Abstract
Structural genomics has as its goal the provision of structural information for all possible ORF sequences through a combination of experimental and computational approaches. The access to genome sequences and cloning resources from an ever-widening array of organisms is driving high-throughput structural studies by the New York Structural Genomics Research Consortium. In this report, we outline the progress of the Consortium in establishing its pipeline for structural genomics, and some of the experimental and bioinformatics efforts leading to structural annotation of proteins. The Consortium has established a pipeline for structural biology studies, automated modeling of ORF sequences using solved (template) structures, and a novel high-throughput approach (metallomics) to examining the metal binding to purified protein targets. The Consortium has so far produced 493 purified proteins from >1077 expression vectors. A total of 95 have resulted in crystal structures, and 81 are deposited in the Protein Data Bank (PDB). Comparative modeling of these structures has generated >40,000 structural models. We also initiated a high-throughput metal analysis of the purified proteins; this has determined that 10%-15% of the targets contain a stoichiometric structural or catalytic transition metal atom. The progress of the structural genomics centers in the U.S. and around the world suggests that the goal of providing useful structural information on most all ORF domains will be realized. This projected resource will provide structural biology information important to understanding the function of most proteins of the cell.
The complete genomes of a number of organisms have been sequenced and many more are underway. This progress in gene sequencing has shifted the landscape of biology, such that goals related to understanding the structure and function of each gene product, as well as their interactions within the cellular environment that lead to the behavior of complex systems are within reach, or at least to be contemplated. The sequencing of model organisms from bacterial species to human has allowed the identification of genes both essential to function, as well as genes that give rise to the diversity of life forms. Although the exact numbers and natures of the genes is still open to question, recent estimates place the numbers at <20,000 for Caenorhabditis elegans and Caenorhabditis briggsae and ∼30,000 for humans (Waterston et al. 2002; Stein et al. 2003). Our ability to recognize genes, their exons and introns, and their potential splice variants, has matured dramatically. This progress has driven highly successful attempts to develop resources to make available ORFs for rapid and highly parallel structural and functional studies of genes (Reboul et al. 2003). The success of these efforts are outlined in this issue, and the leveraging of these ORF sequences to examine protein activity, localization, protein structure, and protein-protein interactions are examples of the value of these resources. Structural biology faces the task of characterizing the shapes and dynamics of the encoded proteins to facilitate the understanding of their functions and mechanisms of action. These ORF resources will ultimately be critical to the success of the nascent structural genomics initiatives that are underway, both in the U.S. and worldwide (Burley et al. 1999; Chance et al. 2002; Lesley et al. 2002; Burley and Bonanno 2003; Gerstein et al. 2003; Goulding et al. 2003; Shi et al. 2003; Terwilliger et al. 2003; Zhang and Kim 2003).
The Protein Structure Initiative (PSI) funded by the National Institute of General Medical Sciences (www.nigms.nih.gov/psi) includes the structural genomics efforts of nine centers in the United States. In the so-called phase 1 of the PSI (Editorial 2004) these multi-institutional collaborations, along with six additional structural genomics centers in Europe and Japan, are building and developing infrastructure to provide integrated pipelines such that in phase 2, beginning in 2005, the goal of providing useful three-dimensional models for most of the known protein sequences can be vigorously pursued. The goal is to be accomplished by selecting and experimentally determining 10,000-15,000 protein structures using X-ray crystallography or NMR spectroscopy. The ORF target selection will be carried out such that at least one representative structure will be solved for most protein families. This solved structure will be used as a structural template to generate all atom-comparative protein models for the other members of the ORF family (Baker and Sali 2001; Vitkup et al. 2001). Each center in the PSI is responsible for developing and testing an integrated structural genomics effort that includes ORF target selection, cloning, expression testing, protein purification, structure solution, and modeling. This effort has required close coordination within the centers in terms of task assignment and monitoring progress, as well as coordination among the centers to maximize the effectiveness of target selection. The latter has been facilitated by the development of a database, Target DB (http://targetdb.pdb.org), which lists the selected targets and progress for all of the centers in a queryable form (Westbrook 2003). This database allows the centers and the NIH to monitor overlap in target selection and has allowed many scientists outside of the centers to access information on proteins of interest to their research programs.
As a rule, the ORF targets selected for the structural genomics efforts are <30% identical (across a reasonable length) to proteins already deposited in the PDB (Sali 1998; Burley et al. 1999; Baker and Sali 2001; Vitkup et al. 2001; Chance et al. 2002; Sali et al. 2003; Shi et al. 2003). This general rule originates from the observation that reliable models can usually be constructed from structural templates that have >30% identity to the sequence of interest. In this initial phase of structural genomics projects, a major emphasis has been placed on throughput, and most structures have arisen from prokaryotic and lower eukaryotic genomes for two major reasons. First, for bacterial and yeast genes, the cloning strategies can be easily executed by each of the centers. Second, the gene products from lower organisms are to a greater degree comprised of small, soluble domains that are easily expressed and purified from bacterial systems. As progress continues, and reliable models for a large fraction of sequence space accessed through structural solution for lower organisms appear, pressure to apply high-throughput methods to gene products from higher organisms will increase. Clearly, comprehensive cloning strategies for higher organisms are more complicated. Thus, the availability of cloning vectors for orthologs from a variety of organisms will remove an important limiting factor in the progress of the overall PSI. Thus, ORF projects such as those outlined in this issue will become increasingly important to structural genomics.
The major benefit from structural genomics efforts is the provision of structural models for biologists to understand gene function. In addition, the wealth of structural information will be used to address issues of protein folding, protein structure prediction, and protein evolution. In terms of biomedical impact, the structural data will facilitate design of therapeutic agents by comparing functionally similar protein structures of pathogens and hosts, or proteins in diseased and normal tissues. The structural genomics efforts have facilitated technical developments in structure determination and the establishment of high-throughput facilities for the use of a wide community of scientists. Also, the structural genomics projects are providing reagents and materials for spin-off projects that examine function in vivo and in vitro. Lastly, retrospective analyses using the unprecedented volume of high-throughput experiments are helping to establish methods to predict experimental outcomes for protein production and crystallization. In this report, we outline the progress of the New York Structural Genomics Consortium (NYSGXRC, www.nysgxrc.org) in implementing and developing its structural genomics pipeline. We emphasize the coordination of bioinformatics efforts with the experimental methods of the consortium, including the development of an integrated consortium database to manage the workflow, the overall progress from cloning to modeling, the impact of the modeling of NYSGXRC structures, and novel experimental and bioinformatics approaches to examining the structure of metalloproteins, termed metallomics (Hasnain 2004; Szpunar 2004).
RESULTS AND DISCUSSION
Design and Use of an Online Experimental Database
One of the key features in the successful internal functioning of the NYSGXRC (and any large multi-task project) has been the development of a database for effective communication among the participants. The Integrated Consortium Experimental Database (IceDB) has been set up to facilitate data management among the various research groups in the NYSGXRC. IceDB fulfills several roles; it serves as a Laboratory Information Management System (LIMS) for exchanging, querying, displaying, and archiving experimental and bioinformatics data; it is used as an automated and versatile bioinformatics tool for bioinformatics screening and analysis; and finally, it is an interface and data exchange platform for users, other centers, and external resources. The system technically is a MySQL relational database that is organically interconnected with a series of locally implemented bioinformatics programs and external databases. The relational database can be accessed through a Web interface at www.nysgxrc.org. It is coded in HTML and Perl CGI languages.
IceDB is composed of two main parts, Target List and Progress Report. Target List contains the potential targets and their annotations in order to aid target selection. Several bioinformatics programs have been implemented for screening, such as calculating peptide statistics, predicting secondary structure, membrane immersed, and disordered regions from the sequences. Progress Report collects and displays the experimental data, and tracks the progress for all the selected targets. The collected experimental data include fields such as cloning, expression, biophysical characterization, crystallization, X_Ray data collection, X_Ray refinement, X_Ray structure, and PDB deposition. Users can insert comments and actual data (graphs, images) as appropriate for each class of field. IceDB automatically generates weekly progress statistics and XML-formatted progress reports for TargetDB, the centralized database of the PSI. IceDB also compares regularly and systematically all the active targets in the internal pipeline with the ones in TargetDB, and identifies potential overlapping cases, the extent of their sequential overlap and similarity, and the stages of experimental progress toward these structures.
