Abstract
An important aspect of the development of systems biology approaches in metazoans is the characterization of expression patterns of nearly all genes predicted from genome sequences. Such “localizome” maps should provide information on where (in what cells or tissues) and when (at what stage of development or under what conditions) genes are expressed. They should also indicate in what cellular compartments the corresponding proteins are localized. Caenorhabditis elegans is particularly suited for the development of a localizome map since all its 959 adult somatic cells can be visualized by microscopy, and its cell lineage has been completely described. Here we address one of the challenges of C. elegans localizome mapping projects: that of obtaining a genome-wide resource of C. elegans promoters needed to generate transgenic animals expressing localization markers such as the green fluorescent protein (GFP). To ensure high flexibility for future uses, we utilized the newly developed MultiSite Gateway system. We generated and validated “version 1.1” of the Promoterome: a resource of ∼6000 C. elegans promoters. These promoters can be transferred easily into various Gateway Destination vectors to drive expression of markers such as GFP, alone (promoter::GFP constructs), or in fusion with protein-encoding open reading frames available in ORFeome resources (promoter::ORF::GFP).
Biological processes result from complex networks of interactions between proteins, nucleic acids, and metabolites (Barabasi and Oltvai 2004). Although conventional approaches aimed at discovering the function of genes one-at-a-time have been successful at elucidating many biological principles, they are not well suited to unravel the structure, function, and dynamics of complex molecular networks. With the availability of (nearly) complete genome sequences, approaches have been developed recently to investigate the systems properties of biological networks (Ideker et al. 2001; Vidal 2001; Kitano 2002).
The C. elegans genome annotation provided the first draft of a comprehensive set of metazoan genes (The C. elegans Sequencing Consortium 1998). Subsequently, genome-wide expression profiling experiments (transcriptome mapping; Reinke et al. 2000; Kim et al. 2001), high-throughput (HT) RNAi analyses (phenome mapping; Fraser et al. 2000; Gonczy et al. 2000; Piano et al. 2000; Kamath et al. 2003; Simmer et al. 2003) and HT yeast two-hybrid projects (interactome mapping; Walhout et al. 2000a, 2002; Davy et al. 2001; Boulton et al. 2002; Reboul et al 2003; Li et al. 2004) were used to initiate studies of various biological processes in the worm at a systems level. However, the approaches mentioned above mainly probe the structure of the proteome network and the function of its components, without providing precise information on dynamic aspects. Thus additional information is needed about gene expression patterns at the cellular and/or subcellular levels (“localizome” mapping). Ultimately, localizome maps should provide information on where (in what cells or tissues) and when (at what stage(s) of development or under what conditions) genes are expressed. They should also indicate in what cellular compartments the corresponding proteins are localized.
Gene expression pattern information is being collected for multiple species, including human (http://bodymap.ims.utokyo.ac.jp/), mouse (http://genex.hgu.mrc.ac.uk/, http://www.informatics.jax.org/menus/expression_menu.shtml), Xenopus (http://www.dkfz-heidelberg.de/abt0135/axeldb.htm), and C. elegans (http://129.11.204.86:591/, http://nematode.lab.nig.ac.jp/db/index.html/). These projects employ different techniques, such as antibody staining, mRNA collection from specific tissues, reporter genes strategies and in situ mRNA localization to generate gene expression pattern data.
Each approach has specific advantages and disadvantages, and provides information on different aspects of gene expression. For example, antibody staining provides direct information about protein localization in vivo, but the time and cost involved in generating antibodies for each predicted protein currently precludes application of this technique on a proteome-wide scale. Expression patterns can also be determined by in situ hybridization against specific mRNAs, as currently employed for C. elegans (Tabara et al. 1996). The drawbacks of this technique include potentially high background levels and potential cross-hybridization to homologous mRNAs. Finally, expression patterns can be determined by generating transgenic animals carrying promoters fused in vitro to LacZ (Hope 1991) or GFP (Chalfie et al. 1994) reporter genes.
The GFP reporter strategy has important advantages for localizome studies. First, GFP expression can be examined in living animals, thus changes in GFP expression levels and patterns can be followed over time during development. Second, GFP can also be expressed as a fusion with an endogenous ORF to investigate subcellular localization in vivo. Such GFP-based localizome data have already proven extremely valuable in yeast (Kumar et al. 2002; Huh et al. 2003). Finally, once a GFP reporter strain has been created, it can be used repeatedly to examine changes in expression in response to external stimuli, pharmacological agents or alterations of gene function. For these reasons, we chose to utilize the GFP reporter strategy to generate expression pattern data for C. elegans. This approach requires the production of C. elegans transgenic strains expressing GFP under the control of a variety of worm promoters (Hope et al. 1998). Here we describe the cloning of the initial version of a C. elegans promoter resource or “promoterome,” and how this resource can be used to generate gene expression pattern and protein localization data.
