Abstract
The recently completed Caenorhabditis elegans genome sequence allows application of high-throughput (HT) approaches for phenotypic analyses using RNA interference (RNAi). As large phenotypic data sets become available, “phenoclustering” strategies can be used to begin understanding the complex molecular networks involved in development and other biological processes. The current HT-RNAi resources represent a great asset for phenotypic profiling but are limited by lack of flexibility. For instance, existing resources do not take advantage of the latest improvements in RNAi technology, such as inducible hairpin RNAi. Here we show that a C. elegans ORFeome resource, generated with the Gateway cloning system, can be used as a starting point to generate alternative HT-RNAi resources with enhanced flexibility. The versatility inherent to the Gateway system suggests that additional HT-RNAi libraries can now be readily generated to perform gene knockdowns under various conditions, increasing the possibilities for phenome mapping in C. elegans.
Thirty years of classical genetics in Caenorhabditis elegans identified and characterized ∼800 genes (∼4% of all genes; Brenner 1974; Jorgensen and Mango 2002). With the availability of the worm genome sequence (The C. elegans Sequencing Consortium 1998), it is now possible to apply genome-wide reverse genetic approaches for functional characterization. One way to uncover the functions of genes is to study their loss-of-function phenotypes after deletion (or gene knockout). Given the lack of homologous recombination in C. elegans, gene deletions are not easily amenable to high-throughput (HT) generation, although efforts are currently underway to achieve this goal. Instead, RNA interference (RNAi; Fire et al. 1998), which relies on the introduction of dsRNA in the worm, readily allows genome-wide loss-of-function screens by gene “knockdowns” (Fraser et al. 2000; Gonczy et al. 2000; Piano et al. 2000, 2002; Hanazawa et al. 2001; Maeda et al. 2001; Zipperlen et al. 2001; Ashrafi et al. 2003; Kamath et al. 2003; Lee et al. 2003; Pothof et al. 2003; Simmer et al. 2003; Vastenhouw et al. 2003).
An RNAi-by-feeding (Timmons and Fire 1998) library was generated by using PCR-amplified genomic DNA fragments as a template (Kamath et al. 2003). This resource targets ∼78% of the 19,920 genes currently predicted in Worm Sequence 112 (WS112; WormBase, http://www.wormbase.org; see Methods for calculations; Kamath et al. 2003; Harris et al. 2004). This library has been successfully used for several genome-wide RNAi screens, for example, in wild-type (WT) N2 animals for systemic functional analysis (Kamath et al. 2003) and for the analysis of genes involved in the regulation of body fat storage (Ashrafi et al. 2003), as well as in the hypersensitive rrf-3 C. elegans strain (Simmer et al. 2003). These data represent a significant step forward in the phenotypic characterization of most C. elegans genes.
Ultimately, the combination of loss-of-function data obtained in numerous, different conditions could lead to the classification of worm genes by “phenotypic signatures.” Phenoclustering methods have been applied on a moderate scale in C. elegans (Boulton et al. 2002; Piano et al. 2002) and in Drosophila (Boutros et al. 2004). The C. elegans “phenome” map resulting from such genome-wide analyses would increase our understanding of a variety of biological processes.
However, several technical points need to be resolved before a comprehensive phenome map can be generated for C. elegans. First, such a map relies upon the generation of RNAi data for all genes. Today, not all worm genes are yet targeted by an RNAi clone. Second, RNAi screens should be performed in different environmental conditions or various genetic backgrounds, applying different procedures. For example, experiments leading to the loss-of-function of two genes, simultaneously, can be carried out in the worm (Simmer et al. 2003; Tewari et al. 2004). Furthermore, RNAi can be applied by different methods of delivery, such as injection (Fire et al. 1998), feeding (Timmons and Fire 1998), or soaking (Tabara et al. 1998), at different times during the development of the worm such as different larval stages, and with different DNA templates for the synthesis of dsRNA, such as genomic DNA fragments, cDNAs, or ORFs. Finally, a new RNAi technology, referred to as inducible hairpin RNAi (Tavernarakis et al. 2000; N.M. Johnson, C.A. Behm, and S.C. Trowell, unpubl.), opens new possibilities for targeting genes. Indeed, genes can be knocked-down at specific times during the development and specific cell types such as neurons can now be targeted. Other improved technologies for loss-of-function assays will likely be developed soon. To accommodate these numerous applications, RNAi resources should thus be generated by using cloning tools that are as flexible as possible (Fig. 1).
Figure 1.
Generating RNAi resources from flexible Gateway ORFeome and promoterome collections. AntR1 and AntR2 refer to antibiotic resistance markers number 1 and 2.
In the infancy of the genomic era (Collins et al. 2003), resources need to be dynamic and flexible. Ideally, a single, comprehensive set of cloned ORFs, that is, an “ORFeome” (Walhout et al. 2000b; Reboul et al. 2003; Rual et al. 2004), should be usable for various HT applications. An ORFeome resource should be updated (ORFs corrected, removed, or added) regularly as new genome annotations become available. An ORFeome should be cloned in a format that allows facile transfer of the ORFs to any desired vector as new technologies become available (Fig. 1). Recently developed recombinational cloning technologies (Liu et al. 1998; Hartley et al. 2000; Walhout et al. 2000a; Zhang et al. 2002) allow such versatile cloning approaches at HT (Reboul et al. 2003; Dricot et al. 2004).