IceDB interfaces with three major external resources and several public databases. This cross-linking is essential to consortium communication, as specific tasks in the structural genomics pipeline are distributed among various independent laboratories. For example, ORF target-selection bioinformatics tasks are primarily carried out at UCSF in the Sali laboratory. A list of curated ORF targets is then transmitted to the large-scale cloning and protein production facilities at Structural Genomix (SGX) in San Diego, where the overall Consortium's effort is directed by Stephen Burley. IceDB regularly exchanges data with the LIMS of SGX. Thus, data generated at SGX on cloning, expression, solubility, and purification of protein targets is automatically uploaded. Purified ORF targets are shipped from SGX to the four crystallographic laboratories in New York for automated crystallization, and these labs use IceDB to track progress in generating crystals and assessing diffraction quality upon preliminary synchrotron data collection. In this way, the crystallography laboratories receive necessary information on the targets from SGX, and SGX can determine which ORF targets are showing progress through the pipeline.
To keep track of structure solution activity at the National Synchrotron Light Source, IceDB automatically communicates with the Automated Structure Determination Platform (ASDP). ASDP is used for high-throughput X-ray structure determination subsequent to data collection (Chance et al. 2002). As the crystallography laboratories complete refinements and deposit their structures in the PDB, they periodically update IceDB. Finally, IceDB is connected to the external resource MODBASE, a comprehensive database of comparative protein-structure models, where the computational model building takes place using the newly solved ORF target structures (Eswar et al. 2003; Pieper et al. 2004). Beside these major platform hubs, which are directly responsible for facilitating major steps in the experimental pipeline, sequence and structure-based functional annotations are implemented for analysis. Target proteins in IceDB are also linked to their entries in SWISS-PROT, GenBank, and Pfam databases.
Output of NYSGXRC Pipeline to Date and Worldwide Progress in Structural Genomics
The current progress report of the NYSGXRC as of May 2004 is shown in Table 1 and can be seen on the first page of the Web site. IceDB currently contains information about 40,000 potential ORF targets, of which 1869 are active along various stages in the experimental pipeline. To date, 1077 ORF targets have been cloned, and expression is observed for 787 of the targets. From these expressing vectors, 493 proteins have been purified and initial crystallization screens have been attempted for each. A total of 235 of the purified proteins (or 48%) have produced some form of crystal with 141, or 29%, producing diffraction quality crystals. Of these diffraction quality crystals, 95 (or 19%) have been solved to date, and 81 have been deposited to the PDB as of May 11, 2004.
Table 1.
Progress of NYSGXRC as of May 2004, Updates Available at www.nysgxrc.org
| Targets selected | 1869 |
| Cloned | 1077 |
| Expression successful | 787 |
| Soluble | 581 |
| Purified | 493 |
| Crystallized | 235 |
| Diffraction-quality Crystals | 141 |
| Native diffraction-data | 98 |
| Phasing diffraction-data | 98 |
| Crystal Structure Complete | 95 |
| Deposited in PDB | 81 |
Among the first 65 NYSGXRC target structures solved, 53 have been classified by SCOP (Murzin et al. 1995; Andreeva et al. 2004). Of the total, 13 have segregated α and β structure, 23 have alternating α and β, 11 are all-β, and six are all-α protein classes. At the fold level, the 53 structures are distributed among 36 fold types. The solved targets were also compared with already known structures using the DALI program (Holm and Sander 1995, 1996). A new fold was assumed if DALI reported a Z-score of <10 for the best hit. Using this cutoff value, 15 of the 65 targets (or 23%) were new folds at the time of submission. This fraction of new folds is several times higher than is generally seen for recently submitted PDB structures. (In the last 5 yr, 3%-5% of all submitted structures were classified as new folds according to SCOP). Of the 25 functionally uncharacterized proteins among the targets, seven are not classified at all in SCOP, whereas the remaining 18 are distributed among four different SCOP classes and 16 fold types.
On the basis of our current protein production rates, we now have sufficient statistics to reliably estimate the NYSGXRC output in the immediate future. The above statistics argue that ∼20% of the soluble proteins delivered to the four crystallography laboratories in New York are producing crystal structures. There is, of course, a delay between the delivery of proteins and structure solution, thus, the current figure of 19% is likely to be adjusted upward as progress on targets in the pipeline continues to accrue. Nevertheless, in the fourth year of the Consortium's operation, SGX has delivered 192 soluble targets as of March 2004 and will deliver 350 additional targets by September 2004, the end of the fourth year. On the basis of our progress to date, we expect that >100 new structures will be solved from these ORF targets. SGX plans to supply the crystallography laboratories with ∼50 soluble targets per month throughout the fifth year of the project; 120 structures are ultimately expected to be produced from these targets. On the basis of this productivity level, the NYSGXRC is poised to achieve its initial goal of producing at least 100 structures in its fifth year of operation.
The production statistics for the 15 structural genomics centers located around the world as of May 2004 include 28,293 proteins cloned with expression observed in 16,468 of the vector targets (or 58%). A total of 6177 targets have been seen to produce soluble protein, from which 5924 proteins have been purified. Thus, the overall experience is that purified protein has been obtained from 36% of the vectors for which expression has been observed. A total of 2162 of the purified proteins formed crystalline material, and 1034 (17% of the purified target set) resulted in diffraction quality crystals, whereas 715 structures have been deposited to the PDB. These outcomes are expected to improve, as some of the proteins are still at some intermediate stage in the various pipelines. Compared with the goal of producing 10,000-15,000 new structures to provide completeness in structural genomics (Vitkup et al. 2001), this is merely a down payment, but represents promising initial progress. If we can rely on the above metrics that suggest 20% of purified proteins will produce diffraction quality crystals, then 50,000-75,000 proteins will need to be purified in order to achieve an overall goal exceeding 10,000 structures. However, the calculation that one-third of the expressing vectors may provide easily purifiable protein may not hold for multidomain proteins from higher organisms (Burley et al. 1999; Chance et al. 2002), at least not without methodological improvements in expression systems. However, it sets a likely lower limit of 200,000 expression vectors that will need to be constructed to complete the overall project.
Modeling NYSGXRC Sequences: How Structural Models Are Informing New Biology
Recent developments in the techniques of structure determination at atomic resolution, X-ray diffraction, and nuclear magnetic resonance spectroscopy, have enhanced the quality and speed of structural studies (Zhang and Kim 2003). Nevertheless, current statistics still show that the known protein sequences (∼1,500,000; Boeckmann et al. 2003) vastly outnumber the available protein structures (∼25,000; Westbrook et al. 2002). Fortunately, domains in protein sequences are gradually evolving entities that can be clustered into a relatively small number of families with similar sequences and structures (i.e., folds; Vitkup et al. 2001). These evolutionary relationships enable the use of computational methods, such as threading and comparative protein structure modeling (Fiser et al. 2001), to predict the structures of protein sequences on the basis of their similarity to known protein structures. The NYSGXRC is combining experimental structure determination methods with computational modeling techniques. This effort, combined with that of other structural genomics centers worldwide, aims to determine a sufficient number of appropriately selected structures, so that most ORF sequences can be placed within modeling distance of at least one known structure (Sali 1998; Sanchez and Sali 1998; Baker and Sali 2001; Vitkup et al. 2001).
A suite of bioinformatics programs and databases is at the foundation of the NYSGXRC's computational efforts. MODBASE (http://salilab.org/modbase) is a comprehensive database of annotated comparative protein structure models (Pieper et al. 2004). MODBASE models are calculated by MODPIPE (Eswar et al. 2003), a fully automated comparative protein structure modeling pipeline. MODPIPE relies on various modules of the comparative modeling software MODELLER (Sali 1995) for its functionality, and is streamlined for large-scale operations on a cluster of PCs. The modeling process comprises the following steps: fold assignment, sequence-structure alignment, model building, and model assessment. MODBASE is updated regularly to reflect the growth in sequence and structure databases, as well as improvements in the software for calculating the models.