Ideally, a promoterome resource should have many other downstream applications, such as the study of physical interactions between promoters and the transcription complexes that regulate them. A global map of protein-DNA interactions would greatly benefit our understanding of the regulatory mechanisms that give rise to the expression patterns observed in vivo. Approaches used to investigate protein-DNA interactions include the yeast one-hybrid system (Deplancke et al. 2004) and chromatin immunoprecipitation followed by DNA microarray analysis (ChIP-CHIP or chip2). The latter has been successfully used to generate a map of the yeast transcriptional regulatory network (Lee et al. 2002). The availability of a cloned C. elegans promoterome will facilitate the production of both yeast one-hybrid reporter constructs and promoters microarray.
To generate a promoterome resource that can be easily transferred to a variety of vectors we decided to use a novel implementation of the Gateway technology (Hartley et al. 2000; Walhout et al. 2000b). In addition to the versatility of the original Gateway recombinational cloning system, MultiSite Gateway (Cheo et al. 2004) allows the transfer of two or more DNA fragments from different Entry clones into a single Destination vector. Promoters are thus flanked by attL4 and attR1 sites, so that promoterome Entry clones can be cloned in fusion with ORFeome Entry clones, which are flanked by attL1 and attL2 recombination sites (Reboul et al. 2003). In summary, thanks to Multisite Gateway, both resources can be used either independently or together (Fig. 1).
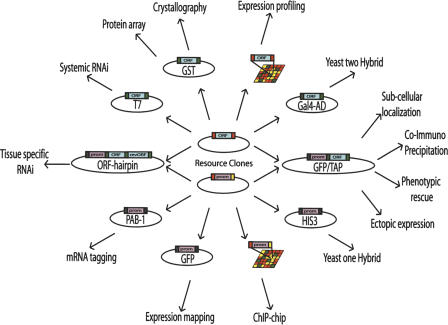
Figure 1.
Downstream application of cloned ORFeome and promoterome clones. MultiSite Gateway Recombinational cloning technology allows transfer of attL1-ORF-attL2 and attL4-promoter-attR1 Entry clones either independently, to attR1-ccdB-attR2 and attR4-ccdB-attL1 destination vectors respectively, or together into a single attR4-ccdB-attR2 destination vector, while preserving the reading frame and orientation. The availability of a cloned C. elegans “promoterome” and the corresponding “ORFeome” should facilitate a vast number of in vivo assays in the worm, such as ectopic expression, subcellular localization, tissue specific RNAi, and tissue specific rescue experiments. Green: attB site; red: attL site; yellow: attR site.
RESULTS
Use of MultiSite Gateway to Generate promoter::GFP and promoter::ORF::GFP Transgenic Worms
The flexibility of the MultiSite Gateway recombinational cloning system described in this issue (Cheo et al. 2004) allows HT generation of various types of gene fusion constructs needed for different genome-wide localizome projects. Since our goals are to generate and analyze both promoter::GFP and promoter::ORF::GFP fusions, we confirmed that promoter sequences flanked by the Gateway sites attB4 and attB1 (B4-promoter-B1-GPF-B2) can faithfully recapitulate known expression patterns (see also, Hope et al. 2004). Furthermore, we demonstrated that promoter fusions containing attB sites can appropriately drive the expression of proteins fused to GFP (B4-promoter-B1-ORF-B2-GFP).
We selected three genes with well-described expression patterns: myo-2 is expressed in pharyngeal muscular cells (Okkema et al. 1993); mec-7 in a subset of sensory neurons (touch cells; Hamelin et al. 1992); and hlh-8 in the M lineage and in the adult vulval muscle cells (Eimer et al. 2002). For each gene, we cloned into a MultiSite Entry vector a 2.5 kb DNA fragment located directly upstream of the predicted start codon. PCR amplicons of these “promoter” sequences were obtained using worm genomic DNA as template with primers flanked by attB sites (attB4 for the forward primer and attB1R for the reverse primer), and then cloned into the pDONR P4-P1R vector to generate attL4-promoterattR1 Entry clones. The promoters were subsequently transferred into pDEST-DD03 Destination vector to generate promoter::GFP fusions (see Methods). Transgenic lines were generated with each construct using micro particle bombardment (Praitis et al. 2001). The three resulting expression patterns matched previous observations obtained with clones not containing any Gateway sites (Fig. 2). We therefore concluded that the presence of attB sites in the final construction does not interfere noticeably with proper expression.
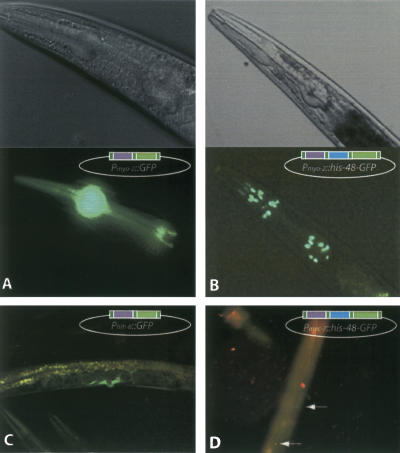
Figure 2.