Upon the release of the C. elegans genome sequence (The C. elegans Sequencing Consortium 1998), a genome-wide approach was initiated to clone all predicted ORFs of the worm ORFeome (Walhout et al. 2000b; Reboul et al. 2001, 2003; Vaglio et al. 2003; WorfDB, http://worfdb.dfci.harvard.edu) using the Gateway recombinational cloning system (Hartley et al. 2000; Walhout et al. 2000a). Version 1.1 of the C. elegans ORFeome resource contains 11,942 cloned ORFs (Reboul et al. 2003) and constitutes a platform for proteomic and functional genomic approaches (Reboul et al. 2003; Rual et al. 2004). The C. elegans ORFeome resource is dynamic, with new ORFs added or corrected continuously (Lamesch et al. 2004). Phenome mapping, as well as other “omic” approaches, should benefit from the flexibility of this resource.
By using the C. elegans ORFeome v1.1 resource (Reboul et al. 2003), we began generating a companion resource for RNAi screening. Herein we describe this ORFeome-RNAi v1.1 library, as well as the results from using this library for a genome-wide RNAi-by-feeding screen of WT C. elegans hermaphrodites. We conclude that the use of an ORFeome approach is feasible for the generation of an alternative RNAi library, and we suggest that this cloning strategy could be applied to other functional genomic approaches.
RESULTS
Generation of the ORFeome-RNAi v1.1 Library
The C. elegans ORFeome v1.1 library contains 11,942 ORFs cloned as Gateway Entry clones, comprising 10,623 ORFs cloned “in frame” plus 1319 ORFs cloned “out of frame” (Reboul et al. 2003). ORFs were cloned out of frame because of mispredictions of their ATG or STOP codons. Only the in-frame ORFs can be used for protein expression, but both sets of clones can be used for RNAi. HT-recombinational cloning protocols were used to transfer ORFs from the pDONR201 Entry plasmid into the pL4440-dest-RNAi Destination vector by using the LR Gateway reaction (see Methods). In all, 11,804 RNAi clones were generated (Supplemental Table A). These clones were archived as glycerol stocks of transformed Escherichia coli strain HT115(DE3) used in RNAi-by-feeding and as miniprep plasmid DNAs (Timmons and Fire 1998). These DNA preps can be used as templates for in vitro dsRNA synthesis before RNAi application by soaking or injection. The ORFeome-RNAi v1.1 library is available to the community upon request (Open Biosystems, http://www.openbiosystems.com; MRC geneservice, http://www.rfcgr.mrc.ac.uk/geneservice; WorfDB, http://worfdb.dfci.harvard.edu).
Bioinformatic analysis revealed that the ORFeome-RNAi v1.1 library contains 11,511 RNAi clones, each expected to target a single gene (see Methods; Supplemental Table A). Several of these RNAi clones target the same genes, so the RNAi library in theory can knockdown the expression of 10,953 (∼55%) of the 19,920 unique protein-encoding genes predicted in WS112 (Supplemental Table B). Of these 10,953 genes, 485 are targeted by two or more RNAi clones. There are 1736 genes, not previously targeted by other existing RNAi-by-feeding libraries, that now have a clone available in the ORFeome-RNAi v1.1 library (Supplemental Table B). This represents an increase of ∼9% in the total number of genes that can be targeted by RNAi-by-feeding. In combination with the previous RNAi-by-feeding library (Kamath et al. 2003) a total of 17,201 predicted worm genes (∼86%) can now be targeted by an RNAi-by-feeding clone.
Genome-Wide Phenotypic Analysis Using the ORFeome-RNAi v1.1 Library
We first used the ORFeome-RNAi v1.1 library for a synthetic lethal screen by conducting a genome-wide RNAi screen simultaneously in WT N2 C. elegans hermaphrodites and in a lin-35/Rb mutant background (see Methods; Fig. 2). The results in the N2 animals are presented here, and the results of the lin-35/Rb synthetic phenotype screen will be reported elsewhere (J. Ceron, J.-F. Rual, M. Vidal, and S. van den Heuvel, unpubl.). Worms were subjected to RNAi-by-feeding at the first larval stage (L1). We identified a variety of phenotypic defects, including lethality and abnormal body morphology, occurring during the embryonic, larval, and adult stages.
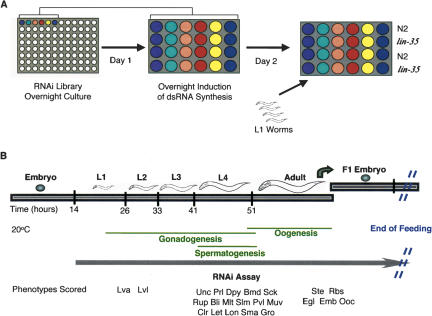
Figure 2.
Overview of the RNAi screening procedure. (A) Overnight cultures of 96 bacterial clones are inoculated in sixteen 24-wells plates (six clones per plate and four wells per clone). After overnight induction of the dsRNA synthesis, three to 10 worms synchronized at the L1-stage (N2 or lin-35 strain) were deposited into the wells. (B) RNAi experiments were performed at 20°C. We observed a wide range of phenotypes across development.
The results from this phenotypic analysis of 10,953 WS112 predicted genes are described in Table 1 and in Supplemental Table B. We observed phenotypes different from WT for 1066 WS112 predicted genes, representing ∼10% of the total ORFs tested. This percentage is similar to that obtained in previous genome-wide RNAi-by-feeding screens (Kamath et al. 2003; Simmer et al. 2003).
Table 1.