MODBASE is organized into several model data sets. The largest contains models for domains in 659,495 sequences of 1,182,126 unique protein sequences in the complete SWISS-PROT/TrEMBL (Boeckmann et al. 2003) database (August 25, 2003). These models correspond to all known protein sequences in SWISS-PROT/TrEMBL that can be matched to at least one known protein structure. The second largest group of model data sets includes MODPIPE models for the SWISS-PROT/TrEMBL sequences that were modeled on the basis of the NYSGXRC structures. We run MODPIPE using all NYSGXRC structures as templates to contribute to their annotation. When a new consortium structure is deposited in the PDB, a MODPIPE run using this new structure as a template is automatically triggered, and models for all sequences in SWISS-PROT/TrEMBL that are related to this structure are calculated. These calculations are repeated periodically for all template structures. All protein sequences in SWISS-PROT/TrEMBL that are related to the NYSGXRC structures can be viewed in MODBASE.
Relying on the first 63 unique NYSGXRC solved structures, MODPIPE produced models for domains in 33,340 sequences in SWISS-PROT/TrEMBL (Table 2). The modeled sequences come from 2676 different organisms, with a kingdom distribution of 41% Prokaryota, 2% Archaea, and 57% Eukaryota. This organism classification has been derived from the NCBI taxonomy database, where all protein sequences are matched with a taxonomy id (Wheeler et al. 2000; Benson et al. 2002). The average ORF target-template sequence identity was 18.6%. Only 10% of the sequences are modeled on the basis of >30% sequence identity over more than 75 residues; 81% of the sequences have models that are predicted to have the correct fold on the basis of the model score (John and Sali 2003) or the PSI-BLAST E-value (Schaffer et al. 2001). Using these data sets, all amino acid sequences in SWISS-PROT/TrEMBL that are related to the NYSGXRC structures can be viewed in MODBASE, which easily facilitates the detection of remote relationships and the annotation of function to proteins previously annotated as hypothetical proteins.
Table 2.
MODBASE Model Data Sets Using the PDB Structures of the First 63 Released and Unique NYSGXRC Targets as Templates
| No. of Sequences
|
|||||||
|---|---|---|---|---|---|---|---|
| PDB code | Target Id | Database Accession | Annotation | Total | FM | M | F |
| 1b54 | P007 | P38197 | Hypothetical UPF0001 protein YBL036C | 151 | 132 | 17 | 2 |
| 1ci0 | P008 | P38075 | Pyridoxamine 5′-phosphate oxidase (EC 1.4.3.5) | 99 | 93 | 0 | 6 |
| 1dfc | P119 | 11513471 | Fascin (Singed-like protein) (p55) | 81 | 27 | 32 | 22 |
| 1f89 | P018 | P49954 | Hypothetical 32.5 kDa protein YLR351C | 547 | 488 | 10 | 55 |
| 1fi4 | P100 | P32377 | Diphosphomevalonate decarboxylase (EC 4.1.1.33) | 154 | 64 | 88 | 5 |
| 1g61 | P111a | Q60357 | Translation initiation factor 6 (alF-6) | 49 | 46 | 2 | 1 |
| 1g62 | P111 | Q12522 | Eukaryotic translation initiation factor 6 (elF-6) | 51 | 50 | 1 | 0 |
| 1hqz | T138 | 113000 | ABP1_YEAST actin binding protein | 175 | 50 | 124 | 2 |
| 1i9a | P109a | 6225535 | IDI_ECOLI isopentenyl-diphosphate delta-isomerase | 1140 | 510 | 11 | 619 |
| 1jd1 | P003 | P40037 | HMF1 protein (High dosage growth inhibitor) | 382 | 354 | 3 | 26 |
| 1jf9 | T129 | P77444 | Selenocysteine lyase (EC 4.4.1.16) | 1669 | 1616 | 0 | 54 |
| 1jfi | P048a | 7513394 | S70618 transcription regulator NC2 alpha chain | 86 | 15 | 0 | 71 |
| 1jg8 | P044a | 4982322 | L-allo-threonine aldolase | 1611 | 1461 | 69 | 123 |
| 1jr7 | T130 | P76621 | Hypothetical protein ygaT | 11 | 10 | 1 | 0 |
| 1jss | T526 | 13542895 | Similar to RIKEN cDNA 2310058G22 gene | 254 | 176 | 2 | 76 |
| 1jsx | T35 | 121191 | GIDB_ECOLI glucose inhibited division protein B | 1583 | 1064 | 27 | 496 |
| 1jyh | T473 | 465566 | GYRI_ECOLI DNA gyrase inhibitory protein Hypothetical 27.5 kDa protein in SPX19-GCR2 | 144 | 97 | 0 | 47 |
| 1jzt | P097 | P40165 | intergenic region | 1058 | 39 | 13 | 1006 |
| 1k47 | T27 | 9937409 | phosphomevalonate kinase | 539 | 385 | 33 | 124 |
| 1k4z | T139 | 399184 | CAP1_HUMAN adenylyl cyclase associated protein | 44 | 34 | 8 | 2 |
| 1k8f | T140 | 134897 | CAP_YEAST adenylyl cyclase associated protein | 48 | 36 | 10 | 2 |
| 1kag | T535 | P24167 | Shikimate kinase I (EC 2.7.1.71) (SKI) | 1005 | 250 | 51 | 706 |
| 1kcx | T45 | 2342488 | dihydropyrimidinase related protein 1 | 701 | 378 | 20 | 312 |
| 1ku9 | T136 | 3025177 | YF63_METJA HYPOTHETICAL PROTEIN MJ1563 | 572 | 131 | 253 | 214 |
| 1l9g | T299 | Q9WYY1 | Hypothetical protein TM0511 | 170 | 152 | 3 | 15 |
| 1la2 | T23 | P11986 | Inositol-3-phosphate synthase (EC 5.5.1.4) (IPS) | 102 | 86 | 0 | 16 |
| 1lnz | T131 | P20964 | Spo0B-associated GTP-binding protein | 1620 | 960 | 40 | 678 |
| 1lx7 | T24 | P12758 | Uridine phosphorylase (EC 2.4.2.3) (UDRPase) | 506 | 384 | 1 | 121 |
| 1m0t | P102 | Q08220 | Glutathione synthetase (EC 6.3.2.3) (GSH-S) | 38 | 37 | 1 | 1 |
| 1m0w | P102a | Q08220 | Glutathione synthetase (EC 6.3.2.3) (GSH-S) | 39 | 38 | 0 | 1 |
| 1n10 | T467 | 28373838 | Phl P 1, A Major Timothy Grass Pollen Allergen | 358 | 336 | 9 | 13 |
| 1ne8 | T503 | P96622 | YDCE protein | 111 | 84 | 21 | 6 |
| 1ni3 | T9 | O13998 | Similar to putative GTP-binding protein | 1058 | 103 | 1 | 955 |
| 1ni5 | T132 | P52097 | Putative cell cycle protein mesj Hypothetical 32.1 kDa protein in ADH3-RCA1 | 920 | 204 | 40 | 689 |
| 1njr | P089 | Q04299 | intergenic region Hypothetical 28.8 kDa protein in PSD1-SKO1 | 4 | 1 | 3 | 0 |
| 1nkq | P096 | P53889 | intergenic region | 379 | 207 | 0 | 172 |
| 1nlx | T746 | P43215 | Pollen allergen Phl p 6 precursor (Phl p VI) | 12 | 12 | 0 | 1 |
| 1nr0 | T745 | Q11176 | Actin interacting protein 1 (AIP1) | 752 | 633 | 33 | 142 |
| 1nvt | T576 | Q58484 | Shikimate 5-dehydrogenase (EC 1.1.1.25) | 543 | 189 | 348 | 7 |
| 1omi | T143 | P22262 | Listeriolysin regulatory protein | 1094 | 301 | 7 | 798 |
| 1p1l | T835 | O28301 | Periplasmic divalent cation tolerance protein (CUTA) | 68 | 63 | 0 | 5 |
| 1p1m | T834 | Q9X034 | Hypothetical protein TM0936 | 780 | 354 | 24 | 404 |
| 1pb6 | T803 | P75899 | Hypothetical transcriptional regulator ycdC | 1364 | 1152 | 74 | 155 |
| 1pqw | T109 | 7448840 | A70984 probable polyketide synthase | 1496 | 1426 | 39 | 35 |
| 1pqy | T783 | P77407 | Hypothetical protein yfdW | 607 | 579 | 11 | 23 |
| 1psq | T817 | P72500 | Probable thiol peroxidase | 950 | 664 | 23 | 263 |
| 1psu | T820 | O28020 | Hypothetical protein AF2264 | 662 | 244 | 26 | 393 |
| 1psw | T832 | Q51063 | ADP-heptose:LPS heptosyltransferase II | 642 | 258 | 229 | 194 |
| 1pug | T5 | P17577 | Hypothetical UPF0133 protein ybaB | 118 | 112 | 6 | 0 |
| 1pui | T16 | P24253 | Probable GTP-binding protein engB | 1380 | 925 | 52 | 438 |
| 1puj | T18 | O31743 | YLQF protein Hypothetical 33.9 kDa esterase in SMC3-MRPL8 | 1128 | 96 | 20 | 1012 |
| 1pv1 | P068 | P40363 | intergenic regionDE (EC 3.1.1.-) | 143 | 36 | 14 | 93 |
| 1q2y | T804 | O31628 | YJCF protein | 1676 | 1123 | 121 | 479 |
| 1q6w | T805 | O28346 | Monoamine oxidase regulatory protein, putative | 566 | 279 | 14 | 285 |
| 1q98 | T1429 | Q57549 | Probable thiol peroxidase (EC 1.11.1.-) | 1218 | 763 | 19 | 436 |
| 1q9j | T760 | P96208 | Hypothetical protein papA5 | 694 | 32 | 3 | 671 |
| 1r3d | T920 | Q9KQM4 | Hypothetical protein VC1974 | 1602 | 563 | 20 | 1030 |
| 1rc6 | T1521 | 16128499 | Hypothetical protein ylbA | 474 | 50 | 46 | 382 |
