Abstract
Microarray transcript profiling and RNA interference are two new technologies crucial for large-scale gene function studies in multicellular eukaryotes. Both rely on sequence-specific hybridization between complementary nucleic acid strands, inciting us to create a collection of gene-specific sequence tags (GSTs) representing at least 21,500 Arabidopsis genes and which are compatible with both approaches. The GSTs were carefully selected to ensure that each of them shared no significant similarity with any other region in the Arabidopsis genome. They were synthesized by PCR amplification from genomic DNA. Spotted microarrays fabricated from the GSTs show good dynamic range, specificity, and sensitivity in transcript profiling experiments. The GSTs have also been transferred to bacterial plasmid vectors via recombinational cloning protocols. These cloned GSTs constitute the ideal starting point for a variety of functional approaches, including reverse genetics. We have subcloned GSTs on a large scale into vectors designed for gene silencing in plant cells. We show that in planta expression of GST hairpin RNA results in the expected phenotypes in silenced Arabidopsis lines. These versatile GST resources provide novel and powerful tools for functional genomics.
Exhaustive gene lists are now available for numerous species including several multicellular eukaryotes, and biological research is moving from the study of single genes toward the parallel study of many genes, taking advantage of genome sequences for large-scale functional surveys. Two key methods have greatly enhanced our ability to assign functions to genes in systematic analyses: microarray transcript profiling (Holloway et al. 2002) and silencing via RNA interference or RNAi (Hannon 2002; Waterhouse and Helliwell 2003). Full transcriptome microarrays give information on when, where, and at what level each and every gene in an organism is expressed. RNAi knockdown lines give information on gene function by showing the effects of a drastic reduction in expression of the target gene. Both methods exploit the molecular hybridization of complementary nucleic acid strands either in vitro, in the case of microarrays, to quantify the level of transcripts in a given RNA sample or in vivo, in the case of RNAi, to target the degradation of transcripts by the Dicer/RISK pathway.
The choice of appropriate sequences, with which to construct these tools, is complicated by the significant similarity present within gene families. It is important for each probe to be specific to a cognate target gene. Stringent criteria must be implemented to restrict, as far as possible, the hybridization between related but different nucleic acid segments. Bioinformatics tools are available for the design of such probes either synthesized as oligonucleotides (e.g., Li and Stormo 2001; Chen and Sharp 2002; Rouillard et al. 2002) or as PCR-amplified DNA segments (Varotto et al. 2001; Xu et al. 2002; Nielsen et al. 2003; Thareau et al. 2003). These tools are particularly relevant for model species characterized by a high level of genetic redundancy. As in many other land plants, the Arabidopsis genome has undergone multiple rounds of polyploidization and gene amplification events leading to extensive duplication of large regions and many small tandem duplications. Consequently, two-thirds of its nuclear genes belong to gene families and, out of these, one-third to families with more than five members (The Arabidopsis Genome Initiative 2000; Simillion et al. 2002).
But probe design is only the first step in functional genomics initiatives, and the production of probe repertoires (oligonucleotide, cDNA, or genomic DNA fragments) required for microarrays or RNAi is cumbersome and expensive. Ideally, this type of reference material should be formatted as robust and versatile resources, readily available to the research community. Using these criteria as a standard, we have built a collection of Arabidopsis gene-specific sequence tags (GSTs). It was created in the context of the Complete Arabidopsis Transcript MicroArray (CATMA) initiative, combining the efforts of laboratories in eight European countries (Hilson et al. 2003). Here, we describe the design and synthesis of GSTs representing at least 21,500 protein-encoding genes. Each GST is a segment of genomic DNA derived from the corresponding gene and is selected to avoid cross-hybridization with other sequences. The format of the GST collection has been optimized to facilitate its subsequent amplification and dissemination and to permit inexpensive quality controls. The CATMA GSTs constitute an excellent source of probes for the production of spotted microarrays. In an extension of this concept, the GSTs have also been cloned in bacterial plasmids and are presently being used for the systematic knockdown of Arabidopsis genes by RNAi. We present here the first results of transcript profiling and reverse genetics applications with the Arabidopsis GSTs to illustrate the potential of this resource for the functional analysis of genes in this model species.
RESULTS
Design
The GSTs were selected with the Specific Primer and Amplicon Design Software, SPADS (Thareau et al. 2003), based on gene models extracted from the structural annotation of the Arabidopsis thaliana nuclear genome (January 2001) and using the eukaryotic gene finder EuGene (Schiex et al. 2001). The results of this fully automated structural annotation tool are available online (http://www.psb.ugent.be/bioinformatics/genomes_ath_index.php).