Summary of RNAi Data
|
|
Genome
|
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genome annotation | ORFeome-RNAi v1.1 WS112a | Kamath et al. (2003) WS61b | |||||||||||||||||||
| Genes targeted | 10,953 | 55.0% | 16,757 | 86.2% | |||||||||||||||||
| Phenotypes | 1066 | 9.7% | 1722 | 10.3% | |||||||||||||||||
| Nonviable phenotypes | 928 | 8.5% | 1170 | 7.0% | |||||||||||||||||
| Viable phenotypes only | 138 | 1.3% | 552 | 3.2% | |||||||||||||||||
| Emb | 342 | 3.1% | 929 | 5.5% | |||||||||||||||||
| Rbs/Ste/Stp | 602 | 5.5% | 474 | 2.8% | |||||||||||||||||
| Chromosome 1
|
Chromosome II
|
Chromosome III
|
|||||||||||||||||||
| Genome annotation
|
ORFeome-RNAi v1.1 WS112a
|
Kamath et al. (2003) WS61b
|
ORFeome-RNAi v1.1 WS112a
|
Kamath et al. (2003) WS61b
|
ORFeome-RNAi v1.1 WS112a
|
Kamath et al. (2003) WS61b
|
|||||||||||||||
| Genes targeted | 1670 | 58.6% | 2445 | NDb | 1979 | 57.2% | 2978 | NDb | 1533 | 58.1% | 2132 | NDb | |||||||||
| Phenotypes | 272 | 16.3% | 334 | 13.7% | 202 | 10.2% | 354 | 11.9% | 232 | 15.1% | 395 | 18.5% | |||||||||
| Nonviable phenotypes | 239 | 14.3% | 240 | 10.3% | 182 | 9.2% | 240 | 8.1% | 197 | 12.8% | 298 | 14.0% | |||||||||
| Viable phenotypes only | 33 | 2.0% | 81 | 3.3% | 20 | 1.0% | 114 | 3.9% | 35 | 2.3% | 97 | 4.6% | |||||||||
| Emb | 88 | 5.3% | 226 | 9.2% | 60 | 3.0% | 204 | 6.9% | 70 | 4.6% | 220 | 10.3% | |||||||||
| Rbs/Ste/Stp | 149 | 8.9% | 98 | 4.0% | 122 | 6.2% | 76 | 2.5% | 139 | 9.1% | 150 | 7.0% | |||||||||
| Chromosome IV
|
Chromosome V
|
Chromosome X
|
|||||||||||||||||||
| Genome annotation
|
ORFeome-RNAi v1.1 WS112a
|
Kamath et al. (2003) WS61b
|
ORFeome-RNAi v1.1 WS112a
|
Kamath et al. (2003) WS61b
|
ORFeome-RNAi v1.1 WS112a
|
Kamath et al. (2003) WS61b
|
|||||||||||||||
| Genes targeted | 1848 | 57.0% | 2693 | NDb | 2490 | 50.1% | 4152 | NDb | 1433 | 51.9% | 2357 | NDb | |||||||||
| Phenotypes | 166 | 9.0% | 293 | 10.9% | 145 | 5.8% | 215 | 5.2% | 49 | 3.4% | 131 | 5.6% | |||||||||
| Nonviable phenotypes | 143 | 7.7% | 187 | 6.9% | 126 | 5.1% | 145 | 3.5% | 41 | 2.9% | 47 | 2.0% | |||||||||
| Viable phenotypes only | 23 | 1.2% | 106 | 3.9% | 19 | 0.8% | 70 | 1.7% | 8 | 0.6% | 84 | 3.6% | |||||||||
| Emb | 55 | 3.0% | 143 | 5.3% | 53 | 2.1% | 108 | 2.6% | 16 | 1.1% | 28 | 1.2% | |||||||||
| Rbs/Ste/Stp | 93 | 5.0% | 72 | 2.7% | 77 | 3.1% | 63 | 1.6% | 22 | 1.5% | 15 | 0.6% | |||||||||
The total number of predicted genes in WS112 is 19,920; 2850 on chromosome I; 3457 on chromosome II; 2639 on chromosome III; 3242 on chromosome IV; 4970 on chromosome V; and 2762 on chromosome X.
The total number of predicted genes in WS61 is 19,427; the number of predicted genes for each chromosome could not be found; ND means not determined.
We observed an RNAi phenotype for 220 WS112 predicted genes for which no phenotype was previously reported in WormBase (Supplemental Table B). For 79 of these 220 genes, no RNAi assay was listed in WormBase. The other 141 genes, targeted in previous RNAi screens, had showed a WT phenotype before. Altogether, our results increase the total number of genes associated with any RNAi phenotype by ∼7%, from 3259 to 3479. Finally, an additional 1145 genes for which no RNAi assay was reported in WormBase did not show any detectable phenotype and were scored as WT in our screen.
A global analysis of the data verifies observations made previously regarding the distributions of viable postembryonic phenotypes (Vpep) and nonviable phenotypes (Nonv) ascribed to various genes located on either the autosomes or the X chromosome (Piano et al. 2000; Maeda et al. 2001; Kamath et al. 2003). In particular, there appears to be a low incidence of genes located on the X chromosome that are associated with any phenotype (Table 1).
Gene inactivation by RNAi-by-feeding is often incomplete and may be too limited to cause fully penetrant loss-of-function phenotypes, thereby generating false-negative results. For example, RNAi is less effective in neurons (Tavernarakis et al. 2000), and genes encoding proteins with long half-lives are more resistant to RNAi (Hannon 2002). Previous genome-wide RNAi screens had various rates of false negatives (Kamath et al. 2003; Simmer et al. 2003), based on comparison of the RNAi results to the loss-of-function phenotypes caused by genetic mutations. Nearly 45% of the genes with a reported mutant phenotype did not show any phenotype in each of these previous studies. More precisely, rates of ∼25% and ∼55% of false negatives were observed for Nonv mutant phenotypes and for Vpep mutant phenotypes, respectively. Matching our RNAi data to the phenotypes described for 269 genetic mutants (resembling the control set of genetic mutants used in Kamath et al. [2003] and Simmer et al. [2003]), we observed an overall false-negative rate of ∼59% (∼43% for Nonv mutant phenotypes and ∼83% for Vpep mutant phenotypes; Supplemental Table C). This somewhat higher frequency of false negatives can be explained mostly by our HT RNAi protocol, in which animals were followed only for one generation and not transferred to a second RNAi plate (see Discussion).