GFP expression in transgenic worms generated with MultiSite Gateway constructed transgenes. (A) GFP expression observed with the Pmyo-2::GFP construct. (B) GFP expression observed with the Pmyo-2::his-48::GFP construct. (C) GFP expression observed with the Phlh-8::GFP construct. (D) GFP expression observed with the Pmec-7::his-48::GFP construct.
In addition, we established our ability to fuse promoter Entry clones with ORF Entry clones to analyze the subcellular localization patterns of protein::GFP fusion constructs. We produced six distinct promoter::ORF::GFP constructs by combining the mec-7 or myo-2 promoters with each of three ORFs (Y71H2B.4, T26A5.9, and B0035.8) from ORFeome v1.1. Out of 18 clones examined by restriction analysis (three for each construct), 15 displayed the correct fragment pattern on an agarose gel (not shown). One of each construct was sequenced and found to encode the expected ORF fused in frame to the GFP encoding sequence, downstream of the expected promoter.
To examine whether MultiSite-generated fusions recapitulate subcellular localization of the ORF, the two constructs harboring B0035.8, the histone H2B HIS-48 encoding ORF, were used to generate transgenic worms (Pmyo-2::his-48::GFP and Pmec-7::his-48::GFP). In both cases expression was observed in the expected cells, with fluorescence signal located in the nuclei of these cells as expected for a histone-GFP fusion protein (Fig. 2B,D). This experiment demonstrates the potential of combining the Gateway cloned promoters with ORFeome clones to produce promoter::ORF::reporter constructs that accurately reflect both gene expression pattern and subcellular localization of a protein.
Defining the C. elegans Promoterome
Promoters are commonly defined as DNA regions located upstream of transcription start sites and containing the necessary binding elements for proper transcriptional regulation. However, gene expression can be regulated by additional cis-acting elements, or enhancers, that can be located distantly upstream and/or in the introns, and/or downstream of genes (e.g., Conradt and Horvitz 1999). In the context of a genome-wide project it would not be possible to tackle at once all the complexity of gene regulation. We thus decided, as a first pass, to focus on the role of the proximal upstream regions in gene expression.
We decided to apply the definition that was used for S. cerevisiae promoters (Lee et al. 2002). For each gene, the “promoter” is defined here as the upstream intergenic region (IGR), specifically the region of DNA that spans from the ATG of the ORF to the extremity of the closest 5′ ORF (see Fig. 3A).
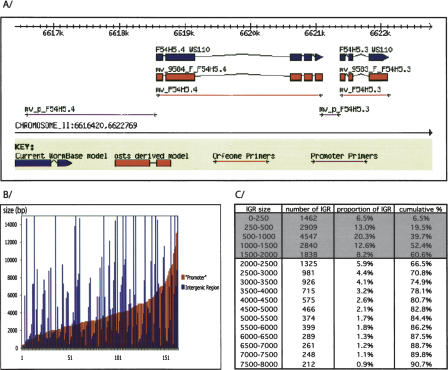
Figure 3.
Defining C. elegans promoterome. (A) Illustration of our Intergenic region (IGR) cloning strategy. Primers have been designed to amplify the entire IGR when smaller than 2 kb (F54H5.3), the first 2 kb of the IGR when bigger than 2 kb (F54H5.4), or 300 bp when smaller than 300 bp. (B) Analysis of previously published “promoters.” We randomly selected 200 publications containing a gene expression pattern determined by translational or transcriptional fusion. When available (164/200) we retrieved the length of the upstream DNA fragment (“promoter”) that was used to drive the observed expression. The sizes of the analyzed region range from 300 bp to 13 kb and mostly represent the distance to the closest convenient restriction site. In many cases a fraction of the IGR was successfully used. (C) IGR sizes in C. elegans genome. For each predicted ORF of the C. elegans genome (WS93) the IGR size was calculated as the distance between the ORF's predicted ATG and the closest extremity of the next upstream ORF. For each range of sizes the number of IGR in that range, its proportion in the genome and the cumulated proportion are indicated.
During the cloning of the ORFeome, we observed that cloning efficiency decreases as PCR fragment size increases above 2 kb (Reboul et al. 2001). Therefore, we set an upper size limit of 2 kb for the PCR fragments to ensure high cloning efficiency. This size restriction implies that regulatory elements located more than 2 kb upstream of the ATG will not be included.
We inspected ∼200 articles describing expression patterns obtained by a promoter::reporter-approach in C. elegans. The size of the promoter regions employed varied from a few hundred base pairs to several kilobasepairs and was often dictated by the location of convenient restriction sites used in cloning (Fig. 3B). We noted that many of the published expression patterns derived from small “promoters” (red bars in Fig. 3B) encompassing only a fraction of the actual IGR (blue bars in Fig. 3B), suggesting that the inclusion of the entire IGR is not always necessary to obtain specific expression. Moreover, sequence comparison between C. elegans and C. briggsae shows a dramatic decrease in the level of homology 1500 bp upstream of the predicted ATG for most genes with a long IGR (G. Achaz, pers. comm.). Thus our strategy should result in the inclusion of most upstream cis-regulatory elements.