| 1ri6 | T1479 | 16128735 | Hypothetical protein ybhE | 1458 | 350 | 1126 | 77 |
| 1rvk | T1522 | 17937161 | isomerase/lactonizing enzyme | 1115 | 864 | 139 | 124 |
| 1s7j | T1581 | 29374770 | Phenazine biosynthesis protein PhzF family | 361 | 236 | 124 | 1 |
| 1ub4C | T1468 | 126777 | Peml-like protein 1 (MazE protein) | 40 | 9 | 8 | 23 |
| 1ub4A | T1469 | 464357 | PemK-like protein 1 (MazF protein) | 112 | 99 | 8 | 5 |
The PDB code, Database Accession, and Annotation columns define the template structure. (No. Sequences) The number of sequences in SWISS-PROT/TrEMBL that could be modeled reliably using the NYSGXRC structure as a template. (Total) The total number or sequences, (F) the number of sequences that have a reliable PSI-BLAST E-value of ≤ 10−4 but a low model reliability score (<0.7), (M) the number of sequences with a model score ≥ 0.7 (reliable model), but with insignificant PSI-BLAST E-value (>10−4), (FM) the number of sequences that have both a reliable model score and a significant PSI-BLAST E-value. The most reliable models have both a reliable PSI-BLAST E-value and a reliable model score (FM). For the models classified as F, the fold assignment is considered reliable, even though the model score is bad. Models classified as M have only a remote relationship to the template, but the good model score suggests that the modeled sequences indeed have the same fold as the template structure. The full table can be viewed at http://salilab.org/modbase/models_nysgxrc.html.
Considering that the target sequences for NYSGXRC were selected to have <30% sequence identity to a known experimental structure, most of the modeled ORF sequences have been characterized structurally for the first time. Thus, these data sets indicate the increased coverage of the sequence-structure space by the NYSGXRC structures. In fact, the experience so far for the U.S. centers is that 70% of their PDB deposits in 2002-2003 are for proteins containing unique sequences, (i.e, sequences with <30% sequence identity to the closest known structure) compared with only 10% of the deposits overall during the same time period (Editorial 2004). The large number of new models that can be calculated on the basis of the newly determined structures illustrates and justifies the premise of structural genomics.
The most interesting cases for functional analysis would be proteins for which sequence-based methods failed to establish a meaningful connection to a protein of known function or structure. On the basis of our current experience, every third target solved in the NYSGXRC pipeline remains functionally uncharacterized. These proteins are ripe for experimental investigation using biochemical or genetic approaches. Although funds are available from the NIH for the study of functionally characterized structures solved by the PSI centers, no mechanism exists to systematically study the uncharacterized proteins (Editorial 2004).
Another way to glean functional insight for unannotated protein structures is through the comparative modeling pipeline. Structure-based search and confirmation of protein relationship is usually more reliable and sensitive than sequence-only based approaches. Such structural (and potentially functional) assignments are called “nontrivial hits” (summarized in Table 2 in the M column), and are usually based on very low (<20%) sequence identity between aligned regions of the target and template sequences. An example is the model of a protein sequence annotated in the TrEMBL database (Boeckmann et al. 2003) as “Hypothetical protein SCP1.152” (Bentley et al. 2002; TrEMBL accession no. Q9ACZ9, organism: Streptomyces coelicolor) that was modeled using the PDB structure 1rvk (NYSGXRC target T1522). The sequence identity between 1rvk and Q9ACZ9 is 19%, the PSI-BLAST E-value is 0.1, and the model score 0.71. The model covers 91 of 104 amino acid residues. 1rvk has been annotated as isomerase/lactonizing enzyme. The modeling results suggest that Q9ACZ9 has a similar function. This specific example illustrates that structure modeling can identify functional similarities between ORF sequences that lack any detectible sequence similarity.
High-Throughput Annotation of Metal-Binding Targets
An interesting conclusion from the production statistics above is that if ∼20% of the soluble ORF targets are ultimately amenable to structural analysis, ∼80% of the proteins are not, and represent ORF targets that are likely to be abandoned. However, these proteins present a potentially valuable resource for spectroscopic and biochemical analysis to better understand structure and function. In general, the consortium has pursued a limited number of approaches to provide characterization of target proteins. This has included light scattering and limited proteolysis mass spectrometry (Burley et al. 1999; Shi et al. 2003). The former is to determine whether the protein preparations are mono-disperse, as such a preparation is much more amenable to crystallization. The latter is to determine whether the purified ORF target represents a compact globular domain, which also crystallizes much more efficiently. This information is used to inform protein-purification strategies in the case of poly-disperse samples, and is used to direct recloning efforts in the case of exposed protease sensitive sites. However, these analyses do not give major insights into structure and function. To provide additional annotation to ORF targets, we have implemented an automated system to analyze all purified ORF targets for transition metal content. This effort has multiple purposes. First, identification of metal binding can be used to make the protein purification and protein-crystallization strategies more efficient by supplementing the buffers with the metal in question. Second, identification of metal binding can be used to aid in annotation of protein function, especially for so-called “hypothetical” proteins. Third, for proteins that crystallize, the intrinsic transition metal can, in favorable cases, be used for anomalous scattering phasing of the structure (Hendrickson 1991; Rajashankar et al. 2001).
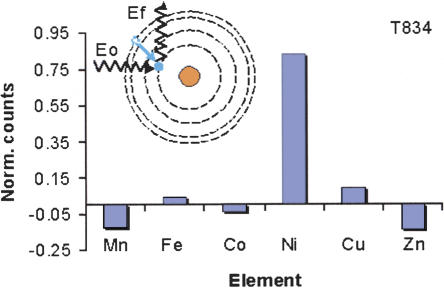
Up to one-third of proteins contain metal atoms (Hasnain 2004; Szpunar 2004) with iron and zinc being the most common among the transition metals (Lujan et al. 1995). Protein samples illuminated with high-energy synchrotron X-rays eject a 1s electron from the first electron shell surrounding a metal nucleus (Fig. 1). Passage of another electron from a higher shell to fill the hole in the first shell yields an emitted X-ray photon (fluorescence emission) with energy characteristic for the individual elements (Chance et al. 1992; Summers et al. 1992; Lujan et al. 1995). We have developed an automated system to scan and detect transition metal content of protein target samples in 16-well plates with a multiplate rail and precise and automated alignment of the samples in a synchrotron X-ray beam. Detection of fluorescent emitted photons is accomplished with a multi-element, fast count rate, high-resolution Germanium detector (Summers et al. 1992; Lujan et al. 1995). Although all elements in the sample will emit fluorescence given sufficiently high energy X-rays incident on the sample, practical considerations of sample thickness and air absorption limit the analysis, in this case, to the following transition elements: Mn, Fe, Co, Cu, Ni, and Zn; however, data collection times are only a few minutes per sample for nanogram detection efficiencies.