The SPADS GST selection process can be divided into four steps. Firstly, the software searched for divergent regions. For each gene model, exons were sequentially matched using BLASTn (Altschul et al. 1997) against the entire genome sequence and segments with significant homology hits were discarded. Secondly, the Primer3 software (Rozen and Skaletsky 2000) selected PCR oligonucleotide pairs in the remaining divergent exonic regions. Thirdly, these oligonucleotide pairs were filtered using BLASTn to eliminate any sequences with significant matches in the surrounding window (minimum 400 kb). This filter diminished the risk of producing spurious PCR products carrying nontarget portions of the Arabidopsis genome. Finally, the remaining potential amplicons were matched using BLASTn against the full genome sequence to determine the presence of potential paralogous genes: for every gene model, SPADS scanned sequentially each potential amplicon, starting with 3′-sequences, until one was identified that met the chosen constraints. For this study, the GST length ranges between 150 and 500 bp, large enough to be purified from contaminants such as PCR primers or primer-dimer PCR artifacts, but small enough to be amplified very efficiently and to retain a sufficient level of specificity. The selected GSTs share no more than 70% identity with any other sequence in the nuclear genome, because above that threshold cross-hybridization is detected in microarray experiments (Richmond et al. 1999; Girke et al. 2000). For other parameters in SPADS, Primer3 and BLASTn used in this study, see Methods and Thareau et al. (2003).
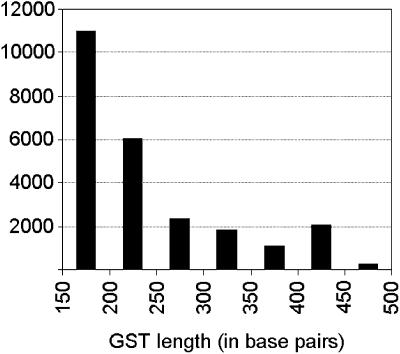
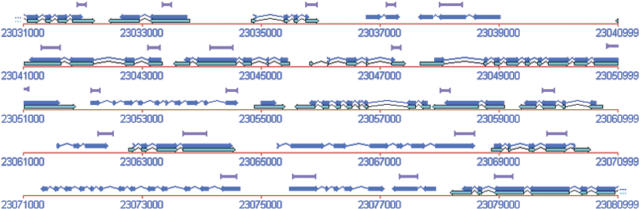
The SPADS algorithm was applied to 29,787 gene models of which 20,981 (70%) resulted in an in silico designed GST. The majority of sequences are in the lower portion of the length spectrum (Fig. 1). Starting the selection from the 3′-end of genes resulted in a distribution of 24%, 15%, and 61% of the probes in the 5′, central, and 3′ third of the gene models, respectively. Although all primer pairs delimiting the GST sequences lie in annotated or predicted exons, SPADS may also select GST sequences overlapping with intron(s). In this study, 606 (2.9%) GSTs were designed with intron sequences, whereas they still contained at least 150 bp matching exons and less than half of the GST matching intron sequence. In addition to GSTs extracted from annotated protein-encoding genes, 139 tags were picked from intergenic regions distributed across the five chromosomes. These intergenic tags can serve as negative internal controls in microarray transcript profiling experiments. An example of the distribution of GST sequences along the annotated genome sequence is shown in Figure 2.
Figure 1.
GST length distribution. The calculated length does not include the extensions added to the gene-specific portion of the tags.
Figure 2.
Graphical representation of GST distribution. The represented genomic segment is 50 kb long and centered around gene At5g57890. Light blue and dark blue boxes represent AGI mRNA and coding sequences, respectively. GSTs are shown as purple brackets. The display was extracted from a screenshot of the FLAGdb++ Arabidopsis genome database (Samson et al. 2002).
Detailed GST information is available from a database at http://www.catma.org (Crowe et al. 2003). For each GST, the CATMA database provides the sequences and corresponding PCR amplification oligonucleotides, their length, genome location, GC%, the GST primary amplification results (with associated gel images), the gene model from which each GST is derived and additional technical information. It also shows links between GSTs and corresponding Arabidopsis Genome Initiative (AGI) gene names together with the Gene Ontology (GO) information about gene function. GST information will also be available via other Arabidopsis resource Web sites (see Web Site References).
The quality of the Arabidopsis genome annotation improves continuously thanks to the refinement of in silico prediction tools and to the increasing number of experimentally documented transcription units (Haas et al. 2002; Seki et al. 2002; Yamada et al. 2003; Castelli et al. 2004). The GST collection is being enlarged incrementally to take into consideration annotation updates. The first version described above, CATMA v1, contained 21,120 in silico designed tags and was based exclusively on gene models from the 2001 EuGene genome annotation (http://www.psb.ugent.be/bioinformatics/genomes_ath_index.php). The second version, CATMA v2, contains an additional 3456 in silico tags (24,576 in total). These new tags were selected according to alternative criteria: (1) the minimum primer GC% was lowered from 40% to 30%; (2) gene models only defined by a coding sequence and that failed to yield a tag in v1 were extended with the 150-bp segment immediately following their stop codon; and (3) AGI gene models located in regions in which no genes were predicted by EuGene were included. A third additional GST set is currently in preparation.
