) followed by Bonferroni’s post-hoc test with one categorical independent variable and one continuous variable (the independent variable can consist of a number of groups). The level of significance was set as P<0.05. The results are reported as the means ± SD. All statistical analyses were performed using IBM SPSS Statistics (Base, ver. 23) (New York, NY, USA).
Ethics
All procedures involving mice were reviewed and approved by the Nippon Medical Science Animal Resource Laboratory Committee of the Nippon Dental University (No. 15-31).
Results
In situ hybridization in the mandible and tibia
General observations
Haematoxylin and eosin (HE)-stained sagittal sections of mouse heads (Figure 1 a-d, m-p) are shown at E12.5 (Figure 1 a,m), E14.5 (Figure 1 b,n), E17.5 (Figure 1 c,o) and P1 (Figure 1 d,p). Meckel’s cartilage (MC) was located in the lateral and posterior region around the condylar process of the mandible from E12.5 to P1 (Figure 1 e-l, Figure 2 a-h, and Figure 3 a-h). In the sagittal section of the mandible at E12.5, mesenchymal cells and oral epithelial cells were aggregated, forming a cell clot. MC appeared in the mesenchymal cells of the posterior region of the mandible (Figure 1 a,e). The dental lamina appeared at E14.5, and immature bone trabeculae-like structure was also observed in the lateral-posterior region of the mandible. This immature bone trabeculae-like structure with mesenchymal cells appeared at the supra-surface and basal regions of the mandible above the immature salivary glands at E14.5 (Figure 1 b,f,j, Figure 2 b,f, and Figure 3 b f). In E17.5, bell-stage tooth germs appeared in the mandible. The immature bone trabeculae of the mandible were located at the superior, inferior and anterior regions of MC (Figure 1 g,k). At P1, numerous developed bone trabeculae-like structures appeared along with the MC in the mandible (Figure 1 d,h,l, Figure 2 d,h, and Figure 3 d,h). In contrast, at first, proximally located gathered mesenchyme cells were observed starting at E12.5, and then, chondrogenic cells were observed at E14.5 in the hind limb. In the mouse tibia, a distal developed growth plate and proximal cartilage cell zone were observed from E17.5 to P1 (Figure 1; Table 1).
Figure 1.
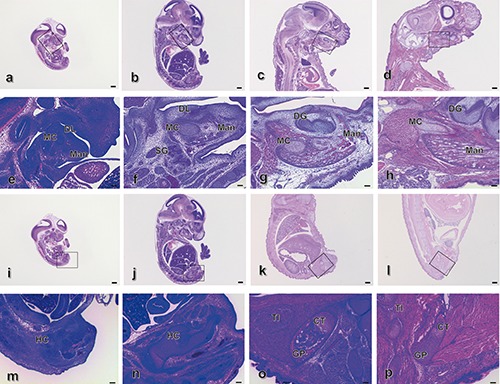
Haematoxylin and eosin (HE) staining of sagittal sections of mouse heads at E12.5 (a,e), E14.5 (b,f), E17.5 (c, ìg) and P1 (d,h) and tibias at E12.5 (i,m), E14.5 (j,n), E17.5 (k,o) and P1 (l,p). e) Magnification of the squared region in (a); f) magnification of the squared region in b); g) magnification of the squared region in (c); h) magnification of the squared region in (d); m) magnification of the squared region in (i); m) magnification of the squared region in (j); o) magnification of the squared region in (k); p) magnification of the squared region in (l). Mad, mandible; BT, bone trabecula-like matrices; MC, Meckel’s cartilage; DL, dental lamina; DG, dental germ; TI, tibia; HC, hyaline cartilage. Scale bars: a-d, i-l), 500 µm, magnification: 0.71x; e-h, m-p), 100 µm, magnification: 4x.
Figure 2.
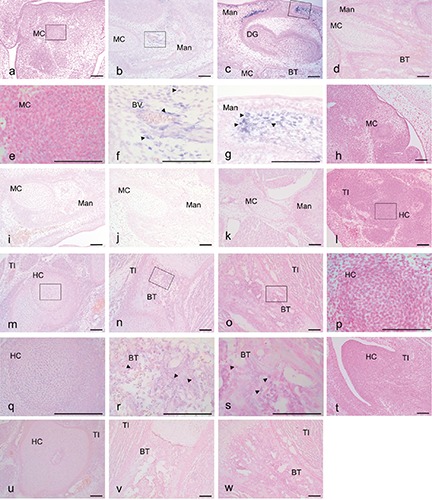
Expression of CGRP mRNA based on in situ hybridization with an anti-sense (a-g, l-o) and a sense (h-k, t-w) probe in mouse heads at E 12.5 (a,e,h), E14.5 (b,f,i), E17.5 (c,g,j) and P1 (d,k) and tibias at E 12.5 (l,p,t), E14.5 (m,q,u), E17.5 (n,r,v) and P1 (o,s,w). The CGRP reaction was localized in (c) and (g) (arrowheads); the CGRP reaction was scattered in (f,r,s) (arrowheads). e) Magnification of the squared region in (a); f) magnification of the squared region in b); g) magnification of the squared region in (c); p) magnification of the squared region in (l); q) magnification of the squared region in (m); r) magnification of the squared region in (n); s) magnification of the squared region in (o). Mad, mandible; B T, bone trabecula-like matrices; MC, Meckel’s cartilage; DG; dental germ; TI, tibia; HC, hyaline cartilage. Scale bars: 100 µm. a-d, h-o, t-w) magnification: 10x; e-g, p-s) magnification: 40x.