Figure 1.
The corrected fluorescence counts for each metal atom are shown in histogram format (i.e., sample well counts minus counts from a blank well). The corrected counts for nickel are very far above background. The inset shows electron orbitals in schematic form with incident X-ray, the transition from higher to lower energy orbitals, and emission of X-ray fluorescence illustrated. The incident X-ray (Eo) knocks out a 1s electron, the unstable core hole is filled by the subsequent transition (seen in color), and a fluorescent X-ray of energy characteristic for the metal atom is emitted (Ef).
We analyzed 143 proteins from prokaryotic sources recently delivered by SGX to the crystallography laboratories for crystallization testing. For each protein, 200 μg of sample were loaded onto the sample plates and dried under controlled conditions. The results in terms of corrected counts for T834, which was annotated as a hypothetical protein (Table 3), are shown in Figure 1. The sample showed significant nickel fluorescence counts, but minimal amounts of the other metals were detected. Of the 143 samples examined, >20 indicated some transition metal content (data not shown). To limit the analysis to likely cases of structural or functional metal atoms, the metal-to-protein stoichiometry was determined by comparison of the corrected counts with an appropriately chosen set of standards for each metal at the same experimental conditions; thus, the number of moles of each metal was accurately measured. The results for the 16 proteins that showed a metal/protein ratio of 0.7 or greater are shown in Table 3; the error in this analysis is ±0.2, such that we report data only for metal binding that is likely to be stoichiometric, and therefore relevant. Of these, two proteins contain two or more metal atoms per protein molecule, and 14 proteins contain one or more metal per molecule (metal/protein ratios 0.7-1.6), including T834. Zinc was observed in eight cases, copper and nickel in three each, and iron and manganese once.
Table 3.
Metal Atoms Found in NYSGXRC Target Proteins and Annotations of Targets and Closely Related Genes
| Target ID | Metal | NMA | Target Annotation | Clusters of Orthologous groups | BLAST-PDB | Related Structure |
|---|---|---|---|---|---|---|
| T763 | Zn | 1.3 | AC: Q9PHR1 | Ev = 2e-95 | No closely related structure | — |
| Putative amidohydrolase | Metal-dependent amidase/aminoacylase/carboxypeptidase | |||||
| OS: Campylobacter jejuni | ||||||
| SIMILARITY-Peptidase Family M40. | ||||||
| T773 | Zn | 0.9 | AC: O34974 | Ev = e-144 | Ev = 7e-10 | |
| YTNJ | Coenzyme F420-dependent | Alkanesulfonate | PDB ID: 1NQK | |||
| OS: Bacillus subtilis | NS, N10-methylene | Monooxygenase. | Identity = 28% | |||
| SIMILARITY-Ntaa/Snaa/Soxa(Dsza) Family of Monooxygenases. | tetrahydromethanopterin reductase | Metal ions = no | ||||
| T788 | Zn | 4.6 | AC: Q9WYG6 | Ev = e-103 | Ev = 3e-06 | PDB ID: 1O57 |
| Orotate phosphoribosyltransferase | 2-keto-4-pentenoate hydratase/2-oxohepta-3-ene-1, 7-dioic acid hydratase | Purine Operon | Identity = 24% | |||
| OS: Thermotoga maritima | Repressor of Bacillus Subtilis. | Metal ions = no | ||||
| SIMILARITY-Purine/pyrimidine phosphoribosyltransferase family. | ||||||
| T790 | Cu | 0.9 | AC: O06156 | Ev = 6e-57 | Ev = 3e-10 | PDB ID: 1IUJ |
| Hypothetical protein Rv3592 | Uncharacterized enzyme; polysaccharide synthesis | Tt1380 Protein | Identity = 39% | |||
| OS: Mycobacterium tuberculosis | Metal ions = Zn | |||||
| T797 | Zn | 0.7 | AC: P44321 | Ev = e-108 | Ev = 7e-67 | PDB ID: 1NKU |
| DNA-3-methyladenine glycosylase | 3-methyladenine DNA glycosylase | DNA/Glycosylase I | Identity = 62% | |||
| OS: Haemophilus influenzae | Metal ions = Zn | |||||
| T813 | Fe | 1.0 | AC: Q58465 | Ev = 2e-74 | There is no closely related structure | — |
| Hypothetical protein MJ1065 | Sialic acid synthase | |||||
| OS: Methanococcus jannaschii | ||||||
| T818 | Zn | 0.7 | AC: O34790 | Ev = e-128 | Ev = e-125 | PDB ID: 1VIZ |
| PcrB protein homolog | Predicted phosphate-binding enzymes, TIM-barrel fold | Hypothetical Protein. | Identity = 99% | |||
| OS: Bacillus subtilis | Metal ions = no | |||||
| T823 | Cu | 1.1 | AC: P24216 | Ev = e-124 | No closely related structure | — |
| Flagellar hook-associated protein 2 | Flagellar capping protein | |||||
| OS: Escherichia coli | ||||||
| T824 | Cu | 2.4 | AC: Q9PNP0 | Ev = 2e-24 | No closely related structure | — |
| Restriction modification enzyme | Type I restriction-modification system methyltransferase subunit | |||||
| OS: Campylobacter jejuni | ||||||
| T830 | Mn | 1.6 | AC: Q9K2H0 | Ev = 6e-18 | No closely related structure | — |
| Hypothetical protein | ADP-ribose pyrophosphatase | |||||
| OS: Streptococcus pneumoniae | ||||||
| SIMILARITY-Nudix Hydrolase | ||||||
| T834 | Ni | 0.8 | AC: Q9X034 | Ev = 1e-79 | Ev = 0.0 | PDB ID: 1P1M |
| Hypothetical protein TM0936 | Cytosine deaminase and related metal-dependent hydrolases | Hypothetical | Identity = 100% | |||
| OS: Thermotoga maritima | Protein Tm0936 | Metal ions = Ni | ||||
| SIMILARITY - Atz/Trz Family | ||||||
| T1403 | Zn | 1.0 | AC: Q9S3S2 | Ev = 8e-93 | No closely related structure | — |
| Beta lactamase regulatory protein homolog | Predicted pyrophosphatase | |||||
| OS: Vibrio cholerae | ||||||
| T1404 | Zn | 0.8 | AC: P33646 | Ev = e-148 | No closely related structure | — |
| MazG protein | Predicted pyrophosphatase | |||||
| OS: Escherichia coli | ||||||
| SIMILARITY-S. CACOI ORF in BLAB 3'Region | ||||||
| T1405 | Zn | 0.7 | AC: O33341 | Ev = e-180 | No closely related structure | — |
| Hypothetical protein Rv2859c | Predicted glutamine amidotransferases | |||||
| OS: Mycobacterium tuberculosis | ||||||
| T1407 | Ni | 1.0 | AC: P11549 | Ev = 1e-82 | Ev = 7e-32 | |
| Lactaldehyde reductase | Fucose permease | Alcohol | PDB ID: 1O2D | |||
| OS: Escherichia coli | Dehydrogenase, | Identity = 30% | ||||
| SIMILARITY-iron-containing alcohol dehydrogenase family. | Metal ions = 2Fe | |||||
| T1421 | Ni | 1.5 | AC: P09151 | Ev = e-170 | There is no closely related structure | — |
| 2-isopropylmalate synthase | Isopropylmalate/homocitrate/citram alate synthases | |||||
| OS: Escherichia coli | ||||||
| SIMILARITY-alpha-IPM synthetase |
(NMA) Number of Metal Atoms per protein molecule; (OS) Organism/Species; (AC) Accession number; (Ev) E-value.