Several predicted intergenic regions are extremely small (the smallest IGR is only 3 bp), prompting us to set a lower size limit of 300 bp. Overall, 60% of predicted IGRs are fully included within these limits (Fig. 3C). For each predicted gene in the C. elegans genome, we determined the IGR and designed PCR primers using the OSP primer prediction software (Hillier and Green 1991).
For the sake of simplicity we refer to the individual clones constituting the promoterome as “promoter Entry clones”, although they also include the 5′ untranslated region and they might not contain all relevant regulatory sites (Fig. 3A). As ORFeome clones do not contain the original start codon (Reboul et al. 2001), our promoter Entry clones also add a translation start codon to allow proper translation of promoter::ORF::reporter constructs.
For this project's design we used WormBase release WS93, which predicts 21,583 ORFs in the genome. This number includes internal splice variants, so the actual number of predicted start codons is 19,540. The number of actual promoters is further decreased by the presence of operons in the C. elegans genome, where a unique promoter drives the expression of several ORFs (Blumenthal et al. 2002; Lercher et al. 2003). The final count of unique promoters is 18,151. As a first pass we focused on cloning the promoters of the genes whose translation start prediction has: (1) not been questioned by the C. elegans ORFeome v1.1 (Reboul et al. 2003), and (2) not changed between WormPep versions WS9 (the reference for the ORFeome v1.1) and WS93. A total of 8964 genes fit these criteria, and the corresponding collection of cloned IGRs then constitutes the C. elegans promoterome version 1.1.
Generation of the C. elegans Promoterome 1.1
We PCR-amplified each target promoter from C. elegans genomics DNA, adding attB4 and attB1R Gateway recombination sites at their 5′ and 3′ end, respectively (Cheo et al. 2004). We then introduced the resulting PCR products into the donor vector pDONR P4-P1R (Invitrogen) using Gateway as described (Walhout et al. 2000b; Reboul et al. 2001). We successfully isolated a clone of the expected size for 6538 promoters (73%). As shown in Figure 4A, the initial PCR amplification gave a visible product 95% of the time. Five percent (5%) of the cloning failures can thus be attributed to the lack of genomic PCR amplification due either to failed primer synthesis or failed amplification under standard conditions. The remaining 22% of failures results from the cumulative loss at each step of the process (Gateway recombination, transformation and/or PCR verification, see Fig. 4B and Methods). Consistent with previous findings during the ORFeome project, the cloning efficiency inversely correlates with the size of the initial PCR product (Table 1). In our analysis, there was no size bias in the genomic PCR success rate or in the efficiency of transferring fragments from Entry clones to Destination vectors (not shown). We therefore concluded that an enhanced competitive advantage towards cloning primer-dimers was the cause of our lower success rate for fragments bigger than 1.5 kb (see Methods).
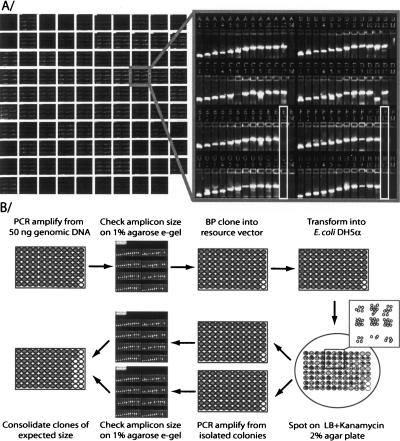
Figure 4.
Overview of promoterome cloning procedure. (A) Electrophoretic analysis of the genomic PCR products. The 8929 PCR reactions were analyzed on 1% agarose E-Gels. Ninety-five percent (95%) of the reactions gave visible PCR products of the expected size (primer pairs were arranged so that for each row the product size increases from 300 to 2000 bp). (B) Schematic representation of large-scale cloning of the C. elegans promoterome. All cloning steps are performed in a 96-well format. Clones of the expected size are consolidated using a Tecan Gemini robot, and stored both as miniprep DNA and bacterial glycerol stock.
Table 1.
HT Cloning Efficiency Is Related to the Size of the Target Fragment
| IGR size | Attempted | Cloned | Success rate |
|---|---|---|---|
| 0-500 | 1548 | 1454 | 94 |
| 500-1000 | 1901 | 1644 | 86 |
| 1000-1500 | 1214 | 909 | 75 |
| 1500-2000 | 791 | 474 | 60 |
| >2000 | 3510 | 2057 | 59 |
| Total | 8964 | 6538 | 73 |
The success rate of the cloning procedure was estimated based on the detection of the correct size insert in at least one of the two colonies examined.
The identity of each cloned promoter was established by sequencing. Of 6018 good-quality traces (Ewing et al. 1998), 5526 corresponded to the expected DNA fragment (92%). Most of the erroneous sequences corresponded to the Donor vector, which carries an ∼2-kb sized ccdB cassette that resembles a large promoter clone by electrophoretic analysis.