The overlap between CATMA v2 and the latest AGI annotation performed by The Institute for Genomic Research (TIGR; release 5.0) was calculated by searching all GST sequences that aligned over a segment of at least 150 bp with at most two mismatches with the gene models including untranslated regions and introns. Out of 29,993 AGI-annotated protein-encoding genes, including sequences labeled as pseudogenes but not taking into account alternative splicing variants, 21,566 matched at least one GST, and 2173 genes more than one GST. Conversely, out of 24,576 GSTs, including controls, 23,280 match at least one AGI gene.
All the data presented below on microarray transcript profiling and large-scale cloning relate to the CATMA v1 collection. All functional and structural annotation refers to the TIGR release 5.0 genome annotation.
Synthesis
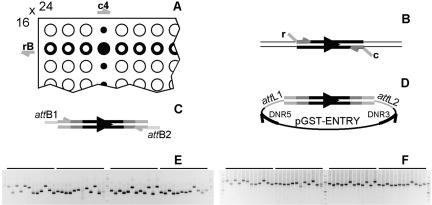
The GSTs were PCR-amplified from genomic DNA. The gene-specific oligonucleotides used for PCR synthesis each contained a 17-nt 5′-extension added to the gene-specific portion selected by SPADS. In total, 40 arbitrary, unique extension sequences had been assigned to the 24 columns (c1-c24) and 16 rows (rA-rP) of a 384 multiwell plate (Fig. 3A; Supplemental Table S1). The column and row extensions were located upstream and downstream, respectively, of the tag sequence with respect to the direction of transcription (Fig. 3B). Based on this structured arrangement, the secondary amplification of the entire GST collection was performed with only 40 oligonucleotides, each carrying the appropriate selective extension at their 3′-end. Well-to-well cross-contaminations that commonly plague cDNA collections maintained for printing microarrays (Knight 2001) are thus resolved in secondary PCR with the set of 40 extension primers because each GST in a multiwell plate is amplified by a unique combination of one column primer and one row primer, depending on its position in that plate (e.g., c4 and rB in Fig. 3A).
Figure 3.
GST amplicon structure. (A) Schematic representation of the GST format, with the 40 extensions allocated to the 24 columns, c, and 16 rows, r, in a 384-multiwell plate. (B) Primary amplification of a GST (thick double line with large arrow) from genomic DNA (thin double line) with a pair of primers each containing a gene-specific portion (horizontally striped) and 5′-extension (gray). The large arrow indicates the orientation of the GST with respect to the direction of transcription. Elements are not drawn to scale. (C) Secondary amplification of a GST with PCR addition of Gateway attB1 and attB2 sites. (D) Structure of the GST entry clone after Gateway recombinational cloning of the amplicon in pDONR207. (E) Secondary amplification results illustrated for 48 GSTs (see Supplemental Table S2). (F) Tertiary PCR validation of BP reactions. GST insert size verification is carried out by amplification of the attL1-GST-attL2 cassette with the DNR5 and DNR3 primers and results in the synthesis of a DNA fragment carrying 167 and 487 bp beyond the original 5′- and 3′-extensions of the primary GST, respectively (see Supplemental Table S2).
Individual bacterial artificial chromosomes (BACs) were chosen as templates for the primary PCR amplification of the GSTs, wherever possible. Reducing the template complexity by three orders of magnitude when compared with the Arabidopsis nuclear genome increased DNA yield and diminished the synthesis of spurious products. For this purpose, we used IGF (Mozo et al. 1999) and TAMU (Choi et al. 1995) BAC clones for which bacterial strains were available, together with complete or end sequence information. Out of the 24,576 GSTs in CATMA v2, 22,614 (92.0%) were amplified from BAC templates. Besides faulty primers, failures probably represent incomplete BAC genome coverage, incorrect mapping of BAC clones on reconstructed chromosome sequences, BAC instability, or faulty BAC tracking. The 1013 remaining successful GSTs (4.1%) were amplified from purified genomic DNA (Col-4 ecotype). In total, 20,388 (96.1%) GSTs were produced as determined by the detection of a single PCR product of the expected size in agarose gel electrophoresis. All primary amplification results including gel images can be studied online via the CATMA database. As previously described (Thareau et al. 2003), a random subset of 956 (3.9%) GST amplicons were sequence-validated (579 amplified from BACs and 377 amplified from total DNA template) as an additional quality control. In all cases, the sequence determined experimentally is identical to the expected GST, indicating that the collection is of very high quality. The sequence validation data are also available in the CATMA database.
Transcript Profiling With CATMA v1 Arrays: Performance Study
GSTs can easily be synthesized in large amounts using small aliquots of the diluted primary amplicons as templates, then purified and printed on glass microarrays. Platforms using the GST collection for microarray production recovered 94.3% to 98.3% of the amplicons in secondary PCRs. We investigated the performance of arrays printed with CATMA v1 GSTs in a control experiment and in a biological experiment assessing differential expression. The first experiment focuses on the dynamic range, sensitivity, and specificity of glass microarrays printed with Arabidopsis GSTs in a series of spiked hybridizations.