In the following section, we examine the known annotations for these 16 proteins. Our analysis is likely to emphasize false negatives, as some metalloproteins may lose a metal atom in the purification step. We have already excluded one false positive, where a stoichiometry of 0.5 Zn/protein was observed for T1429 (data not shown, AC:Q57549). This target was solved by the NYSGXRC (1q98). An anomalous difference Fourier analysis showed no evidence of a metal atom signature. However, this protein does have exposed Cys residues that may be able to coordinate adventitious Zn during the purification. This is one factor leading to the choice of a cutoff of 0.7 metal/protein for the annotation of metalloprotein identity.
Functional Annotation of Metal-Binding Proteins
The 16 NYSGXRC target protein sequences that are strongly indicated to be metalloproteins were retrieved from IceDB, and bioinformatics analysis was carried out to analyze the consistency of the metal binding with known annotations. Table 3 shows the target IDs, the SWISS-PROT or GenBank identifier, the annotation as provided by IceDB, additional annotations from relevant databases, and the organism from which the target is derived (Boeckmann et al. 2003). A BLAST search was carried out against SWISS-PROT to identify closely related homologs. For T834 (0.8 Ni/protein), the BLAST search revealed a related hypothetical protein and similarity to members of the Atz/Trz family. Clusters of Orthologous Groups (COG) was examined; these results are shown in Table 3. For T834, this search indicated a close relationship with cytosine deaminases and related metal-dependent hydrolyases. The crystal structure of this protein was solved within the NYSGXRC (1p1m) and a single Ni ion was confirmed as part of the structure (PDB data in Table 3). Thus, in this case, the metal analysis was supported by bioinformatics comparisons and direct structure determination.
For T763, a zinc/protein stoichiometry of 1.3 was measured; the protein was annotated as a putative amidohydrolyase. The BLAST search indicated a close relationship with a zinc-containing carboxypeptidase and an overall similarity with the M40 peptidase family. The COG analysis indicated that the target belongs to a metal-dependent amidase family. A search against PDB found no significant homologies. The annotation of this protein as a metalloprotein is very strongly confirmed by the bioinformatics analysis, although the crystal structure of this protein remains unsolved.
T830 is annotated as a hypothetical protein with similarity to an ADP-ribose pyrophosphatase, which is indicated to have a magnesium cofactor. The COG database also indicates that this target belongs to the same enzyme family. No similarity to any structure in the PDB was found. The annotation of T830 as a manganese-containing enzyme is reasonable, as active sites that bind magnesium generally can be exchanged for manganese. Thus, the metal analysis provides evidence that this target is a metal-dependent hydrolyase.
For T1407, T797, T1403, and T1404, the identification as metalloproteins is well supported by bioinformatics, which, in each case, provides a functional annotation (in terms of enzyme activity) consistent with metal binding by the target. T1407 (binding Ni) is annotated in the alcohol dehydrogenase family (the presence in this target of a metal-binding motif was also seen in PROSITE). A related structure in the PDB is seen to contain Fe. Zinc-containing T797 is a DNA-glycosylase closely related to PDB entry 1nku, which also contains zinc. T1404, the MazG protein and the related T1403 are indicated to have pyrophosphatase motifs consistent with zinc binding.
In several cases, the metal binding provides a new annotation for protein of unknown or not-well-understood functions. T790, indicated to contain copper, was annotated a hypothetical protein and COG indicated an uncharacterized enzyme. A related structure in the PDB is seen to contain Zn. T1405 also is listed as a hypothetical protein predicted to be related to glutamine amidotransferases; the metalloprotein annotation may assist in better understanding its function. In other cases, the proteins have good annotations, but no indication of metal binding, and the metal content may suggest important structural or functional information. For example, T773 is annotated as a monooxygenase and the zinc ion may be related to the protein's catalytic function, or may serve as a structural metal. T788 has over 4 Zn/protein indicated; it is unclear as to how this may be related to its annotated enzyme function. However, T824, which has over 2 Cu/protein and is annotated as type-I restriction enzyme, may have metal functions directly related to the DNA cleavage mechanism of this protein. In the case of T818, the indicated zinc atom may represent a false positive, in that the structure of a nearly identical sequence shows no metal atom or indication of a likely metal-binding site.
Overall, the metallomics analysis found many metalloproteins among the 143 proteins examined so far. On the basis of the observed annotations, the metal content was, in most cases, very reasonable, and in other cases, potentially informative with respect to protein function. Using the cutoff of measured metal/protein stoichiometry of 0.7, the rate of false positives may be in the range of from 5% to 10%. The range of false negatives cannot be estimated yet without more data. Over the next 18 mo, we expect to screen over 900 additional proteins provided by SGX, such that we can better refine these numbers.
Conclusion: Opportunities and Limitations of the Protein Structure Initiative and the Next Challenge for Structural Biology
The NYSGXRC has assembled a robust pipeline for structural genomics research that is part of an international initiative to provide structural models for all possible ORF sequences. On the basis of current structural information in the PDB, domains in ∼57% of the known protein sequences can be modeled using MODPIPE and are available in MODBASE (Sanchez and Sali 1998; Sanchez et al. 2000; Pieper et al. 2002, 2004). As the PSI expands into its second phase in 2005, the expansion of this sequence coverage will rapidly increase, providing a valuable resource for biologists worldwide.
Although this sequence coverage and the number of modeled proteins may look impressive, usually only one domain within the ORF sequence of each protein is modeled. On average, proteins have two or three domains. That is, an average yeast ORFs codes for 472 amino acid residues, whereas the average size of domains in CATH (Orengo et al. 1997), a database of structural domains, is 175. The average model size in MODBASE is 192 residues, very similar to this domain size (Pieper et al. 2004). This limitation on comparative modeling is a direct consequence of the available structural templates. An additional problem is that membrane protein structures are poorly represented in PDB, whereas 15%-30% of proteins in various genomes are predicted to contain transmembrane helices (Liu and Rost 2001). It is also suggested that up to 20% of proteins contain unstructured regions, at least in the absence of their binding partners (Tompa 2002), which often makes them unsuitable for structure determination experiments. These limitations are not likely to be overcome, even on completion of the PSI. The experience of the NYSGXRC is that the average size of the proteins solved to date is ∼250 residues (www.nysgxrc.org/nysgxrc/result.html), slightly larger than the average domain size seen in CATH, but much smaller than the average protein size in yeast. In addition, we have few examples of transmembrane segments and unstructured regions in our solved structures; such domains are often excluded during target selection. However, ∼40% of the solved targets to date are eukaryotic in origin. Thus, the expectation is that domains from a wide variety of targets may be solved in the PSI-2.
The next challenge involves understanding the domain interactions and the assembly of proteins into complexes, Figure 2 (Gavin et al. 2002; Sali et al. 2003). Whereas structural genomics aims to provide atomic resolution models for the domains that make up the proteins and complexes that are functionally relevant to cell biology, it does not explicitly address how these structures interact with each other. The interacting surfaces of the domains dock in functionally relevant ways that are amenable to experimental tools such as cryo-EM, cross-linking, footprinting, and genetic analysis (Sali et al. 2003; Guan et al. 2004; Tong et al. 2004). A next phase of structural genomics efforts will be a gradual transition to structural proteomics, when the experimental information on organization of protein complexes and domain interactions, combined with computational modeling, will be used to understand the structure and dynamics of macromolecular assemblies. Structural genomics efforts are imperative prerequisites to these future efforts.
Figure 2.
Protein domains will be solved by structural genomics, the docking of domains in protein structures or the structures of assemblies will be a challenging next step for structural biology to be solved by a combination of structure modeling of domains combined with experimental data from techniques such as cryo-EM, cross-linking, footprinting, and genetic knockout analysis.
METHODS
Metallomics Analysis
We irradiated samples with synchrotron X-rays produced by the NSLS X-ray ring (the ring operates at the constant energy of 2.8 GeV and current decaying with time from 280 to ∼200 mA). The beamline configuration is similar to that used for focused beam X-ray absorption spectroscopy measurements (Chance et al. 1996), with the monochromator set to 10 keV, and harmonic rejection using a Ni-coated mirror. The setup consists of a multiplate rail positioned at 45° with respect to the beam that brings a sample plate in a position close to the synchrotron X-ray source, an x-z stage and two ionization chambers placed before and after the sample plate for precise alignment of a sample in the plate to the beam, and a multi-element, fast count rate, high-resolution Germanium detector placed perpendicular to the beam path, that captures the X-ray fluorescence. The detector has 13 separate elements, whose counts can be summed for signal averaging. Sufficient electronics exists, such that three metals can be analyzed using single channel analysis at a time (Lujan et al. 1995). We focus on collecting signals from the following transition elements: Mn, Fe, Co, Cu, Ni, and Zn, such that two runs are required to collect all of the required data.