We developed a C. elegans promot erome database (PromoteromeDB; http://vidal.dfci.harvard.edu/promoteromedb) to integrate and disseminate the data collected in the promoterome-cloning project. The interface to this database displays IGR size, primer sequences, cloning and sequencing information, as well as links to corresponding entries in WorfDB (http://worfdb.dfci.harvard.edu; Vaglio et al. 2003) and WormBase (http://www.wormbase.org; Stein et al. 2001) for each promoter.
DISCUSSION
The first version of the promoterome generated here contains a third of the predicted C. elegans “promoters” in a cloning format that allows their transfer into various destination vectors in HT settings.
Through the production of promoter::reporter and promoter::ORF::reporter constructs the promoterome can be used to generate gene expression pattern and protein localization data on a genome-wide scale. However, to date, the generation of transgenic strains carrying low-copy integrated constructs is a very tedious and time-consuming process that constitutes the bottleneck of such a project. It will thus take a collaborative effort from the C. elegans community to generate gradually evolving versions of the worm localizome.
As mentioned earlier, the scale of the project described herein led us to disregard regulatory elements contained in introns and the 3′ UTR, or located far upstream of the transcription start. As a result, a fraction of the expression patterns derived from the promoterome may not accurately reflect the native expression pattern of the corresponding proteins as could be observed with antibodies. However, despite these limitations, the construction of a localizome data set should provide insights into the function of numerous otherwise uncharacterized genes (Werner 2003). Moreover, the generation of a standardized localizome map should allow integration of this map with other functional genomics data sets. For example, integrating the spatio-temporal component derived from localization studies with the interactome map (Li et al. 2004) should allow the generation of tissue-specific interaction maps that would be more relevant to biological processes. Indeed, for an interaction to occur in vivo both partners have to be present in the same cell/tissue and at the same time during development and, so far, the C. elegans protein-protein interaction map fails to take into account the complex manner in which gene expression is regulated.
Loss-of-function phenotypes obtained by RNAi are commonly utilized to infer the role of a given gene. Although this approach is very powerful, only 10.3% of the ORFs assayed gave rise to an observable phenotype in N2 animals; the use of the RNAi sensitive mutant strain rrf-3(pk1426) only increased this number to 12.8% (Simmer et al. 2003). The promoterome could be used to deliver hairpin RNAi constructs to specific tissues or cells (Wesley et al. 2001). This may facilitate the identification of phenotypes for genes or tissues that are refractory to traditional RNAi delivery.
The availability of the promoterome should also allow for the production of a wide variety of GFP reporter strains. These can be used to characterize phenotypes otherwise too subtle to be observed, as recently demonstrated in a screen for genes involved in C. elegans fat accumulation (Ashrafi et al. 2003).
Finally, the promoterome will constitute a tool to study transcriptional regulatory networks through protein-DNA interactions studies. Its potential to be applied in a large-scale yeast one-hybrid setting is demonstrated elsewhere in this issue (Deplancke et al. 2004). Overlapping the localizome map and the data from protein-DNA interaction maps should allow better understanding of the molecular mechanisms that drive development. The use of a common resource of promoters to generate both data sets should greatly facilitate their integration and specifically the identification of transcription-factor binding elements (TFBEs). The information gathered about TFBEs in the IGRs should allow the identification of TFBEs located in other regions such as the introns, 3′ untranslated regions or even within the ORFs themselves.
The promoterome cloning strategy described herein can be easily adapted to other organisms of interest provided that its annotated genome sequence is available. We believe that the combined use of ORFeomes and promoteromes resources will prove very powerful to unravel the complexity of gene regulatory networks in various model organisms.
METHODS
Large-Scale Cloning of Promoters
Individual PCR reactions were performed in a 50-μl reaction volume using 50 ng of wild-type (N2) C. elegans genomic DNA as template and one unit of High Fidelity Platinum Taq polymerase (Invitrogen). To facilitate the analysis of the size of PCR products, primers (Illumina) were organized in 96-well plates by increasing size of the IGR. PCR products were analyzed on 1% agarose E-Gels (Invitrogen; Fig. 4A). To clone the PCR products into the MultiSite Entry vector pDONR P4-P1R (Cheo et al. 2004), 2 μl of non-purified PCR product was transferred to 8 μl of BP reaction mix containing 100 ng of Entry vector, 0.5 μl of BP clonase and 2 μl of 5X BP reaction buffer. After overnight incubation at 25°C, 2 μl of BP reaction was transformed into 20 μl of E. coli DH5α thermocompetent cells (Inoue et al. 1990), using ABgene Thermo-Fast 96-well plates. Transformed cells were spotted on LB agar+kanamycin plates using a Genmate workstation (Tecan). In pilot experiments, we isolated eight colonies for each of 94 promoters, to evaluate our background level. We observed that 86.6% of promoters under 1500 bp display the expected size in at least six out of eight colonies. In contrast, bigger promoters could be split into two categories: 32% showed at least six out of eight correctly sized clones, while 68% showed less than two out of eight correct clones. In most cases the clones of the wrong size corresponded to primer-dimers. We therefore decided to isolate two colonies per clone, and select one that displayed the expected insert size by PCR. All clones displaying the correct insert size were rearrayed and archived as both bacterial glycerol stocks and plasmid DNA mini-preps (a schematic of the protocol is shown in Fig. 4B).