Figure 3.
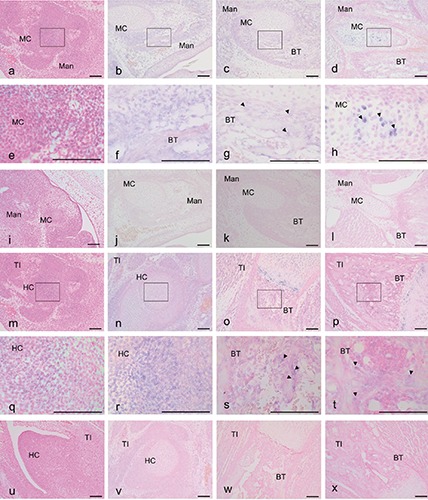
Expression of VEGF-A mRNA based on in situ hybridization with an anti-sense (a-h, m-t) and a sense (i-l, u-x) probe in mouse heads at E 12.5 (a,e,i), E14.5 (b,f,j), E17.5 (c,g,k) and P1 (d,h,l) and tibias at E 12.5 (m,q,u), E14.5 (n,r,v), E17.5 (o,s,w) and P1 (p,t,x). The VEGF-A reaction was localized in (g,h,s,t) (arrowheads); the VEGF-A reaction was scattered in (f,g,r) (arrowheads). e) Magnification of the squared region in (a); f) magnification of the squared region in (b); g) magnification of the squared region in (c); h) magnification of the squared region in (d); q) magnification of the squared region in (m); r) magnification of the squared region in (n); s) magnification of the squared region in (o); t) magnification of the squared region in (p). Mad, mandible; BT, bone trabecula-like matrices; MC, Meckel’s cartilage; TI, tibia; HC, hyaline cartilage. Scale bars: 100 µm. a-d, i-p, u-x) magnification: 10x; e-h, q-t) magnification: 40x.
Table 1.
In situ hybridization activity of CGRP, VEGF and OPN in the mandible and tibia.
| CGRP | VEGF | OPN | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E12.5 | E14.5 | E17.5 | P1 | E12.5 | E14.5 | E17.5 | P1 | E12.5 | E14.5 | E17.5 | P1 | |
| Mandible | ||||||||||||
| Numerous mesenchymal cells at MC | + | + | - | - | + | + | ++ | ++ | - | - | - | - |
| Mesenchymal cells surrounding bone matrix | - | + | - | - | - | - | - | - | - | + | + | + |
| Mesenchymal cells surrounding blood vessels | - | + | - | - | - | - | - | - | - | - | - | - |
| Supra-surface | - | - | + | - | - | - | - | - | - | - | - | - |
| Basal region | - | + | - | - | - | - | - | - | - | - | - | - |
| Bone matrix | - | - | - | - | - | + | + | + | - | + | +++ | +++ |
| Nuclei | - | - | - | - | - | ++ | - | - | - | - | - | - |
| Cell membranes | - | - | - | - | - | ++ | - | - | - | - | - | - |
| Chondrocyte of MC | - | - | - | - | - | - | - | - | - | - | + | + |
| Tibia | ||||||||||||
| Numerous mesenchymal cells at HC | + | + | - | - | + | + | - | - | - | - | - | - |
| Hypertrophic zone | - | - | + | + | - | - | ++ | ++ | - | - | ++ | ++ |
| Bone matrix | - | - | - | - | - | - | + | + | - | - | +++ | +++ |
| Pre-hypertrophic zone | - | - | - | - | - | - | ++ | ++ | - | - | - | - |
MC, Meckel’s cartilage; HC, hypertrophic cartilage; -, not detected; +, scatter; ++, concentrate; +++, condense.
In situ hybridization of CGRP
Hybridization with an anti-sense probe for CGRP was detected in numerous mesenchymal cells of both the mandible (Figure 2 a,e) and the tibia (Figure 2 l,p) at E12.5. In mesenchymal cells surrounding bone matrix and blood vessels at E14.5, hybridization signals were clearly detected in the mandible (Figure 2 b,f) but not the tibia (Figure 2 m,q). mRNA for CGRP was not detected in the MC of the mouse mandible at E14.5 (Figure 2 b,f). However, mRNA for CGRP was detected in the supra-surface and basal regions of the mandible (Figure 2 c,g) at E17.5. Furthermore, mRNA for CGRP was not detected in the mandible (Figure 2d) at P1. In the tibia, mRNA for CGRP was clearly detected at the hypertrophic zone from E17.5 (Figure 2 n,r) to P1 (Figure 2 o,s), and no specific signals using sense probes were detected in the mandible (Figure 2 h-k) or the tibia (Figure 2 u-x).