A total of 16 sample wells were bored in a Teflon plate, and three plates can be simultaneously loaded onto a multiplate rail. The synchrotron beam is shaped by slits to match the size of the sample well (2.5 × 6.5 mm). After loading samples in sample wells and drying them in a controlled manner, the plates are placed into the rail. The first run consists of selecting the characteristic energies for three metals using the detector software and starting an automated program that positions sample wells in front of the beam and collects the data. A total of 60, 1-sec-long counting intervals are summed. The second run screens the same set of 48 samples for another three metals. The total time to complete both runs is about 4 h, or about 4 min/sample.
The validity of the metal determinations was evaluated as follows. We have previously published methods of quantitation for metal atoms in biological samples using X-ray absorption spectroscopy (Chance et al. 1992; Lujan et al. 1995). For this experimental setup, the metal-to-protein stoichiometry was derived from standards measured before the sample data collection. Standard sets, prepared with water-soluble chlorides or nitrates of transition metals in the weight range from 0.4 to 5 μg show linear dependence of metal mass with measured fluorescence counts. Subsets of samples spiked with defined amounts of cytochrome-c were also measured to confirm the validity of the above standards on real protein samples. However, these experiments only define the measurement error and detection limit for the analysis. The decision to use a specific cutoff (0.7 metal/protein used here) for assigning a valid metalloprotein is entirely arbitrary. Setting the criteria high increases the false negatives, setting it lower (e.g., <0.5) increases the false positives. As we analyze more samples, we will get a better idea of the expected percent of each kind of error associated with a specific cutoff value.
Target sequences were retrieved from IceDB in the NYSGXRC Web site (www.nysgxrc.org/nysgxrc-cgi/search_progress_report.cgi), and were analyzed by PSI-BLAST searches against SWISS-PROT (Altschul et al. 1997; Boeckmann et al. 2003), Cluster of Orthologous Groups (COG; www-archbac.u-psud.fr/genomics/COG_Guess.html), and the Protein Data Bank (Berman et al. 2000). The protein having the lowest E-value (if a candidate was found at <10-4) is selected, and the annotations are included in Table 3.
Acknowledgments
We thank Stephen Burley and Steve Almo for advice on this project and Jeff Bonnano for coordinating sample delivery from SGX. Chris Lima kindly analyzed T1429 for presence of metal atoms by anomalous difference Fourier. This research is supported primarily by a grant from the National Institute for General Medical Sciences under the PSI Program (P50-GM-62529). Additional funding is provided under R01-GM-54762 (A.S.), R33-CA-84699 (A.S.), and the National Institute for Biomedical Imaging and Bioengineering and its Biomedical Technology Centers Program under P41-EB-01979 (M.R.C.). Support from the Sander Family Supporting Foundation, Sun Academic Equipment Grant EDUD-7824-020257-US, an IBM SUR grant, and an Intel computer hardware gift are also acknowledged (A.S.).
Footnotes
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2537904.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva, A., Howorth, D., Brenner, S.E., Hubbard, T.J., Chothia, C., and Murzin, A.G. 2004. SCOP database in 2004: Refinements integrate structure and sequence family data. Nucleic Acids Res. 32: D226-D229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, D. and Sali, A. 2001. Protein structure prediction and structural genomics. Science 294: 93-96. [DOI] [PubMed] [Google Scholar]
- Benson, D.A., Karsch-Mizrachi, I., Lipman, D.J., Ostell, J., Rapp, B.A., and Wheeler, D.L. 2002. GenBank. Nucleic Acids Res. 30: 17-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley, S.D., Chater, K.F., Cerdeno-Tarraga, A.M., Challis, G.L., Thomson, N.R., James, K.D., Harris, D.E., Quail, M.A., Kieser, H., Harper, D., et al. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417: 141-147. [DOI] [PubMed] [Google Scholar]
- Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, I.N., and Bourne, P.E. 2000. The Protein Data Bank. Nucleic Acids Res. 28: 235-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckmann, B., Bairoch, A., Apweiler, R., Blatter, M.C., Estreicher, A., Gasteiger, E., Martin, M.J., Michoud, K., O'Donovan, C., Phan, I., et al. 2003. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 31: 365-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley, S.K. and Bonanno, J.B. 2003. Structural genomics. Methods Biochem. Anal. 44: 591-612. [PubMed] [Google Scholar]
- Burley, S.K., Almo, S.C., Bonanno, J.B., Capel, M., Chance, M.R., Gaasterland, T., Lin, D., Sali, A., Studier, F.W., and Swaminathan, S. 1999. Structural genomics: Beyond the human genome project. Nat. Genet. 23: 151-157. [DOI] [PubMed] [Google Scholar]
- Chance, M.R., Sagi, I., Wirt, M.D., Frisbie, S.M., Scheuring, E., Chen, E., Bess Jr., J.W., Henderson, L.E., Arthur, L.O., South, T.L., et al. 1992. Extended x-ray absorption fine structure studies of a retrovirus: Equine infectious anemia virus cysteine arrays are coordinated to zinc. Proc. Natl. Acad. Sci. 89: 10041-10045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chance, M.R., Miller, L.M., Fischetti, R.F., Scheuring, E., Huang, W.X., Sclavi, B., Hai, Y., and Sullivan, M. 1996. Global mapping of structural solutions provided by the extended X-ray absorption fine structure ab initio code FEFF 6.01: Structure of the cryogenic photoproduct of the myoglobin-carbon monoxide complex. Biochemistry 35: 9014-9023. [DOI] [PubMed] [Google Scholar]
- Chance, M.R., Bresnick, A.R., Burley, S.K., Jiang, J.S., Lima, C.D., Sali, A., Almo, S.C., Bonanno, J.B., Buglino, J.A., Boulton, S., et al. 2002. Structural genomics: A pipeline for providing structures for the biologist. Protein Sci. 11: 723-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Editorial. 2004. PSI-phase 1 and beyond. Nat. Struct. Mol. Biol. 11: 201. [DOI] [PubMed] [Google Scholar]
- Eswar, N., John, B., Mirkovic, N., Fiser, A., Ilyin, V., Pieper, U., Stuart, A.C., Marti-Renom, M.A., Madhusudhan, M.S., Yerkovich, B., et al. 2003. Tools for comparative protein structure modeling and analysis. Nucleic Acids Res. 31: 3375-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiser, A., Sanchez, R., Melo, F., and Sali, A. 2001. Comparative protein structure modeling. In Computational biochemistry and biophysics (eds. M. Watanabe et al.), pp. 275-312. Marcel Decker, NY.