Low rates of false positives were previously described in genome-wide RNAi-by-feeding screen (Kamath et al. 2003; Simmer et al. 2003). In these studies, a false positive was scored when the RNAi phenotype was lethal, whereas the reported genetic mutant shows a nonlethal phenotype. Published false-positive rates vary from ∼0% to 0.4% (Kamath et al. 2003; Simmer et al. 2003). By applying the same strategy to a similar set of 103 loci, we counted four Nonv phenotypes in our RNAi experiment (Supplemental Table D). After sequence verification, we found one obvious cross-contamination. The remaining three clones have the correct sequence. Two of the three corresponding genes are reported as Nonv by RNAi in WormBase. Overall, our rate of false-positive results is low and in part can be due to cross-contamination.
Reproducibility
The ORFeome-RNAi v1.1 library contains 430 pairs in which both clones target the same gene and should give rise to the same phenotype. These clones were processed independently and therefore constitute a blind control for the reproducibility of scoring. In 78% (337/430), we obtained a WT phenotype for both clones of the pair. In 10% (45/430), we observed a phenotype for both clones. Thus, the results are in agreement regarding the scoring of WT versus phenotype for ∼89% of the pairs of clones (382/430; Supplemental Table E). In ∼11% (48/430), there is disagreement. Variability in the results from repeated RNAi-by-feeding assays was observed previously (Simmer et al. 2003). Variability ranges from ∼10% to 30% between experiments and is likely due to lack of reproducibility from one RNAi assay to another (Simmer et al. 2003). To verify this phenomenon, we performed a second RNAi assay for the 48 pairs of RNAi clones in disagreement. In the second assay, we observed similar phenotypes for both clones ∼52% of the time (25/48; Supplemental Table E). Thirteen pairs of clones now gave rise to a WT phenotype for both clones. Ten pairs of clones were still not in agreement; their correct identities were verified by DNA sequence analysis.
Interestingly, of the 10 pairs of clones in disagreement in the second RNAi assay, six showed large differences in size of the cloned ORF. For these six pairs, the longer ORF clone was the one giving rise to the observed phenotype. These numbers are not statistically significant but suggest that effectiveness of RNAi-by-feeding may be somewhat dependent on the length of the clone, the longer clones being more effective in RNAi. We went on to perform RNAi assay by injection for three of these six pairs of clones and obtained a phenotype in all six (three by two) RNAiby-injection assays. Thus, the output of an RNAi assay depends on the method of delivery of dsRNA.
DISCUSSION
The generation of complete resources of cloned ORFs represents an important and challenging step in the development of proteomics and functional genomics (Reboul et al. 2003; Rual et al. 2004). Within months after the generation of the C. elegans ORFeome v1.1 (Reboul et al. 2003), we transferred >11,000 ORFs into the pL4440-dest RNAi vector, generating a new RNAi resource. The technology that allowed this genome-wide cloning effort is now being used for the production of additional resources, such as Activation Domain and DNA Binding Domain fusion collections for yeast two-hybrid screens.
The C. elegans ORFeome-RNAi v1.1 library contains 11,511 RNAi clones, each expected to target a single gene (Supplemental Table A), and includes 1736 C. elegans genes not targeted previously by any RNAi-by-feeding clones (Supplemental Table B). We used the C. elegans ORFeome-RNAi v1.1 library to perform a genome-wide RNAi screen in WT C. elegans hermaphrodites (Fig. 2). We observed a phenotype for ∼10% of the WS112 genes studied (Supplemental Table A; Table 1). Our RNAi results increase by 220 the number of genes for which an RNAi phenotype has been described. We confirmed phenotypes for numerous genes.
As previously observed (Simmer et al. 2003), we found that the RNAi effect varies from one RNAi-by-feeding screen to another. This lack of reproducibility is likely due to the inability to control the amount of dsRNA animals are exposed to, resulting in a high rate of false negatives. We also suggest that the RNAi effect could depend on the method of dsRNA delivery and on the size of the RNAi clone, with longer clones generally more effective, as described previously for RNAi-by-injection (Parrish et al. 2000). As for most large-scale approaches, results obtained from genome-wide RNAi screens should be interpreted carefully, especially negative results. Several issues need to be addressed before a comprehensive phenome map of C. elegans can be generated, for example, by generating genome-wide RNAi data in different conditions and by developing new RNAi protocols. In this regard, ORFeome-based RNAi resources should help improve phenome mapping by taking advantage of the versatility of recombinational cloning to apply new RNAi technologies to genome scale (Fig. 1).
Cloned ORFs versus cloned genomic DNA fragments, as template DNA could improve the efficiency of RNAi. Cloned ORFs are free of introns and thus provide more template for siRNAs. Moreover, a study based on the RNAi results obtained with the genomic RNAi library (Kamath et al. 2003) showed a lower incidence of RNAi phenotype among loci with a small portion of coding sequence and large introns. This observation suggested that genomic DNA as RNAi template reduces RNAi efficiency (Cutter et al. 2003). Nevertheless, our proportion of hits (Table 1) and rate of false-negative results are comparable to previous results (Kamath et al. 2003; Simmer et al. 2003). Furthermore, an analysis using the current version of the genome annotation demonstrates that the fraction of coding gene sequence exerts a weaker negative effect than initially reported (A.D. Cutter, pers. comm.). Overall, it seems that the presence of introns in the DNA template does not have a dramatic negative impact on the efficiency of RNAi.