Using Multisite Gateway to Express GFP Constructs In Vivo
To generate promoter::GFP and promoter::ORF::GFP clones, we generated two MultiSite Gateway compatible destination vectors: pDEST-DD03 and pDEST-MB14. To generate pDEST-DD03, a HindIII-KpnI fragment containing the attR4-ccdB-attR2 cassette was moved from pDEST-6 (Invitrogen) into the unc-119 rescue vector pDP#MM016b (Praitis et al. 2001). To generate pDESTMB14, a KpnI-ApaI fragment containing the GFP encoding sequence and the unc-54 3′ UTR was moved from pPD95.79 (Fire et al. 1990) into pDP#MM016b. Next, an EcoRV fragment containing the attR4-ccdB-attR2 cassette was moved from pDEST-6 (Invitrogen) into this construct digested with Acc65I and made blunt by fill-in with Klenow. The unc-119 rescuing fragment present in these vectors can be used as a marker to select transgenic worms. Multi-Site LR reactions were performed in 10-μl volumes, but otherwise according to manufacturers specifications (Invitrogen). Transgenic worm-strains were generated using a ballistic transformation protocol as previously described (Praitis et al. 2001).
Acknowledgments
This work was funded by the National Cancer Institute (NCI 4 R33 CA097516-02 awarded to AJMW, IAH, and MV), the Leukemia Research Foundation (MB), the King Baudouin Foundation and the Belgian American Educational Foundation (Henri Benedictus-BAEF Fellowship in Biomedical Engineering awarded to BD), and the Fu Fellowship (QL). Thanks to Tracey Clingingsmith for her indispensable administrative assistance, John Nahory and Judith Hernandez from Illumina for their technical help.
Footnotes
[All clones in promoterome v1.1 are available to the scientific community through Open Biosystems (http://www.openbiosystems.com) and MRC geneservice (http://www.hgmp.mrc.ac.uk/geneservice).]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2497604.
References
- Ashrafi, K., Chang, F.Y., Watts, J.L., Fraser, A.G., Kamath, R.S., Ahringer, J., and Ruvkun, G. 2003. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 421: 268-272. [DOI] [PubMed] [Google Scholar]
- Barabasi, A.L. and Oltvai, Z.N. 2004. Network biology: Understanding the cell's functional organization. Nat. Rev. Genet. 5: 101-113. [DOI] [PubMed] [Google Scholar]
- Blumenthal, T., Evans, D., Link, C.D., Guffanti, A., Lawson, D., Thierry-Mieg, J., Thierry-Mieg, D., Chiu, W.L., Duke, K., Kiraly, M., et al. 2002. A global analysis of Caenorhabditis elegans operons. Nature 417: 851-854. [DOI] [PubMed] [Google Scholar]
- Boulton, S.J., Gartner, A., Reboul, J., Vaglio, P., Dyson, N., Hill, D.E., and Vidal, M. 2002. Combined functional genomic maps of the C. elegans DNA damage response. Science 295: 127-131. [DOI] [PubMed] [Google Scholar]
- The C. elegans Sequencing Consortium. 1998. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science 282: 2012-2018. [DOI] [PubMed] [Google Scholar]
- Chalfie, M., Tu, Y., Euskirchen, G., Ward, W.W., and Prasher, D.C. 1994. Green fluorescent protein as a marker for gene expression. Science 263: 802-805. [DOI] [PubMed] [Google Scholar]
- Cheo, D.L., Titus, S.A., Byrd, D.R.N., Hartley, J.L., Temple, G.F., and Brasch, M.A. 2004. Concerted assembly and cloning of mutiple DNA segments using in vitro site-specific recombination: Functional analysis of multi-segment expression clones. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Conradt, B. and Horvitz, H.R. 1999. The TRA-1A sex determination protein of C. elegans regulates sexually dimorphic cell deaths by repressing the egl-1 cell death activator gene. Cell 98: 317-327. [DOI] [PubMed] [Google Scholar]
- Davy, A., Bello, P., Thierry-Mieg, N., Vaglio, P., Hitti, J., Doucette-Stamm, L., Thierry-Mieg, D., Reboul, J., Boulton, S., Walhout, A.J., et al. 2001. A protein-protein interaction map of the Caenorhabditis elegans 26S proteasome. EMBO Rep. 2: 821-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplancke, B., Dupuy, D., Vidal, M., and Walhout, A.J.M. 2004. A Gateway-compatible yeast one-hybrid system. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Eimer, S., Donhauser, R., and Baumeister, R. 2002. The Caenorhabditis elegans presenilin sel-12 is required for mesodermal patterning and muscle function. Dev. Biol. 251: 178-192. [DOI] [PubMed] [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8: 175-185. [DOI] [PubMed] [Google Scholar]