- Gavin, A.C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J., Rick, J.M., Michon, A.M., Cruciat, C.M., et al. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141-147. [DOI] [PubMed] [Google Scholar]
- Gerstein, M., Edwards, A., Arrowsmith, C.H., and Montelione, G.T. 2003. Structural genomics: Current progress. Science 299: 1663. [DOI] [PubMed] [Google Scholar]
- Goulding, C.W., Perry, L.J., Anderson, D., Sawaya, M.R., Cascio, D., Apostol, M.I., Chan, S., Parseghian, A., Wang, S.S., Wu, Y., et al. 2003. Structural genomics of Mycobacterium tuberculosis: A preliminary report of progress at UCLA. Biophys. Chem. 105: 361-370. [DOI] [PubMed] [Google Scholar]
- Guan, J., Almo, S.C., and Chance, M.R. 2004. Synchrotron radiolysis and mass spectrometry: A probe of the actin cytoskeleton. Acct. Chem. Res. 37: 221-229. [DOI] [PubMed] [Google Scholar]
- Hasnain, S.S. 2004. Synchrotron techniques for metalloproteins and human disease in post genome era. J. Synchrotron. Radiat. 11: 7-11. [DOI] [PubMed] [Google Scholar]
- Hendrickson, W.A. 1991. Determination of macromolecular structures from anomalous diffraction of synchrotron radiation. Science 254: 51-58. [DOI] [PubMed] [Google Scholar]
- Holm, L. and Sander, C. 1995. Dali: A network tool for protein structure comparison. Trends Biochem. Sci. 20: 478-480. [DOI] [PubMed] [Google Scholar]
- ____. 1996. Mapping the protein universe. Science 273: 595-603. [DOI] [PubMed] [Google Scholar]
- John, B. and Sali, A. 2003. Comparative protein structure modeling by iterative alignment, model building, and model Assessment. Nucleic Acids Res. 31: 3982-3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesley, S.A., Kuhn, P., Godzik, A., Deacon, A.M., Mathews, I., Kreusch, A., Spraggon, G., Klock, H.E., McMullan, D., Shin, T., et al. 2002. Structural genomics of the Thermotoga maritima proteome implemented in a high-throughput structure determination pipeline. Proc. Natl. Acad. Sci. 99: 11664-11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. and Rost, B. 2001. Comparing function and structure between entire proteomes. Protein Sci. 10: 1970-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujan, H.D., Mowatt, M.R., Wu, J.J., Lu, Y., Lees, A., Chance, M.R., and Nash, T.E. 1995. Purification of a variant-specific surface protein of Giardia lamblia and characterization of its metal-binding properties. J. Biol. Chem. 270: 13807-13813. [DOI] [PubMed] [Google Scholar]
- Murzin, A.G., Brenner, S.E., Hubbard, T., and Chothia, C. 1995. SCOP: A structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 247: 536-540. [DOI] [PubMed] [Google Scholar]
- Orengo, C.A., Michie, A.D., Jones, S., Jones, D.T., Swindells, M.B., and Thornton, J.M. 1997. CATH—A hierarchic classification of protein domain structures. Structure 5: 1093-1108. [DOI] [PubMed] [Google Scholar]
- Pieper, U., Eswar, N., Stuart, A.C., Ilyin, V.A., and Sali, A. 2002. MODBASE, a database of annotated comparative protein structure models. Nucleic Acids Res. 30: 255-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieper, U., Eswar, N., Braberg, H., Madhusudhan, M.S., Davis, F., Stuart, A.C., Mirkovic, N., Rossi, A., Marti-Renom, M.A., Fiser, A., et al. 2004. MODBASE, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Res. 32: D217-D222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajashankar, K., Chance, M.R., Burley, S.K., Jiang, J.S., Almo, S.C., Bresnick, A.R., Hunag, R., He, G., Chen, H., Sullivan, M., et al. 2001. Structural genomics at the National Synchrotron Light Source. NSLS Activity Report 2002: 2-28 to 2-32. [Google Scholar]
- Reboul, J., Vaglio, P., Rual, J.F., Lamesch, P., Martinez, M., Armstrong, C.M., Li, S., Jacotot, L., Bertin, N., Janky, R., et al. 2003. C. elegans ORFeome version 1.1: Experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat. Genet. 34: 35-41. [DOI] [PubMed] [Google Scholar]
- Sali, A. 1995. Comparative protein modeling by satisfaction of spatial restraints. Mol. Med. Today 1: 270-277. [DOI] [PubMed] [Google Scholar]
- ____. 100,000 protein structures for the biologist. Nat. Struct. Biol. 5: 1029-1032. [DOI] [PubMed]
- Sali, A., Glaeser, R., Earnest, T., and Baumeister, W. 2003. From words to literature in strutural proteomics. Nature Insight 422: 216-225. [DOI] [PubMed] [Google Scholar]
- Sanchez, R. and Sali, A. 1998. Large-scale protein structure modeling of the Saccharomyces cerevisiae genome. Proc. Natl. Acad. Sci. 95: 13597-13602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez, R., Pieper, U., Mirkovic, N., de Bakker, P.I., Wittenstein, E., and Sali, A. 2000. MODBASE, a database of annotated comparative protein structure models. Nucleic Acids Res. 28: 250-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer, A.A., Aravind, L., Madden, T.L., Shavirin, S., Spouge, J.L., Wolf, Y.I., Koonin, E.V., and Altschul, S.F. 2001. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 29: 2994-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, W., Ostrov, D., Gerchman, S., Kycia, H., Studier, W., Edstrom, W., Bresnick, A.R., Ehrlich, J., Blanchard, J., Almo, S.C., et al. 2003. High-throughput structural biology and proteomics. In Protein chips, biochips, and proteomics: The next phase of genomics discovery, Chapter 12, pp. 299-324. Marcel Decker, NY.
- Stein, L.D., Bao, Z., Blasiar, D., Blumenthal, T., Brent, M.R., Chen, N., Chinwalla, A., Clarke, L., Clee, C., Coghlan, A., et al. 2003. The genome sequence of Caenorhabditis briggsae: A platform for comparative genomics. PLoS Biol. 1: E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers, M.F., Henderson, L.E., Chance, M.R., Bess Jr., J.W., South, T.L., Blake, P.R., Sagi, I., Perez-Alvarado, G., Sowder III, R.C., Hare, D.R., et al. 1992. Nucleocapsid zinc fingers detected in retroviruses: EXAFS studies of intact viruses and the solution-state structure of the nucleocapsid protein from HIV-1. Protein Sci. 1: 563-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpunar, J. 2004. Metallomics: A new frontier in analytical chemistry. Anal. Bioanal. Chem. 378: 54-56. [DOI] [PubMed] [Google Scholar]
- Terwilliger, T.C., Park, M.S., Waldo, G.S., Berendzen, J., Hung, L.W., Kim, C.Y., Smith, C.V., Sacchettini, J.C., Bellinzoni, M., Bossi, R., et al. 2003. The TB structural genomics consortium: a resource for Mycobacterium tuberculosis biology. Tuberculosis (Edinb) 83: 223-249. [DOI] [PubMed] [Google Scholar]
- Tompa, P. 2002. Intrinsically unstructured proteins. Trends Biochem. Sci. 27: 527-533. [DOI] [PubMed] [Google Scholar]
- Tong, A.H., Lesage, G., Bader, G.D., Ding, H., Xu, H., Xin, X., Young, J., Berriz, G.F., Brost, R.L., Chang, M., et al. 2004. Global mapping of the yeast genetic interaction network. Science 303: 808-813. [DOI] [PubMed] [Google Scholar]
- Vitkup, D., Melamud, E., Moult, J., and Sander, C. 2001. Completeness in structural genomics. Nat. Struct. Biol. 8: 559-566. [DOI] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Westbrook, J., Feng, Z., Jain, S., Bhat, T.N., Thanki, N., Ravichandran, V., Gilliland, G.L., Bluhm, W., Weissig, H., Greer, D.S., et al. 2002. The Protein Data Bank: Unifying the archive. Nucleic Acids Res. 30: 245-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbrook, J., Feng, Z., Chen, L., Yang, H., and Berman, H.M. 2003. The Protein Data Bank and structural genomics. Nucleic Acids Res. 31: 489-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler, D.L., Chappey, C., Lash, A.E., Leipe, D.D., Madden, T.L., Schuler, G.D., Tatusova, T.A., and Rapp, B.A. 2000. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 28: 10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C. and Kim, S.H. 2003. Overview of structural genomics: From structure to function. Curr. Opin. Chem. Biol. 7: 28-32. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- www.nigms.nih.gov/psi; NIH Web site providing information and relevant links for the Protein Structure Initiative.
- http://targetdb.pdb.org; Web site operated by the Protein Databank to allow searching of targets from the structural genomics centers.
- www.nysgxrc.org; Web site operated by the NYSGRC. Its functions are to provide a public target list and progress as well as to allow consortium members to enter target data.
- http://salilab.org/modbase; MODBASE, a comprehensive database of comparative protein structure models.
- www-archbac.u-psud.fr/genomics/COG_Guess.html; Clusters of Orthologous Groups Database Query Page to perform similarity search in COG database. This provides a function and COG category guess for input sequence.
- http://salilab.org/modbase/models_nysgxrc.html; Summary and statistics of homology modeling results using the NYSGXRC PDB structures as templates.