The objective was to assemble a series of targets for hybridization, spiked with in vitro synthesized and purified polyadenylated RNAs, called spike RNA, covering a range of biologically relevant concentrations. The experimental design and the subsequent interpretation of the results are based on the assumption that polyadenylated messenger RNAs constitute 1% of total RNA, that a cell contains on average 300,000 transcripts, and that the average transcript length is 1500 nt.
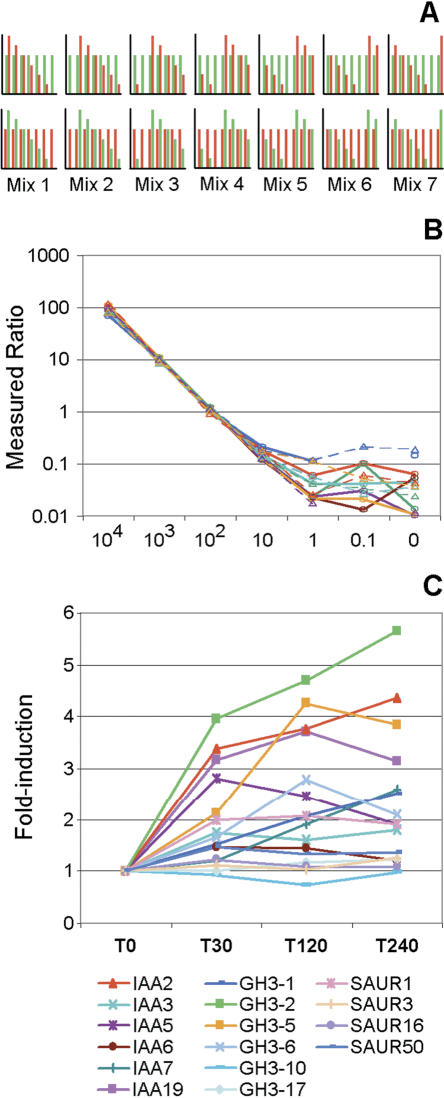
All hybridizations were performed with the same RNA extracted from aerial parts of germinating Arabidopsis seedlings and completed with various amounts of spike RNAs. They were chosen based on the following criteria: (1) the corresponding cDNA clones were available, and (2) their expression had not been previously detected in shoot material. A total of 14 spike RNAs were reverse-transcribed in vitro, although one of them failed quality controls and was removed from further analysis (Supplemental Table S3). The spike RNAs were added to the shoot total RNA to generate seven RNA mixes containing 104, 103, 102, 10, 1, 0.1 or zero copies per cell of each (Fig. 4A). The spike concentrations were staggered to obtain in each of the scaled RNAs the same total amount of spike RNAs in a weight ratio of 1/2520 versus shoot total RNA. The spike RNAs were present in seven pairs that followed the same concentration scale in the RNA mixes (each pair is represented as a separate bar in the 14 graphs; Fig. 4A). They were compared with a unique reference RNA, with all spike RNA species at a concentration of 100 copies per cell, in a total of 14 hybridizations including both dye-swap configurations (Fig. 4A).
Figure 4.
Microarray results. (A) Spiking experiment: Schematic representation of the hybridization series. Each graph shows the spike mRNA content of one microarray hybridization, both in the Cy3 (green) and Cy5 (red) labeled RNA mix. Each bar in a graph represents the concentration of a pair of spike RNAs in an RNA mix (Supplemental Table S3). The numbers indicate the seven scaled RNA mixes containing staggered spike RNAs. The two series of seven graphs represent the reciprocal dye-swap hybridization series in which the reference RNA, containing each of the spike RNA at 100 copies per cell, is labeled either with the Cy3 (top) or Cy5 (bottom) fluorophore. (B) Spiking experiment: Average signal ratios for 13 GSTs corresponding to the spike RNAs across the dilution range expressed as the number of copies per cell. Ratios are represented left to right from the highest to the lowest concentration for all spike RNAs in the seven RNA mixes. As expected, spike RNAs at a 100 copy-per-cell concentration yield a ratio of 1. (C) IAA treatment experiment: Fold induction of known auxin-inducible genes in the Aux/IAA, GH3, and SAUR families.
The dynamic range of the spike RNA signal ratios detected by the GSTs covered three to four logs in close-to-perfect linear dose-response curves. The sensitivity reached in this set of hybridizations was well below 10 copies per cell (Fig. 4B). However, the limit of sensitivity could not be calculated exactly because some of the genes coding for the spike RNAs might have been transcribed at low levels in the shoot sample. Probe specificity was investigated by studying whether the increase in spike RNA concentration significantly drove up the signal associated with GSTs other than the cognate probes. Such patterns could not be detected for any of the 14 genes tested, even though a spike RNA at its highest concentration represented an estimated 3% of the mRNA pool. BLAST searches indicated that the closest nonidentical GSTs that matched spike RNA sequences shared segments only of up to 17 ungapped nucleotides or E-values as low as 2.4 × 10-13. Taking into consideration the entire microarray data set, no correlation was observed between GST length and signal (data not shown).