- Fire, A., Kondo, K., and Waterston, R. 1990. Vectors for low copy transformation of C. elegans. Nucleic Acids Res. 18: 4269-4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser, A.G., Kamath, R.S., Zipperlen, P., Martinez-Campos, M., Sohrmann, M., and Ahringer, J. 2000. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408: 325-330. [DOI] [PubMed] [Google Scholar]
- Gonczy, P., Echeverri, G., Oegema, K., Coulson, A., Jones, S.J., Copley, R.R., Duperon, J., Oegema, J., Brehm, M., Cassin, E., et al. 2000. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature 408: 331-336. [DOI] [PubMed] [Google Scholar]
- Hamelin, M., Scott, I.M., Way, J.C., and Culotti, J.G. 1992. The mec-7 β-tubulin gene of Caenorhabditis elegans is expressed primarily in the touch receptor neurons. EMBO J. 11: 2885-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley, J.L., Temple, G.F., and Brasch, M.A. 2000. DNA cloning using in vitro site-specific recombination. Genome Res. 10: 1788-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier, L. and Green, P. 1991. OSP: A computer program for choosing PCR and DNA sequencing primers. PCR Methods Appl. 1: 124-128. [DOI] [PubMed] [Google Scholar]
- Hope, I.A. 1991. `Promoter trapping' in Caenorhabditis elegans. Development 113: 399-408. [DOI] [PubMed] [Google Scholar]
- Hope, I.A., Arnold, J.M., McCarroll, D., Jun, G., Krupa, A.P., and Herbert, R. 1998. Promoter trapping identifies real genes in C. elegans. Mol. Gen. Genet. 260: 300-308. [DOI] [PubMed] [Google Scholar]
- Hope, I.A., Stevens, J., Garner, A., Hayes, J., Cheo, D.L., Brasch, M.A., and Vidal, M. 2004. Feasibility of genome-scale construction of promoter::reporter gene fusions for expression in Caenorhabditis elegans using a Multisite Gateway recombination system. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Huh, W.K., Falvo, J.V., Gerke, L.C., Carroll, A.S., Howson, R.W., Weissman, J.S., and O'Shea, E.K. 2003. Global analysis of protein localization in budding yeast. Nature 425: 686-691. [DOI] [PubMed] [Google Scholar]
- Ideker, T., Galitski, T., and Hood, L. 2001. A new approach to decoding life: Systems biology. Annu. Rev. Genomics Hum. Genet. 2: 343-372. [DOI] [PubMed] [Google Scholar]
- Inoue, H., Nojima, H., and Okayama, H. 1990. High efficiency transformation of Escherichia coli with plasmids. Gene 96: 23-28. [DOI] [PubMed] [Google Scholar]
- Kamath, R.S., Fraser, A.G., Dong, Y., Poulin, G., Durbin, R., Gotta, M., Kanapin, A., Le Bot, N., Moreno, S., Sohrmann, M., et al. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231-237. [DOI] [PubMed] [Google Scholar]
- Kim, S.K., Lund, J., Kiraly, M., Duke, K., Jiang, M., Stuart, J.M., Eizinger, A., Wylie, B.N., and Davidson, G.S. 2001. A gene expression map for Caenorhabditis elegans. Science 293: 2087-2092. [DOI] [PubMed] [Google Scholar]
- Kitano, H. 2002. Looking beyond the details: A rise in system-oriented approaches in genetics and molecular biology. Curr. Genet. 41: 1-10. [DOI] [PubMed] [Google Scholar]
- Kumar, A., Agarwal, S., Heyman, J.A., Matson, S., Heidtman, M., Piccirillo, S., Umansky, L., Drawid, A., Jansen, R., Liu, Y., et al. 2002. Subcellular localization of the yeast proteome. Genes & Dev. 16: 707-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, T.I., Rinaldi, N.J., Robert, F., Odom, D.T., Bar-Joseph, Z., Gerber, G.K., Hannett, N.M., Harbison, C.T., Thompson, C.M., Simon, I., et al. 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298: 799-804. [DOI] [PubMed] [Google Scholar]
- Lercher, M.J., Blumenthal, T., and Hurst, L.D. 2003. Coexpression of neighboring genes in Caenorhabditis elegans is mostly due to operons and duplicate genes. Genome Res. 13: 238-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., Armstrong, C.M., Bertin, N., Ge, H., Milstein, S., Boxem, M., Vidalain, P.O., Han, J.D., Chesneau, A., Hao, T., et al. 2004. A map of the interactome network of the metazoan C. elegans. Science 303: 540-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkema, P.G., Harrison, S.W., Plunger, V., Aryana, A., and Fire, A. 1993. Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics 135: 385-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piano, F., Schetter, A.J., Mangone, M., Stein, L., and Kemphues, K.J. 2000. RNAi analysis of genes expressed in the ovary of Caenorhabditis elegans. Curr. Biol. 10: 1619-1622. [DOI] [PubMed] [Google Scholar]