In our screen, animals were exposed to RNAi-by-feeding during the L1 stage (Fig. 2), as opposed to the L4 stage in previous screens (Kamath et al. 2003; Simmer et al. 2003). Consequently, we observed phenotypes linked to sterility (sterile, sterile progeny, and reduced brood size) for a larger number of genes (∼5.5% of those analyzed) than reported before (∼2.8%; Table 1; Kamath et al. 2003). On the other hand, Kamath et al. (2003) observed an embryonic lethal phenotype (Emb) for ∼5.5% of assayed genes, whereas we observed 3.1%. This difference might arise because many genes are essential for both the development of the adult germ line and the embryos. For these genes, scoring Emb versus sterile phenotypes could depend on the time of initiation of the RNAi assay, larval stage L1 versus L4.
Several factors are likely to contribute to a reduced frequency of phenotypes as well as to phenotypes that differ from those listed in WormBase for a given gene. First, we used a different RNAi protocol, and the criteria for scoring phenotypes were slightly more stringent than those used previously. To increase the throughput of the screening, we initiated the screening on L1 stage worms, we followed the animals for one generation and did not transfer them to a second RNAi plate. Phenotypes occurring late in the screen were difficult to score because food was limited. Subtle phenotypes may have been overlooked because we were scoring for visually obvious phenotypes.
Despite the higher rate of false negatives, we observe a comparable rate, ∼10%, of hits for all genes targeted (Table 1). This is probably due to higher representation in the ORFeome v1.1 resource of genes that can give rise to a phenotype. Highly expressed genes that are more represented in the ORFeome1.1 resource (Reboul et al. 2003) have more chance to be essential (Cutter et al. 2003).
The C. elegans ORFeome v1.1 and ORFeome-RNAi v1.1 libraries represent ∼55% of the predicted worm ORFeome. Importantly, these collections should be considered as dynamic resources. New Gateway Entry clones, generated in the ongoing ORFeome project (Lamesch et al. 2004), will be subsequently transferred into the RNAi vector in the near future. If a new improved RNAi vector is made available to the community, the Gateway cloning system will allow quick transfer of all cloned ORFs into it. For example, a hairpin RNAi method (Tavernarakis et al. 2000) was developed by using the ORFeome v1.1 resource (Reboul et al. 2003) to generate hairpin RNAi constructs in the pWormgate system (N.M. Johnson, C.A. Behm, and S.C. Trowell, unpubl.). In addition, such hairpin constructs could be fused to any specific promoter cloned by MultiSite Gateway (Fig. 1; Dupuy et al. 2004).
The complete ORFeome-RNAi v1.1 library (Supplemental Table A) is available upon request from Open Biosystems (http://www.openbiosystems.com) and MRC geneservice (http://www.rfcgr.mrc.ac.uk/geneservice; WorfDB, http://worfdb.dfci.harvard.edu). The data generated in the WT RNAi screen (Supplemental Table B) will be available in WormBase shortly (http://www.wormbase.org; Harris et al. 2004) and in RNAiDB (http://nematoda.bio.nyu.edu; Gunsalus et al. 2004). Integrated with other functional genomic data, these RNAi resources should contribute to a deeper understanding of the biology of C. elegans.
In summary, we demonstrated the value of an ORFeome-based approach for the generation of alternative HT-RNAi resources and ultimately, for the improvement of phenome mapping in C. elegans.
METHODS
Generation of the ORFeome-RNAi v1.1 Library
ORFs were transferred into pL4440-dest-RNAi, a Gateway-compatible RNAi vector adapted from the original pL4440-RNAi vector (Timmons and Fire 1998). Gateway recombinational cloning reactions were performed as described by the manufacturer (Invitrogen) with minor changes. The pL4440-dest-RNAi vector was digested with EcoRI and NcoI before the LR reaction, which significantly improved (>10-fold) the efficiency of the LR reaction. LR reaction plates were incubated overnight at 25°C. By using 96 well plates and HT-liquid handling systems, up to 1500 LR reactions were carried out by one person per day.
It is not possible to transform LR products directly into the RNAi-by-feeding bacterial strain HT115 (DE3), an RNase III-deficient strain of E. coli. Indeed, HT115 (DE3) bacteria are resistant to the ccdB toxic gene used in the Gateway cloning system. Therefore, LR products were first transformed into E. coli DH5α strain. Bacterial transformations were performed with 100 μg/mL ampicillin selection in liquid cultures. Plasmid DNA minipreps were prepared in a 96-well format for each pool of transformants by using a Qiagen 9600 Robot. DNA preparations were subsequently transformed into the RNAi-by-feeding bacterial strain HT115 (DE3). Out of the 11,942 ORFs cloned as Entry clones in the C. elegans ORFeome v1.1 resource, we obtained HT115 (DE3) transformants for 11,804 ORFs (Supplemental Table A).
All the Entry clones used in the LR reaction were verified by DNA sequencing analysis (Reboul et al. 2003). We also verified the identity of 96 RNAi clones picked at random from the library by sequencing. The universal primers used for PCR amplification and sequencing are pL4440-dest-RNAi-FOR (5′GTTTTCCCAGT CACGACGTT3′) and pL4440-dest-RNAi-REV (5′TGGATAACCG TATTACCGCC3′). We observed the expected sequences in 96% of the time. Three of the clones contained the wrong ORFs, and one clone did not contain any insert. We conclude that a tiny portion of the phenotypes may have been attributed to the wrong genes due to cross-contamination. The identity of the 220 RNAi clones corresponding to genes for which no phenotype was previously reported in WormBase has been verified by DNA sequence analysis.