Overall, these results indicate that CATMA arrays provide an excellent dynamic range, a sensitivity that is comparable to the performance of other platforms in routine experiments and a good specificity of the signal, not confounded by cross-hybridization.
Transcript Profiling With CATMA v1 Arrays: A Biological Test Case
To demonstrate the analysis of differential gene expression based on CATMA v1 arrays, we studied the changes induced by the treatment of Arabidopsis seedlings with the phytohormone indole-3-acetic acid (IAA) at physiological concentration. Seedlings were germinated and grown in liquid medium, then treated with the addition of IAA at 1 μM, 10 d after germination for zero, 30, 120, and 240 min. In total, nine microarray hybridizations were performed according to a complete experimental loop design (Supplemental Fig. S1). The statistical analysis of the hybridization data indicated that 1123 GSTs showed significant expression differences (p < 2.37 × 10-6) between time points following treatment (Supplemental Table S4). The transcript profiles of well-documented auxin-responsive genes were surveyed. Fold-induction was analyzed for members of the Aux/IAA, SAUR, and GH3 gene families represented in the array (Hagen and Guilfoyle 2002; Liscum and Reed 2002). Among these 60 genes, 16 appeared as differentially expressed, and all but one had an upward trend (Fig. 4C), confirming the transcriptional up-regulation of the three selected gene families upon application of exogenous IAA, even at the 1 μM level.
In addition, we investigated whether the global interpretation of the transcript profiles provided relevant information about the biological mechanisms at play in response to the auxin treatment. For that purpose, clusters of genes were constructed that correlated with theoretical up- or down-regulation patterns. Certain biological themes associated with these genes and defined in the controlled GO vocabulary (Rhee et al. 2003) were significantly overrepresented (Table 1): among them, as expected, the GO term “response to auxin stimuli,” but also categories indicative of an increase in protein synthesis characterized by up-regulated genes in the later time points, and of modification in energy metabolism characterized by down-regulated genes. Similarly, root segments in which lateral root formation was induced synchronously by treatment with an auxin transport inhibitor (N-1-naphthylphthalamic acid [NPA]) followed by transfer to growth medium containing auxin 1-naphthalene acetic acid (NAA) had an increased expression of genes involved in translation initiation, ribosome biogenesis, and assembly within 4 h following the transfer (Himanen et al. 2003).
Table 1.
Functional Characterization of Gene Clusters
| GO term | EASE score | Gene symbol |
|---|---|---|
| Cluster A. Up and early genes | ||
| BP response to auxin stimulus | 1.91e-5 | AT1G04240; AT1G15580; AT1G52830; AT3G23030; AT4G34760; AT4G34770; AT4G37390 |
| MF heat shock protein activity | 5.99e-3 | AT3G09440; AT5G02490; AT5G56010; AT5G56030 |
| BP protein folding | 3.42e-2 | AT3G09440; AT3G44110; AT5G02490; AT5G56010; AT5G56030 |
| MF transcription factor activity | 4.74e-2 | AT1G04240; AT1G15580; AT1G21910; AT1G25440; AT1G52830; AT2G31280; AT3G02790; AT3G15540; AT3G19360; AT3G23030; AT3G58120; AT3G60630; AT5G25190; AT5G45980; AT5G65320 |
| Cluster B. Up and late genes | ||
| BP response to auxin stimulus | 3.14e-11 | AT1G04240; AT1G09700; AT1G15580; AT1G28130; AT2G14960; AT3G07390; AT3G23030; AT4G27260; AT4G34760; AT4G34770; AT4G37390; AT4G38860; AT5G47370; AT5G54510 |
| MF alcohol dehydrogenase activity | 6.60e-4 | AT2G47130; AT2G47140; AT3G26760; AT4G05530; AT4G13180 |
| BP response to stimulus | 4.05e-3 | AT1G04240; AT1G05010; AT1G09530; AT1G09700; AT1G15580; AT1G28130; AT1G70940; AT1G74670; AT2G13540; AT2G14960; AT2G22490; AT2G47860; AT3G07390; AT3G12710; AT3G23030; AT3G24500; AT4G27260; AT4G34760; AT4G34770; AT4G36010; AT4G37390; AT4G37590; AT4G38410; AT4G38860; AT5G47370; AT5G54510 |
| CC cytosolic small ribosomal subunit (sensu Eukarya) | 1.42e-2 | AT1G58380; AT4G34670; AT5G10360; AT5G35530 |