In situ hybridization of VEGF-A
Low levels of hybridization with an anti-sense probe for VEGF-A were detected in numerous mesenchymal cells in the mandible (Figure 3 a,e) and tibia (Figure 3 m,q) at E12.5. In the mesenchymal cells and bone matrices at E14.5, cytoplasmic positivity for the hybridization signals was observed in the mandible (Figure 3 b,f). However, almost no localized signal was detected in the tibia (Figure 3 n,r). mRNA for VEGF-A was also not detected at the MC of the mouse mandible at E14.5 (Figure 2 b,f). In contrast, mRNA for VEGF-A was detected in numerous mesenchymal cells around MC and bone matrices in the mandible at E17.5 (Figure 3 c,g). mRNA for VEGF-A was also not detected in the MC of the mouse mandible at E17.5 (Figure 3 c,g). mRNA for VEGF-A was strongly detected in some chondrocytes of the MC in the mandible (Figure 3 d,h). In the tibia, mRNA for VEGF-A was clearly detected in the hypertrophic zone at E14.5 (Figure 3 n,r) and then clearly shifted between the prehypertrophic and hypertrophic zones from E17.5 and P1 (Figure 3 o,p,s,t), and no specific signals using sense probes were detected in the mandible (Figure 3 e-h) or the tibia (Figure 3 u-x).
In situ hybridization of OPN
Hybridization with an anti-sense probe for OPN was detected in numerous mesenchymal cells around the bone matrices and MC in the mandible at E14.5 (Figure 4 b,e). The mRNA for OPN was detected around the bone matrices in the mandible at E17.5 (Figure 4 c,f). The mRNA for OPN was strongly detected at P1 around the bone matrices and some chondrocyte cells of the MC in the mandible (Figure 4 d,g). Moreover, the mRNA for OPN was clearly observed in the hypertrophic zone of the tibia at E17.5 and P1 but not at E12.5 and E14.5 (Figure 4 l-q), and no specific signals using sense probes were detected in the mandible (Figure 4 h-k) or the tibia (Figure 4 r-u).
Figure 4.
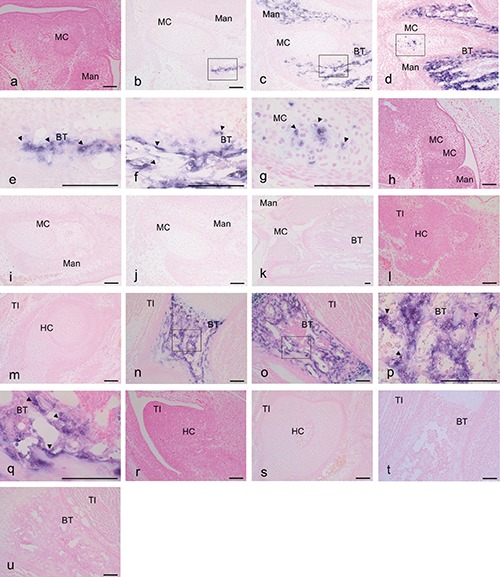
Expression of OPN mRNA based on in situ hybridization with an anti-sense (a-g, n-q) and a sense (h-k, r-u) probe in mouse heads at E 12.5 (a,h), E14.5 (b,e,i), E17.5 (c,f,j) and P1 (d,g,k) and tibias at E 12.5 (l,r), E14.5 (m,s), E17.5 (n,p,t) and P1 (o,q,u). The reaction of OPN was localized in all reaction plates (arrowheads). e) Magnification of the squared region in (b); f) magnification of the squared region in (c); g) magnification of the squared region in (d); p) magnification of the squared region in (n); q) magnification of the squared region in (o). Mad, mandible; B T, bone trabecula-like matrices; MC, Meckel’s cartilage; TI, tibia; HC, hyaline cartilage. Scale bars: 100 µm. a-d, h-o, r-u) magnification: 10x; e-g, p,q) magnification: 40x.
Immunohistochemistry for CGRP and CD31 in the mandible and tibia
At E17.5, the anti-CGRP antibody revealed a slight positive reaction in mesenchymal cells in the bone trabeculae-like structures of the mandible; by contrast, in the tibia, a positive reaction was observed in numerous chondrocytes (Figure 5 a,b,d,e). The anti-CD31 antibody reaction was primarily observed in the mesenchymal cells surrounding the MC of the mandible and subsequently in the hyaline cartilage of the tibia at E14.5 (Figure 5 g,h,j,k).
Figure 5.