Bioinformatic Analysis
For the C. elegans genome sequence and related genome annotation, we used version WS112 available at WormBase (http://www.wormbase.org; Harris et al. 2004). There are 19,920 protein encoding predicted genes in WS112 (Supplemental Table B).
To determine which WS112 genes are targeted by the ORFeome-RNAi v1.1 clones, we first retrieved the sequences from the primer pairs used in the ORFeome project (Reboul et al. 2003; Vaglio et al. 2003) and used these sequences to perform BLAST analysis against version WS112 of the worm genome. For each RNAi clone, the resulting position was then queried for an overlap of >100 nucleotides with the transcript(s) (UTRs plus exonic sequences) of one or more predicted WS112 genes.
With this strategy, we determined that there are 153 RNAi clones that target more than one WS112 predicted gene (Supplemental Table A). Furthermore, there are 140 RNAi clones that do not target any WS112 predicted genes (Supplemental Table A). These results are somewhat surprising because the cloned ORFs were PCR amplified from a worm cDNA library (Reboul et al. 2003). The information related to these cloned ORFs should be corrected in forthcoming releases from WormBase. The results obtained with these 293 RNAi clones were removed from our analysis. We found 10,953 WS112 genes targeted by the collection of 11,511 remaining RNAi clones. Out of the 10,953 genes, 485 were targeted by more than one RNAi clone. Given the low rate of false-positive results and the high rate of false-negative results in RNAi screens (see Results; Kamath et al. 2003; Simmer et al. 2003), all the phenotypes different from WT were scored by default for those 485 genes in the Supplemental Tables B, C, and D.
The data related to the previous RNAi library (Kamath et al. 2003) were retrieved from WormBase WS112 (WormBase, http://www.wormbase.org; Harris et al. 2004). In WS112, there are 16,537 RNAi clones that target 15,465 WS112 genes. However, 16,757 RNAi clones were described as targeting 16,757 WS61 genes (Kamath et al. 2003). The discrepancy is probably caused by altered gene predictions between WS61 and WS112, and missing data for 220 RNAi clones in WormBase WS112.
RNAi Screening
We performed the RNAi screen simultaneously in WT N2 C. elegans hermaphrodites and in a lin-35/Rb mutant background (Fig. 2). Standard methods were used for culturing worms on nematode growth medium (NGM; Brenner 1974). Screening was performed in 24-well format plates containing NGM supplemented with 50 μg/mL ampicillin, 12.5 μg/mL tetracycline, and 1.43 mg/mL IPTG. Unseeded feeding plates were kept in the dark for less than a week at 4°C. Before bacterial inoculation, the feeding plates were dried for 2 to 3 h in a laminar airflow hood or in a 37°C incubator. RNAi bacterial clones were grown in 96-well format for 24 h at 37°C in 600 μL LB with 50 μg/mL carbenicillin and 12.5 μg/mL tetracycline. Each well of the 24-well format plates was inoculated with 115 μL of bacterial culture by using an extendable automated multichannel pipette. The expression of dsRNA was induced overnight in the presence of IPTG (1.43 mg/mL), at room temperature and in the dark. After dsRNA induction, three to 10 worms synchronized in the L1-stage were deposited into the wells (Fig. 2).
Most of the previous RNAi studies were done by initiating RNAi on young adults (Fraser et al. 2000; Gonczy et al. 2000; Piano et al. 2000, 2002; Maeda et al. 2001; Zipperlen et al. 2001; Kamath et al. 2003; Simmer et al. 2003). Instead, we fed worms at the L1 larval stage (Fig. 2). This procedure necessitates fewer worm manipulations and thus allows a higher throughput. A disadvantage of this method is food limitation in the F1 generation. This might explain the high rate of false-negative results in our assay, particularly for Vpep phenotypes.
Phenotypes were scored visually twice on days 4 and 5 of the 20°C culture. We looked for 18 different Vpep phenotypes: Lva (larval arrest), Unc (uncoordinated), Prl (paralyzed), Dpy (dumpy), Bmd (body morphology defect), Sck (sick), Bli (blister), Mlt (molting defect), Slm (slim), Him (high incidence of male), Pvl (protruding vulva), Muv (multivulva), Clr (clear), Lon (long), Sma (small), Gro (growth defect), Egl (egg laying defect), and Soc (social). We looked for eight different Nonv phenotypes: Ste (sterile), Rbs (reduced brood size), Let (Lethal), Emb (embryonic lethal), Ooc (oocytes), Stp (sterile progeny), Rup (ruptured), and Lvl (larval lethal). When the phenotype occurs in the F1 generation, F1 is added before the phenotype name (e.g., F1 Lva). Emb was defined as >20% dead embryos. Phenotypes observed with partial penetrance are indicated with a “p”, for example, pEmb for partially penetrant embryonic lethality. A partial phenotype is scored when the phenotype occurs in <90% of the P0 or F1 progeny.
Acknowledgments
We thank the C. elegans Sequencing Consortium for the genome sequence; members of WormBase for discussions and for making RNAi data available; the participants of the ORFeome meeting for discussions; the members of the Plasterk laboratory, and F. Simmer in particular; the Ahringer laboratory, and R. Kamath in particular; the Piano laboratory, and A. Cutter and G. Achaz for sharing data and discussions; N. Bertin, V. Goidts, and N. Li for technical assistance; C. Fraughton for laboratory support; T. Clingingsmith for administrative assistance; M. Cusick for critical reading of the manuscript; and all the members of the van den Heuvel and Vidal laboratories, and D. Dupuy and the ORFeome team in particular. This work was supported by National Institutes of Health grant CA95281 (S.v.d.H.) and National Cancer Institute grant 7 R33 CA81658-02 (M.V.). J.C was the recipient of a Postdoctoral Fellowship from the Secretaría de Estado de Educación y Universidades (Spain), cofunded by the European Social Fund.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2505604.