| MF structural constituent of ribosome | 1.60e-2 | AT1G25260; AT1G58380; AT2G44860; AT3G53020; AT4G29410; AT4G34670; AT5G10360; AT5G35530; AT5G45800 |
| CC ribosome | 1.90e-2 | AT1G25260; AT1G58380; AT2G44860; AT3G53020; AT4G29410; AT4G34670; AT5G10360; AT5G35530; AT5G45800 |
| MF oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | 2.83e-2 | AT2G45400; AT2G47130; AT2G47140; AT3G26760; AT4G05530; AT4G13180 |
| MF metalloexopeptidase activity | 3.41e-2 | AT3G51800; AT3G59990; AT5G02480 |
| Cluster C. Down and early genes | ||
| MF pyruvate decarboxylase activity | 6.59e-4 | AT4G33070; AT5G17380; AT5G54960 |
| BP aldehyde catabolism | 2.93e-3 | AT1G06130; AT4G33070; AT5G17380 |
| MF carbon-carbon lyase activity | 1.59e-2 | AT4G25290; AT4G33070; AT4G38970; AT5G17380; AT5G54960 |
| BP glyoxylate catabolism | 2.09e-2 | AT4G33070; AT5G17380 |
| BP metal ion transport | 2.84e-2 | AT1G14660; AT1G70300; AT2G36950; AT3G05220; AT3G25410; AT3G47780; AT5G22830; AT5G41780 |
| BP catabolism | 3.07e-2 | AT1G06130; AT1G11080; AT1G17290; AT1G26560; AT1G50250; AT1G71980; AT1G77120; AT2G35770; AT3G30775; AT4G32840; AT4G33070; AT4G38970; AT5G01720; AT5G17380; AT5G37930; AT5G51750 |
| MF phosphotransferase activity, alcohol group as acceptor | 4.04e-2 | AT1G20650; AT1G35670; AT1G53420; AT1G53510; AT3G17510; AT3G21070; AT3G23150; AT4G23650; AT4G26270; AT4G32840; AT4G33080; AT4G33950; AT5G01810; AT5G20250; AT5G25110 |
| BP amino acid biosynthesis | 4.78e-2 | AT1G17290; AT3G30775; AT3G66658; AT4G33070; AT5G10860; AT5G17380 |
| Cluster D. Down and late genes | ||
| BP disaccharide biosynthesis | 9.72e-3 | AT1G23870; AT3G43190; AT5G20280; AT5G51460 |
| BP aldehyde metabolism | 2.65e-2 | AT1G06130; AT2G36460; AT4G33070 |
| BP glycolysis | 3.08e-2 | AT1G17290; AT2G36460; AT3G24170; AT4G26280; AT4G32840 |
| BP response to wounding | 3.95e-2 | AT1G48420; AT2G30490; AT2G37040; AT2G43710 |
All listed GO terms have an EASE score indicative of biased representation below 5 × 10−2 and are ordered according to decreasing score values. To limit redundancy, parent categories synonymous of child categories were omitted when gene content overlapped by two-thirds or more, within the same GO main category (BP, biological process; MF, molecular function; CC, cellular component).
As usual in genome-scale surveys of transcriptional activity, a large portion of the differentially expressed genes (334; 29.7%) had no known function. More peculiar to our case, 76 (6.8%) of the GSTs showing differential expression did not match with any gene model in the TIGR release 5.0 genome annotation, suggesting that the independent EuGene genome annotation that led to the initial GST design offers useful complementary information (http://www.psb.ugent.be/bioinformatics/genomes_ath_index.php).
These observations demonstrate that CATMA v1 microarrays provide a robust platform for the genome-scale analysis of transcriptional regulation: they allow the detection of known gene expression profiles, and they are useful for the identification of relevant biological themes via the statistical analysis of functional annotations linked to differentially expressed genes. The analytical power of the GST arrays will improve with the expansion of the GST collection and the increase in genome coverage.
Large-Scale Cloning Into Plasmids
The use of GSTs as PCR amplicons fixed to a solid support is limited to hybridization with target nucleic acid in solution. The introduction of these double-strand DNA segments into bacterial vectors allows their convenient distribution as bacterial glycerol stocks in multiwell plates. More importantly, it is the first step for subcloning into vectors designed for functional assays based on gene-specific tags, such as RNAi. For that purpose, the GST collection was introduced into a bacterial entry plasmid using the Gateway recombinational cloning technology (Hartley et al. 2000; Walhout et al. 2000). This system is advantageous because it permits the efficient cloning of large numbers of DNA fragments regardless of their sequences. Furthermore, fragments can be transferred straightforwardly from these entry clones into any destination vectors of interest, provided the appropriate recombination sites and selectable cassettes are combined.