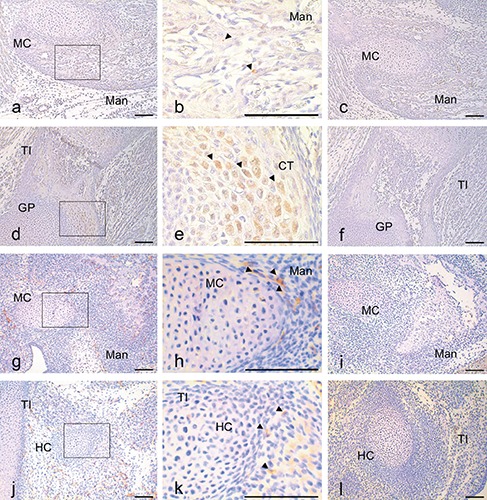
Immunohistochemical staining for anti-CGRP in sagittal sections of E17.5 mice and for anti-CD31 in sagittal sections of E14.5 mice. Positive anti-CGRP reactions (a,b,d,e) and negative controls (c,f) in mouse head at E17.5 (a-c) and tibia at E17.5 (m-s). Positive anti-CD31 reactions (g,h,j,k) and negative controls (i,l) in mouse head at E14.5 (g-i) and tibia at E14.5 (j-l). The reactions of CGRP and CD31 were observed through immunohistochemical localization in all reaction plates (arrowheads). b) Magnification of the squared region in (a); e) magnification of the squared region in (d). Mad, mandible; BT, bone trabecula-like matrices; MC, Meckel’s cartilage; TI, tibia; HC, hyaline cartilage. Scale bars: 100 µm. a,c,d,f) magnification: 10x; b,e) magnification: 40x.
Quantification of mRNA abundance in the mandible and tibia using real-time RT-PCR
The abundance of VEGF-A, CD31, MMP-2, CGRP, Col I, Col II, LYVE-1, and OPN mRNA in the mouse mandible and tibia at 7 different stages ranging from E12.5 to P5 is shown in Figure 4 a-h. The abundance of CGRP mRNA gradually decreased from E12.5 to E17.5 and then remained almost constant from P0 to P5 in the mandible (P<0.001). In contrast, the CGRP mRNA levels in the tibia gradually increased from E12.5 to E17.5 and then decreased at P5. Different levels of CGRP mRNA in the mandible and tibia were observed at E14.5 and E17.5 (P<0.001) (Figure 6a).
Figure 6.
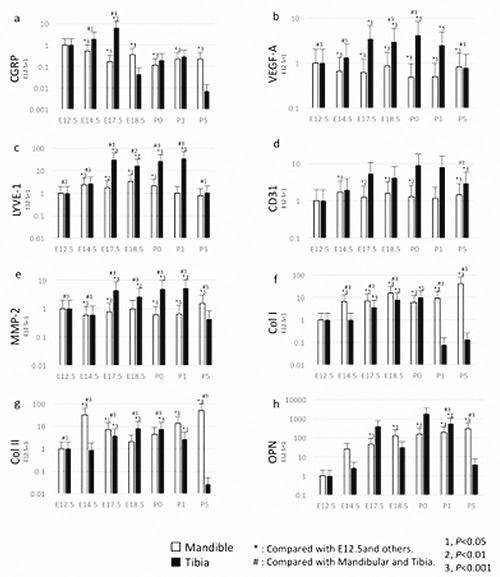
Expression of VEGF-A, CD31, MMP-2, CGRP, Col I, Col II, and OPN mRNA over time (E12.5, E14.5, E17.5, E18.5, P0, P1 and P). The abundance of VEGF-A, CD31, MMP-2, CGRP, Col I, Col II, and OPN mRNA was normalized to GAPDH at E12.5, E14.5, E17.5, E18.5, P0, P1 and P5, followed by normalization to the target gene expression at E12.5. The white bars represent the mandible, and the blacks bar represent the tibia. a, VEGF-A mRNA expression; b, CD31 mRNA expression; c, MMP-2 mRNA expression; d, CGRP mRNA expression; e, Col I mRNA expression; f, Col II mRNA expression; g, LYVE-1 mRNA expression; h, OPN mRNA expression. Differences between E12.5, E14.5, E17.5, E18.5, P0, P1 and P5: 1; P<0.05, 2; P<0.01, 3; P< 0.001. *Compared with E12.5 and others; #compared with the mandible and the tibia. All error bars represent mean ± SD.
The levels of Col I, Col II and OPN mRNA gradually increased in the mandible from E12.5 to P5 (P<0.001), with the exception of Col II at E14.5. The abundance of CD31, MMP2 and LYVE-1 mRNAs remained constant in the mandible from E12.5 to P5, while the levels of VEGF-A were low during this time period. The levels of VEGF-A, CD31, MMP-2, Col I, Col II and OPN mRNA gradually increased in the tibia from E12.5 to P1 (P<0.001). However, the levels of these mRNAs were low at P5, and Col I was also low at P1. Furthermore, different levels of Col I, Col II, and OPN mRNA were observed between the mandible and the tibia at P5 (P<0.001) (Figure 6 e,f,h).