References
- Ashrafi, K., Chang, F.Y., Watts, J.L., Fraser, A.G., Kamath, R.S., Ahringer, J., and Ruvkun, G. 2003. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 421: 268-272. [DOI] [PubMed] [Google Scholar]
- Boulton, S.J., Gartner, A., Reboul, J., Vaglio, P., Dyson, N., Hill, D.E., and Vidal, M. 2002. Combined functional genomic maps of the C. elegans DNA damage response. Science 295: 127-131. [DOI] [PubMed] [Google Scholar]
- Boutros, M., Kiger, A.A., Armknecht, S., Kerr, K., Hild, M., Koch, B., Haas, S.A., Consortium, H.F., Paro, R., and Perrimon, N. 2004. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science 303: 832-835. [DOI] [PubMed] [Google Scholar]
- Brenner, S. 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The C. elegans Sequencing Consortium. 1998. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science 282: 2012-2018. [DOI] [PubMed] [Google Scholar]
- Collins, F.S., Morgan, M., and Patrinos, A. 2003. The Human Genome Project: Lessons from large-scale biology. Science 300: 286-290. [DOI] [PubMed] [Google Scholar]
- Cutter, A.D., Payseur, B.A., Salcedo, T., Estes, A.M., Good, J.M., Wood, E., Hartl, T., Maughan, H., Strempel, J., Wang, B., et al. 2003. Molecular correlates of genes exhibiting RNAi phenotypes in Caenorhabditis elegans. Genome Res. 13: 2651-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dricot, A., Rual, J-F., Lamesch, P., Bertin, N., Dupuy, D., Hao, T., Lambert, C., Hallez, R., Delroisse, J-M., Vandenhaute, J., et al. 2004. Generation of the Brucella melitensis ORFeome version 1.1. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Dupuy, D., Li, Q-R., Deplancke, B., Boxem, M., Hao, T., Lamesch, P., Sequerra, R., Bosak, S., Doucette-Stamm, L., Hope, I.A., et al. 2004. A first version of the Caenorhabditis elegans promoterome. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., and Mello, C.C. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806-811. [DOI] [PubMed] [Google Scholar]
- Fraser, A.G., Kamath, R.S., Zipperlen, P., Martinez-Campos, M., Sohrmann, M., and Ahringer, J. 2000. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408: 325-330. [DOI] [PubMed] [Google Scholar]
- Gonczy, P., Echeverri, C., Oegema, K., Coulson, A., Jones, S.J., Copley, R.R., Duperon, J., Oegema, J., Brehm, M., Cassin, E., et al. 2000. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature 408: 331-336. [DOI] [PubMed] [Google Scholar]
- Gunsalus, K.C., Yueh, W.C., MacMenamin, P., and Piano, F. 2004. RNAiDB and PhenoBlast: Web tools for genome-wide phenotypic mapping projects. Nucleic Acids Res. 32: D406-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanazawa, M., Mochii, M., Ueno, N., Kohara, Y., and Lino, Y. 2001. Use of cDNA subtraction and RNA interference screens in combination reveals genes required for germ-line development in Caenorhabditis elegans. Proc. Natl. Acad. Sci. 98: 8686-8691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon, G.J. 2002. RNA interference. Nature 418: 244-251. [DOI] [PubMed] [Google Scholar]
- Harris, T.W., Chen, N., Cunningham, F., Tello-Ruiz, M., Antoshechkin, I., Bastiani, C., Bieri, T., Blasiar, D., Bradnam, K., Chan, J., et al. 2004. WormBase: A multi-species resource for nematode biology and genomics. Nucleic Acids Res. 32: D411-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley, J.L., Temple, G.F., and Brasch, M.A. 2000. DNA cloning using in vitro site-specific recombination. Genome Res. 10: 1788-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen, E.M. and Mango, S.E. 2002. The art and design of genetic screens: Caenorhabditis elegans. Nat. Rev. Genet. 3: 356-369. [DOI] [PubMed] [Google Scholar]
- Kamath, R.S., Fraser, A.G., Dong, Y., Poulin, G., Durbin, R., Gotta, M., Kanapin, A., Le Bot, N., Moreno, S., Sohrmann, M., et al. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231-237. [DOI] [PubMed] [Google Scholar]
- Lamesch, P., Milstein, S., Hao, T., Rosenberg, J., Li, N., Sequerra, R., Bosak, S., Doucette-Stamm, L., Vandenhaute, J., Hill, D.E., et al. 2004. C. elegans ORFeome Version 3.1: Increasing the coverage of ORFeome resources with improved gene predictions. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Lee, S.S., Lee, R.Y., Fraser, A.G., Kamath, R.S., Ahringer, J., and Ruvkun, G. 2003. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat. Genet. 33: 40-48. [DOI] [PubMed] [Google Scholar]
- Liu, Q., Li, M.Z., Leibham, D., Cortez, D., and Elledge, S.J. 1998. The univector plasmid-fusion system, a method for rapid construction of recombinant DNA without restriction enzymes. Curr. Biol. 8: 1300-1309. [DOI] [PubMed] [Google Scholar]
- Maeda, I., Kohara, Y., Yamamoto, M., and Sugimoto, A. 2001. Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr. Biol. 11: 171-176. [DOI] [PubMed] [Google Scholar]