The attB1 and attB2 recombination sites were added to the respective 5′-end and 3′-end of the original primary amplicons by PCR. To this end, each secondary amplicon was synthesized with one of 24 upstream column oligonucleotides made from the attB1 sequence fused to a “c” extension, and one of 16 downstream row oligonucleotides made of the attB2 sequence fused to an “r” extension (Fig. 3C; Supplemental Table S1). The attB1-GST-attB2 amplicons were then recombined with the attP1-ccdB-attP2 cassette in the pDONR207 vector (Fig. 3D). The successful insertion of a GST as an attL1-GST-attL2 cassette into an entry clone was confirmed by the analysis of the insert size: tertiary validation amplicons were produced by PCR with the DNR5 and DNR3 oligonucleotides anchored on the backbone of the vector, beyond the attL sites, and sized by agarose gel electrophoresis (Fig. 3E). For this purpose, template DNA was extracted from pooled Escherichia coli clones selected in gentamycin liquid medium after the BP clonase reaction and bacterial transformation. Because this reaction is highly efficient, it is not necessary to pick individual colonies grown on selective solid medium to recover the expected entry clones. All reactions (secondary PCR, BP clonase, E. coli transformation and growth, tertiary PCR) were performed in 96-well plates maintaining the same format as for the primary GST amplifications. As this paper goes into press, we confirmed entry clone pools for 17,304 different GSTs. The pooled E. coli strains carrying the GST entry plasmids will be distributed by the Nottingham Arabidopsis Stock Centre (NASC).
Subcloning in hpRNA Expression Vectors
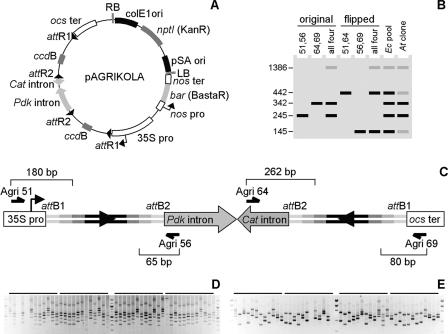
The most efficient method to knock down a plant gene by posttranscriptional silencing is to express in transgenic individuals an hpRNA made of a double-strand stem with a fragment of the transcript targeted for degradation and a loop with intron sequences that are spliced out from the RNA molecule (Smith et al. 2000). Systematic reverse genetics screens based on this method required the construction of large series of recombinant DNA molecules in which, for each gene of interest, a fragment was cloned as an inverted repeat separated by the intron spacer. Because the GSTs were selected to avoid cross-hybridization between distinct genes, they were an ideal substrate for the production of a genome-scale Arabidopsis hpRNA expression plasmid collection. Conventional cloning protocols using restriction and ligation enzymes for the production of such hairpin transgenes are laborious and not conducive to high-throughput approaches. Therefore, vectors were designed for the one-step introduction of two identical inverted repeats into a destination vector, taking advantage of the Gateway technology (Wesley et al. 2001). The vector we developed for the strong expression of hpRNA in plant cells was designated pAGRIKOLA (Fig. 5A). It contained the inverted-repeat structure of pHELLSGATE12, with the 35S CaMV promoter, the octopine synthase (ocs) terminator, and the spacer separating the two inverted ccdB genes carrying head-to-head the intron-2 of the Flaveria Pdk gene and the intron of the castor bean Cat gene (Helliwell and Waterhouse 2003). The backbone of pAGRIKOLA was derived from pGreenII0229, resulting in a smaller vector (10,461 bp) than pHELLSGATE12 and including the bar gene facilitating herbicide selection of transformed plants. The complete sequence of pAGRIKOLA has been experimentally confirmed (GenBank accession no. AY568055).
Figure 5.
Construction of hpRNA expression plasmids. (A) Map of the pAGRIKOLA destination vector. (B) Schematic representation of the multiplex analysis of the expression plasmids. Numbers on the left show how many base pairs were added to the primary GSTs upon amplification with the Agri primers indicated at the top of each lane. The largest band corresponds to the amplification of both GST cassettes with the external Agri51 and Agri69 primers. Profiles in lanes 7 and 8 correspond to results in panels D and E, respectively, and are generated via amplification with all four Agri primers. The profile in lane 8 only shows results obtained with a plasmid in which the spacer intron is initially in its original orientation, as pictured in C. (C) Structure of recombined hairpin cassette with the inverted GST repeats. GST elements are depicted as in Figure 3. Bracket numbers indicate the length of the fragment delimited by the 5′-end of each Agri primer and the 3′-end of the closest attB site, on either strand. Configuration with the flipped spacer intron is not shown. (D) Multiplex PCR validation of double LR clonase reaction products. Each lane shows the amplification results for one E. coli pool after liquid selection (see Supplemental Table S2). (E) Multiplex PCR verification of A. tumefaciens transformants. For each initial pool, four individual Agrobacterium colonies were picked. Most tetrads contain both types of colonies, with the expression plasmid carrying the intron spacer initially either in the original or flipped orientation (see Supplemental Table S2).
Entry clones carrying an attL1-GST-attL2 cassette were recombined with both pAGRIKOLA attR1-ccdB-attR2 cassettes in a double LR clonase reaction. Note that the DNA template carrying the attL1-GST-attL2 cassette recombined in the double LR reaction could either be the purified entry plasmid (Fig. 3D) or the tertiary validation PCR product (Fig. 3E). Both templates yielded >95% subcloning efficiency in 96-well-plate LR clonase reactions.