Discussion
CGRP first appears in mouse at E16.5, localizes around blood vessels and is closely opposed to the cartilaginous bone matrix in the limb.1 Here, we initially detected the anti-sense probe for CGRP in numerous mesenchymal cells of the tibia at E12.5 and then clearly detected it in both the mandible and tibia at E14.5. The appearance of the ossification centre occurs earlier in the mandible than in the human limb.35,36 Moreover, our in situ hybridization faintly detected CGRP in the chondroblasts of the MC and numerous mesenchymal cells around the MC. Additionally, we clearly observed CGRP mRNA in the mesenchymal cells around the blood vessels and immature bone matrices of the developing mouse mandible at E14.5. CGRP mRNA was clearly detected using in situ hybridization in the numerous mesenchymal cells of the tibia at E12.5. Based on these results, CGRP expression regulates bone matrix formation between E12.5 and E17.5 in mandibular bone and tibia. In contrast, the expression of CGRP mRNA is detectable in neuronal cells near trabecular bone structures, and components of the CGRP receptor are expressed in differentiated osteoblast cultures.8 Here, CGRP hybridization with the anti-sense probe was clearly detected in the osteoblast-like cells surrounding the blood vessels in immature bone matrices. These results are consistent with those of a previous study showing that CGRP expression is associated with blood vessels during the formation of the mouse masseter muscle.37 In the present study, during bone matrix formation, CGRP expression around the blood vessels started at E14.5 in the mandible and E17.5 in the tibia. In general, CGRP is widely distributed in the bone tissue and regulates bone remodelling through blood vessels around bone cells during growth and repair.3 CGRP-deficient mice present with low trabecular bone volume, a small number of trabeculae, and large trabecular spacing.16 Therefore, CGRP may be a primary regulator of OPN and of key bone matrix markers in the development of mandibular and tibia trabeculae and bone volume. In the present study, CGRP mRNA was detected at E12.5 and then decreased during development. In contrast, OPN and Col I gradually increased from E12.5 to E17.5 in the mouse mandible, but not the tibia. Under treatment with CGRP and osteocalcin, mRNAs were increased in a co-culture system.8 CGRP plays a role in the initial steps of trabecular bone formation, as it regulates OPN and Col I. Subsequently, OPN leads to the development of trabecular bone mineralization during the membranous ossification stage of bone formation. The results of the present study further revealed that the different levels of CGRP, OPN and Col I mRNA indicated a different mechanism of mandible formation compared to tibia formation with regards to membranous and/or cartilaginous ossification. Notably, OPN mRNA was clearly detected at the trabecular bone starting at E17.5 and then appeared at detectable levels in the mandible MC through in situ hybridization. Thus, there are different systems regulating early bone formation and metabolism between the mandible and tibia. The vertebrate skeleton and mandible are primarily derived from the mesoderma during development; however, the formation of this bone is different from that of skeletal bone and orofacial bone, which are derived from somites and branchial arches. The mandible originates from intramembranous and endochondral tissues, unlike the tibia, which originates from endochondral tissues.38-41
VEGF-A is essential to support blood vessel growth in the developing jaw, which in turn is essential for normal chondrocyte proliferation and therefore jaw extension. Cranial neural crest cell-derived VEGF-A regulates proliferation, vascularization and ossification of cartilage and membranous bone.18-21 Additionally, VEGF-A regulates the growth of the mandibular artery, leading to extension of the MC.19 In contrast, VEGF-A stimulates intramembranous bone formation in the mouse mandible despite a lack of regulated bone ossification in the normal mandible.19 In general, strong VEGF-A expression was observed in human glenoid fossa in cases of malinter-digitation.42 VEGF-A is strongly expressed by NCCs during craniofacial development,19 and mice lacking VEGF-A display craniofacial defects, including cleft palate, unfused cranial sutures, and shorter jaws.22 Here, we detected a VEGF-A mRNA pattern in the mandible similar to that of CD31, MMP-2 and Col II from E12.5 to P1. Conversely, the VEGF-A expression pattern differed from those of Co1, OPN and CGRP. In contrast, the pattern of VEGF-A mRNA in the tibia was similar to those of LYV-1, CD31 and OPN from E12.5 to P1. VEGF-A clearly affects the formation of blood vessels and bone matrices in the postnatal stages of tibia development. In the present study, the pattern of VEGF-A mRNA was similar to that of CGRP in both the mandible and tibia from E12.5 to E17.5. This finding suggests that VEGF-A may promote rapid formation of blood vessels in the tibia and mandible during embryonic development.