- Parrish, S., Fleenor, J., Xu, S., Mello, C., and Fire, A. 2000. Functional anatomy of a dsRNA trigger: Differential requirement for the two trigger strands in RNA interference. Mol. Cell 6: 1077-1087. [DOI] [PubMed] [Google Scholar]
- Piano, F., Schetter, A.J., Mangone, M., Stein, L., and Kemphues, K.J. 2000. RNAi analysis of genes expressed in the ovary of Caenorhabditis elegans. Curr. Biol. 10: 1619-1622. [DOI] [PubMed] [Google Scholar]
- Piano, F., Schetter, A.J., Morton, D.G., Gunsalus, K.C., Reinke, V., Kim, S.K., and Kemphues, K.J. 2002. Gene clustering based on RNAi phenotypes of ovary-enriched genes in C. elegans. Curr. Biol. 12: 1959-1964. [DOI] [PubMed] [Google Scholar]
- Pothof, J., van Haaften, G., Thijssen, K., Kamath, R.S., Fraser, A.G., Ahringer, J., Plasterk, R.H., and Tijsterman, M. 2003. Identification of genes that protect the C. elegans genome against mutations by genome-wide RNAi. Genes & Dev. 17: 443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboul, J., Vaglio, P., Tzellas, N., Thierry-Mieg, N., Moore, T., Jackson, C., Shin-i, T., Kohara, Y., Thierry-Mieg, D., Thierry-Mieg, J., et al. 2001. Open-reading-frame sequence tags (OSTs) support the existence of at least 17,300 genes in C. elegans. Nat. Genet. 27: 332-336. [DOI] [PubMed] [Google Scholar]
- Reboul, J., Vaglio, P., Rual, J.F., Lamesch, P., Martinez, M., Armstrong, C.M., Li, S., Jacotot, L., Bertin, N., Janky, R., et al. 2003. C. elegans ORFeome version 1.1: Experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat. Genet. 34: 35-41. [DOI] [PubMed] [Google Scholar]
- Rual, J.F., Hill, D.E., and Vidal, M. 2004. ORFeome projects: Gateway between genomics and omics. Curr. Opin. Chem. Biol. 8: 20-25. [DOI] [PubMed] [Google Scholar]
- Simmer, F., Moorman, C., Van Der Linden, A.M., Kuijk, E., Van Den Berghe, P.V., Kamath, R., Fraser, A.G., Ahringer, J., and Plasterk, R.H. 2003. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 1: E12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabara, H., Grishok, A., and Mello, C.C. 1998. RNAi in C. elegans: Soaking in the genome sequence. Science 282: 430-431. [DOI] [PubMed] [Google Scholar]
- Tavernarakis, N., Wang, S.L., Dorovkov, M., Ryazanov, A., and Driscoll, M. 2000. Heritable and inducible genetic interference by double-stranded RNA encoded by transgenes. Nat. Genet. 24: 180-183. [DOI] [PubMed] [Google Scholar]
- Tewari, M., Hu, P.J., Ahn, J.S., Ayivi-Guedehoussou, N., Vidalain, P.O., Li, S., Milstein, S., Armstrong, C.M., Boxem, M., Butler, M.D., et al. 2004. Systematic interactome mapping and genetic perturbation analysis of a C. elegans TGF-β signaling network. Mol. Cell 13: 1-20. [DOI] [PubMed] [Google Scholar]
- Timmons, L. and Fire, A. 1998. Specific interference by ingested dsRNA. Nature 395: 854. [DOI] [PubMed] [Google Scholar]
- Vaglio, P., Lamesch, P., Reboul, J., Rual, J.F., Martinez, M., Hill, D., and Vidal, M. 2003. WorfDB: the Caenorhabditis elegans ORFeome database. Nucleic Acids Res. 31: 237-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw, N.L., Fischer, S.E., Robert, V.J., Thijssen, K.L., Fraser, A.G., Kamath, R.S., Ahringer, J., and Plasterk, R.H. 2003. A genome-wide screen identifies 27 genes involved in transposon silencing in C. elegans. Curr. Biol. 13: 1311-1316. [DOI] [PubMed] [Google Scholar]
- Walhout, A.J., Sordella, R., Lu, X., Hartley, J.L., Temple, G.F., Brasch, M.A., Thierry-Mieg, N., and Vidal, M. 2000a. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science 287: 116-122. [DOI] [PubMed] [Google Scholar]
- Walhout, A.J., Temple, G.F., Brasch, M.A., Hartley, J.L., Lorson, M.A., van den Heuvel, S., and Vidal, M. 2000b. GATEWAY recombinational cloning: Application to the cloning of large numbers of open reading frames, or ORFeomes. Methods Enzymol. 328: 575-592. [DOI] [PubMed] [Google Scholar]
- Zhang, P., Li, M.Z., and Elledge, S.J. 2002. Towards genetic genome projects: Genomic library screening and gene-targeting vector construction in a single step. Nat. Genet. 30: 31-39. [DOI] [PubMed] [Google Scholar]
- Zipperlen, P., Fraser, A.G., Kamath, R.S., Martinez-Campos, M., and Ahringer, J. 2001. Roles for 147 embryonic lethal genes on C. elegans chromosome I identified by RNA interference and video microscopy. EMBO J. 20: 3984-3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.wormbase.org; Source for C. elegans genome and RNAi data.
- http://worfdb.dfci.harvard.edu; Source for C. elegans ORFeome data.
- http://www.openbiosystems.com; Source for request of ORFeome-RNAi v1.1 clones.
- http://nematoda.bio.nyu.edu; Source for C. elegans RNAi data.
- http://www.rfcgr.mrc.ac.uk/geneservice; Source for request of ORFeome-RNAi v1.1 clones.