- Praitis, V., Casey, E., Collar, D., and Austin, J. 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157: 1217-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboul, J., Vaglio, P., Tzellas, N., Thierry-Mieg, N., Moore, T., Jackson, C., Shin-i, T., Kohara, Y., Thierry-Mieg, D., Thierry-Mieg, J., et al. 2001. Open-reading-frame sequence tags (OSTs) support the existence of at least 17,300 genes in C. elegans. Nat. Genet. 27: 332-336. [DOI] [PubMed] [Google Scholar]
- Reboul, J., Vaglio, P., Rual, J.F., Lamesch, P., Martinez, M., Armstrong, C.M., Li, S., Jacotot, L., Bertin, N., Janky, R., et al. 2003. C. elegans ORFeome version 1.1: Experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat. Genet. 34: 35-41. [DOI] [PubMed] [Google Scholar]
- Reinke, V., Smith, H.E., Nance, J., Wang, J., Van Doren, C., Begley, R., Jones, S.J., Davis, E.B., Scherer, S., Ward, S., et al. 2000. A global profile of germline gene expression in C. elegans. Mol. Cell. 6: 605-616. [DOI] [PubMed] [Google Scholar]
- Simmer, F., Moorman, C., Van Der Linden, A.M., Kuijk, E., Van Den Berghe, P.V., Kamath, R., Fraser, A.G., Ahringer, J., and Plasterk, R.H. 2003. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 1: E12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, L., Sternberg, P., Durbin, R., Thierry-Mieg, J., and Spieth, J. 2001. WormBase: Network access to the genome and biology of Caenorhabditis elegans. Nucleic Acids Res. 29: 82-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabara, H., Motohashi, T., and Kohara, Y. 1996. A multi-well version of in situ hybridization on whole mount embryos of Caenorhabditis elegans. Nucleic Acids Res. 24: 2119-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaglio, P., Lamesch, P., Reboul, J., Rual, J.F., Martinez, M., Hill, D., and Vidal, M. 2003. WorfDB: The Caenorhabditis elegans ORFeome database. Nucleic Acids Res. 31: 237-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal, M. 2001. A biological atlas of functional maps. Cell 104: 333-339. [DOI] [PubMed] [Google Scholar]
- Walhout, A.J., Sordella, R., Lu, X., Hartley, J.L., Temple, G.F., Brasch, M.A., Thierry-Mieg, N., and Vidal, M. 2000a. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science 287: 116-122. [DOI] [PubMed] [Google Scholar]
- Walhout, A.J., Temple, G.F., Brasch, M.A., Hartley, J.L., Lorson, M.A., van den Heuvel, S., and Vidal, M. 2000b. GATEWAY recombinational cloning: Application to the cloning of large numbers of open reading frames, or ORFeomes. Methods Enzymol. 328: 575-592. [DOI] [PubMed] [Google Scholar]
- Walhout, A.J., Reboul, J., Shtanko, O., Bertin, N., Vaglio, P., Ge, H., Lee, H., Doucette-Stamm, L., Gunsalus, K.C., Schetter, A.J., et al. 2002. Integrating interactome, phenome, and transcriptome mapping data for the C. elegans germline. Curr. Biol. 12: 1952-1958. [DOI] [PubMed] [Google Scholar]
- Werner, T. 2003. Promoters can contribute to the elucidation of protein function. Trends Biotechnol. 21: 9-13. [DOI] [PubMed] [Google Scholar]
- Wesley, S.V., Helliwell, C.A., Smith, N.A., Wang, M.B., Rouse, D.T., Liu, Q., Gooding, P.S., Singh, S.P., Abbott, D., Stoutjesdijk, P.A., et al. 2001. Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27: 581-590. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://bodymap.ims.u-tokyo.ac.jp/; Bodymap Human and Mouse Gene Expression Database. [DOI] [PMC free article] [PubMed]
- http://genex.hgu.mrc.ac.uk; EMAP, the Edinburgh Mouse Atlas Project.
- http://www.informatics.jax.org/menus/expression_menu.shtml; MouseGenome Informatics.
- http://www.dkfz-heidelberg.de/abt0135/axeldb.htm; A Xenopus laevis database.
- http://129.11.204.86:591; The Hope Laboratory Expression Pattern Database.
- http://nematode.lab.nig.ac.jp/db/index.html; NEXTDB, the Nematode Expression Pattern Database.
- http://worfdb.dfci.harvard.edu/; WorfDB, the C. elegans ORFeome Cloning Project Database.
- http://www.wormbase.org/; WormBase Web site.
- http://www.hgmp.mrc.ac.uk/geneservice; MRC geneservice.
- http://www.openbiosystems.com; Open Biosystems.
- http://vidal.dfci.harvard.edu/promoteromedb; PromoteromeDB the C. elegans Promoterome Database.