Localization of VEGF-A hybridization with the anti-sense probe indicated that this molecule is present between E14.5 and E18.5 in the cartilage matrix during endochondral bone formation of long bones, such as the tibia, and during ossification of cartilage and intramembranous ossification of bones, such as the mandible. The in situ hybridization analysis in the present study positively showed that accumulation of CGRP and VEGF-A expression were significantly increased at E14.5 in the mandible. However, weak CGRP and VEGF-A expression was first observed at E17.5 in the tibia. The VEGF-A gene and protein were strongly expressed in foetal tissue of mesenchymal origin derived from the dermis and hypodermis, but not in the head and neck tissues, which are of neuroectodermal and dermal–hypodermal derivation.43 Our results indicated that the localized CGRP expression was similar to that of VEGF-A expression during tibia and mandible ossification. Moreover, a positive reaction for anti-CGRP antibody was detected in mesenchymal cells in the bone trabeculae-like structures of the mandible at E17.5 and then in the hyaline cartilage of the tibia at E14.5. CGRP may regulate VEGF-A expression, and subsequently VEGF-A may be upregulated as an intermediate pathway for vascular endothelial cells and induced CD31.37 Therefore, a time lag of expression was observed between the mandible and tibia during foetal development. These results also showed that the pattern of OPN hybridization with the anti-sense probe was similar to that of CGRP in the tibia and mandible. In general, CGRP regulates the formation of bone.2-9 Therefore, CGRP and OPN may contribute to bone formation during the developmental stages we investigated here. At 12.5 days of age, the expression level of each factor is low, and bone element factors, such as OPN and Co1 I, increased in bone matrices, but no changes in factors other than these two markers were observed in the mandible and tibia. However, the mRNA expression level of vascular genesis markers differed between the mandible and tibia, and the localization of VEGF-A and CGRP mRNA expression shown using in situ hybridization clearly differed over the time course. Indeed, the localization of VEGF-A and CGRP mRNAs was first weakly detected and then spread throughout the blood vessels, affecting osteoblasts and the surrounding connective tissue, ultimately further changing the mesenchymal cells around the MC of mandible at the examined stages. However, we did not observe the expression of these markers at the protein level, and previous studies have reported that these markers are actually synthesized in conjunction with mRNA expression.43 Thus, in the present study, we observed general markers with established factors in bone formation between the mandible and tibia at the mRNA level.
Yuodelis suggested that the MC contributes to the length and width of the mandible and to masticatory muscle and jaw movement in the human temporomandibular joint.44 The intramembranous ossification centre appears anterior to the mandible, thereby associating the role of MC with ossification of the mouse mandible.31 Ishizeki et al. indicated that the posterior portion of MC is involved in rodent bone formation.45 In contrast, based on immunohistochemical analysis of RANKL, OPG and MMP distribution, MC does not regulate the formation of the mouse mandible.32,33 CGRP is a marker of osteoblasts, and CGRP mRNA is observed in chondroblasts of MC.35 Here, we observed low levels of CGRP mRNA in the chondrocytes of MC at E14.5 and E17.5 but not P0. Moreover, OPN mRNA was initially detected in the posterior region of the MC at P1 in the mouse mandible. Therefore, CGRP may regulate MC calcification starting at early postnatal stages. CGRP was also detected in mesenchymal cells of immature bone matrices and blood vessels. Therefore, CGRP likely regulates the formation of the mandible through calcification of MC.
References
- 1.Bidegain M, Roos BA, Hill EL, Howard GA, Balkan W. Calcitonin gene-related peptide (CGRP) in the developing mouse limb. Endocr Res 1995;2:743-55. [DOI] [PubMed] [Google Scholar]
- 2.Li J, Kreicbergs A, Bergström J, Stark A, Ahmed M. A site-specific CGRP innervation coincides with bone formation during facture healing and modeling: a study in rat angulated tibia. J Orthop Res 2007;25:1204-12. [DOI] [PubMed] [Google Scholar]
- 3.Irie K, Hara-Irie F, Ozawa H, Yajima T. Calcitonin gene-related peptide (CGRP) - containing nerve fibers in bone tissue and their involvement in bone remodeling. Microsc Res Tech 2002;58:85-90. [DOI] [PubMed] [Google Scholar]
- 4.Hukkanen M1, Konttinen YT, Santavirta S, Paavolainen P, Gu XH, Terenghi G, et al. Rapid proliferation of calcitonin gene-related peptideimmunoreactive nerves during healing of rat tibial fracture suggests neural involvement in bone growth and remodelling. Neuroscience 1993;54:969-79. [DOI] [PubMed] [Google Scholar]
- 5.Konttinen Y, Imai S, Suda A. Neuropeptides and the puzzle of bone. Acta Orthop Scand 1996;67:632-9. [DOI] [PubMed] [Google Scholar]
- 6.Valentijn K, Gutow AP, Troiano N, Gundberg C, Gilligan JP, Vignery A. Effects of calcitonin gene-related peptide on bone turnover in ovariectomized rats. Bone 1997;21:269-74. [DOI] [PubMed] [Google Scholar]
- 7.Millet I, Vignery A. The neuropeptide calcitonin gene-related peptide inhibits TNF-alpha but poorly induces IL-6 production by fetal rat osteoblasts. Cytokine 1997;9:999-1007. [DOI] [PubMed] [Google Scholar]
- 8.Bo Y, Yan L, Gang Z, Tao L, Yinghui T. Effect of calcitonin gane-related peptide on osteoblast differentiation in an osteoblast and endothelial cell co-culture system. Cell Biol Int 2012;36:909-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang Z, Yang Q, Xiong W, Li GH, Liao H, Xiao J, et al. Effect of CGRP-adenoviral vector transduction on the osteoblastic differentiation of rat adipose-derived stem cells. PLoS One 2013;8:e72738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong J, He Y, Zhang X, Wang L, Sun T, Zhang M, et al. Calcitonin gene-related peptide regulates the glowth of epidermal stem cell in vitro. Peptides 2010;31:1860-65. [DOI] [PubMed] [Google Scholar]
- 11.Yoo YM, Kwag JH, Kim KH, Kim CH. Effects of neuropeptides and mechanical loading on bone cell resorption in vitro. Int J Mol Sci 2014;15:5874-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Norevall LI, Forsgren S, Matsson L. Expression of neuropeptides (CGRP, substance P) during and after orthodontic tooth movement in the rat. Eur J Orthod 1995;17:311-25. [DOI] [PubMed] [Google Scholar]
- 13.Yamashiro T, Fujiyama K, Fujiyoshi Y, Inaguma N, Takano-Yamamoto T. Inferior alveolar nerve transection inhibits increase in osteoclast appearance during experimental tooth movement. Bone 2000;26:663-9. [DOI] [PubMed] [Google Scholar]
- 14.Yu T, Calvo L, Anta B, López-Benito S, López-Bellido R, Vicente-García C, et al. In vivo regulation of NGF-mediated functions by Nedd4-2 ubiquitination of TrkA. J Neurosci 2014;34:6098-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ballica R, Valentijn K, Khachatryan A, Guerder S, Kapadia S, Gundberg C, et al. Targeted Expression of Calcitonin Gene-Related Peptide to Osteoblasts Increases Bone Density in Mice. J J Bone Miner Res 1999; 14(7):1067-74. [DOI] [PubMed] [Google Scholar]
- 16.Schinke T, Liese S, Priemel M, Haberland M, Schilling AF, Catala-Lehnen P, et al. Decreaed bone formation and osteopenia in mice lacking alpha-calcitonin gene-related peptide. J Bone Miner Res 2004;19:2049-56. [DOI] [PubMed] [Google Scholar]
- 17.Hoff AO, Catala-Lehnen P, Thomas PM, Priemel M, Rueger JM, Nasonkin I, et al. Increased bone mass is an unexpected phenotype associated with deletion of the calcitonin gene. J Clin Invest 2002;10:1849-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stalmans I, Lambrechts D, De Smet F, Jansen S, Wang J, Maity S, et al. VEGF: A modifier of the del22q11 (DiGeorge) syndrome? Nat Med 2003;9:173-82. [DOI] [PubMed] [Google Scholar]
- 19.Wiszniak S, Mackenzie FE, Anderson P, Kabbara S, Ruhrberg C, Schwarz Q. Neural crest cell-derived VEGF promotes embryonic jaw extention. Proc Natl Acad Sci USA 2015;112:6086-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill C, Jacobs B, Kennedy L, Rohde S, Zhou B, Baldwin S, et al. Cranial neural crest deletion of VEGFa causes cleft palate with aberrant vascular and bone development. Cell Tissue Res 2015; 361:711-22. [DOI] [PubMed] [Google Scholar]
- 21.Duan X, Bradbury SR, Olsen BR, Berendsen AD. VEGF stimulates intramembranous bone formation during craniofacial skeletal development. Matrix Biol 2016;52-54:127-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zelzer E, McLean W, Ng YS, Fukai N, Reginato AM, Lovejoy S, et al. Skeletal defects in VEGF (120/120) mice reveal multiple roles for VEGF in skeletogenesis. Development 2002;129:1893-904. [DOI] [PubMed] [Google Scholar]
- 23.Nomura S, Wills AJ, Edwards DR, Heath JK, Hogan BL. Development expression of 2ar (osteopontin) and SPARC (osteonectin) RNA as revealed by in situ hybridization. J Cell Biol 1988;106:441-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sugimoto M, Hirota S, Sato M, Kawahata H, Tsukamoto I, Yasui N, et al. Impaired expression of nocollagenous bone matrix protein mRNAs during fracture healing in ascorbic acid-deficient rats. J Bone Miner Res 1998; 13(2):271-8. [DOI] [PubMed] [Google Scholar]
- 25.Shibata S, Fukada K, Suzuki S, Ogawa T, Yamashita Y. In situ hybridization and immunohistochemistry of bone sialoprotein and secreted phosphoprotein 1 (osteopontin) in the developing mouse mandibular condylar cartilage compared with limb bud cartilage. J Anat 2002;200:309-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ichikawa H, Itota T, Nishitani Y, Torii Y, Inoue K, Sugimoto T. Osteopontin-immunoreactive primary sensory neurons in the rat spinal and trigeminal nervous systems. Brain Res 2000;863: 276-81. [DOI] [PubMed] [Google Scholar]
- 27.Shibata S, Fukuoka H, Sato R, Abe T, Suzuki Y. An in situ hybridization study of the insulin-like growth factor system in developing condylar cartilage of the fetal mouse mandible. Eur J Histochem 2012;56:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shibata S, Sakamoto Y, Baba O, Qin C, Murakami G, Cho BH. An immunohistochemical study of matrix proteins in the craniofacial cartilage in midterm human fetuses. Eur J Histochem 2013;57:e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujikawa K, Yokohama-Tamaki T, Morita T, Baba O, Qin C, Shibata S. An in situ hybridization study of perlecan, DMP1, and MEPE in developing condylar cartilage of the fetal mouse mandible and limb bud cartilage. Eur J Histochem 2015;59:2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frommer J, Margolies MR. Contribution of Meckel’s cartilage to ossification of the mandible in mice. J Dent Res 1971;50:1260-7. [DOI] [PubMed] [Google Scholar]
- 31.Sakakura Y, Tsuruga E, Irie K, Hosokawa Y, Nakamura H, Yajima T. Immunolocalization of receptor activator of nuclear factor-κB ligand (RANKL) and osteoprotegerin (OPG) in Meckel’s cartilage compared with developing endochondral bones in mice. J Anat 2005;207:325-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakakura Y, Hosokawa Y, Tsuruga E, Irie K, Nakamura M, Yajima T. Contributions of matrix metalloproteinases toward Meckel’s cartilage resorprion in mice: immunohistochemical studies, including comparisons with developing endochondral bones. Cell Tissue Res 2007;328:137-51. [DOI] [PubMed] [Google Scholar]
- 33.Tsuzurahara F, Soeta S, Kawawa T, Baba K, Nakamura M. The role of macrophages in the disappearance of Meckel’s cartilage during mandibular development in mice. Acta Histochem 2011;113:194-200. [DOI] [PubMed] [Google Scholar]
- 34.Ballica R, Valentijn K, Khachatryan A, Guerder S, Kapadia S, Gundberg C, et al. Targeted expression of calcitonin gene-related peptide to osteoblasts increases bone density in mice. J Bone Miner Res 1999;14:1067-74. [DOI] [PubMed] [Google Scholar]
- 35.Sperber GH. Craniofacial embryogenetics and development. J. Wright & Sons, Bristol, UK; 1976; pp. 147-159. [Google Scholar]
- 36.Tanaka O. Time of the appearance of catilage centers in human embryos with special reference to its individual difference. Okajimas Folia Anat Jpn 1976;53:173-98. [DOI] [PubMed] [Google Scholar]
- 37.Azuma Y, Miwa Y, Sato I. Expression of CGRP in embryonic mouse masseter muscle. Ann Anat 2016;206:34-47. [DOI] [PubMed] [Google Scholar]
- 38.Chung UI, Kawaguchi H, Takato T, Nakamura K. Distinct osteogenic mechanisms of bones of distinct origins. J Orthop Sci 2004;9:410-4. [DOI] [PubMed] [Google Scholar]
- 39.Kuratani S. Cephalic neural crest cells and the evolution of craniofacial structures in vertebrates: morphological and embryological significance of the premandibular-mandibular boundary. Zoology (Jena) 2005;108:13-25. [DOI] [PubMed] [Google Scholar]
- 40.Leucht P, Kim JB, Amasha R, James AW, Girod S, Helms JA. Embryonic origin and Hox status determine progenitor cell fate during adult bone regeneration. Development 2008;135: 2845-54. [DOI] [PubMed] [Google Scholar]
- 41.Reichert JC, Gohlke J, Friis TE, Quent VM, Hutmacher DW. Mesodermal and neural crest derived ovine tibial and mandibular osteoblasts display distinct molecular differences. Gene 2013;525: 99-106. [DOI] [PubMed] [Google Scholar]
- 42.Rabie AB, Shum L, Chayanupatkul A. VEGF and bone formation in the glenoid fossa during forward mandibular positioning. Am J Orthod Dentofacial Orthop 2002;122:202-9. [DOI] [PubMed] [Google Scholar]
- 43.Marini M, Bertolai R, Ambrosini S, Sarchielli E, Vannelli GB, Sgambati E. Differential expression of vascular endothelial growth factor in human fetal skeletal site-specific tissues: Mandible versus femur. Acta Histochem 2015; 117:228-34. [DOI] [PubMed] [Google Scholar]
- 44.Yuodelis RA. The morphogenesis of the human temporomandibular joint and its associated structures. J Dent Res 1966;45:182-91. [DOI] [PubMed] [Google Scholar]
- 45.Ishizeki K, Kubo M, Yamamoto H, Nawa T. Immunocytochemical expression of type I and type II collagens by rat Meckel’s chondrocytes in culture during phenotypic transformation. Arch Oral Biol 1998;43:117-26. [DOI] [PubMed] [Google Scholar]