For each GST, the following steps included (1) heat-shock transformation of E. coli (DH5α) with the double LR reaction products; (2) selection in kanamycin liquid medium; (3) validation of the resulting E. coli bacterial pool containing the hpRNA expression constructs; (4) purification of the pool plasmid DNA; (5) freeze-thaw transformation of Agrobacterium tumefaciens (GV3101) with plasmid DNA; (6) selection of transformed agrobacteria in kanamycin liquid medium; (7) plating of transformants on kanamycin solid medium; (8) picking of individual A. tumefaciens colonies carrying hpRNA plasmids; and (9) PCR verification that these colonies contained a correct hpRNA expression construct. Steps 1 through 6 were all performed in 96-well plates with the same format as that for the primary GST amplifications.
Because the double LR recombination could produce different intermediates depending on which repeats recombined with each other, the intron spacer in the expression clone either retained its original orientation or was flipped. Because the spacer contained two head-to-head introns, a portion of the hpRNA loop would be spliced out in plant cells regardless of its final orientation, which guarantees efficient silencing (Helliwell and Waterhouse 2003). The integrity of the artificial GST hairpin gene was monitored via multiplex PCR (Steps 3 and 9) to confirm that both GST inserts had the expected size and to determine the orientation of the intron spacer. The four primers (Agri51, Agri56, Agri64, and Agri69; Fig. 5C) were positioned in such a way that the GST subunits present in hpRNA expression plasmids were easily distinguished by size in agarose gel electrophoresis. Whether the intron spacer was in its original or flipped orientation, the amplified paired DNA fragments were Agri51-attB1-GST-attB2-Agri56 (adding 245 bp to the primary GST length) and Agri64-attB2-GST-attB1-Agri69 (plus 342 bp) or Agri51-attB1-GST-attB2-Agri64 (plus 442 bp) and Agri56-attB2-GST-attB1-Agri69 (plus 145 bp; Fig. 5B). In Step 3, the pool of E. coli transformants selected in liquid medium contained both constructs with either the original or the flipped intron spacer and gave rise to the four possible fragments in the multiplex PCR (Fig. 5D). In Step 9, individual Agrobacterium colonies originating from the acquisition of a single hpRNA expression plasmid molecule yielded prominently one of the two combinations, but often also the other in lower amounts (Fig. 5E). The latter might result from template-switching during PCR or from recombination events between the inverted GSTs occurring in Agrobacterium. As the plasmids are equally effective with the introns in either orientation, this “intron-flipping” is no more than a cosmetic nuisance. The fifth larger band generally observed in Steps 3 and 9 corresponded to the complete hairpin cassette amplified by the outside primers Agri51 and Agri69. As this paper goes to press, we confirmed GST hpRNA expression clones for 8136 different GSTs. The pooled E. coli strains carrying the hpRNA expression plasmids will be distributed by the Nottingham Arabidopsis Stock Centre (NASC).
Phenotypes of Silenced Arabidopsis Lines
As proof of concept, a medium-scale transformation project is currently underway to analyze randomly selected hpRNA clones. A. tumefaciens containing the hpRNA constructs was used to transform Arabidopsis thaliana (Col-0) by the conventional floral dip method (Clough and Bent 1998). Seeds from transformed plants were germinated under BASTA selection, and a minimum of 10 individual T1 transformants per construct were selected to provide a collection of T2 RNAi lines. Preliminary results were encouraging and suggested that the hpRNA constructs could be effectively deployed to generate an “interference” series in T1 Arabidopsis lines. We noted a varied degree of penetrance in phenotypes from individual lines that might result from different T-DNA copy numbers, positional effects, or from effective interference initiating at different stages of plant development. T2 lines have yet to be tested to evaluate the heritability and reproducibility of the observed phenotypes. We report here three illustrative examples in which phenotypic series were obtained useful for determining the function of the target gene product.
The first example indicated successful silencing of the target gene CesA7 (At5g17420; GST ID: CATMA5a15680) encoding a component of cellulose synthases involved in the production of secondary cell walls. The cesA7 knockout mutant is smaller and grows more slowly than wild type. The cellulose content of secondary cell walls is greatly reduced in the mutant plants, resulting in less rigid stems unable to keep a normal upright position, which leads to a characteristic drooping phenotype (Turner and Somerville 1997; Taylor et al. 1999). The same characteristic phenotype was observed to different degrees of penetrance in each of the 10 T1 transformants obtained with this hpRNA clone (Fig. 6A). This example demonstrates that GST-induced silencing could effectively mimic a null mutant and give the phenotype expected when the target gene is not expressed.
Figure 6.
Phenotypes exhibited by T1 plants carrying a GST hpRNA transgene targeting the indicated endogenous gene. (A) At5g17420 codes for CesA7, a subunit of cellulose synthase. (Top) Five-week-old control wild-type Arabidopsis plants. (Bottom) Eight-week-old T1 plants. The transformed plants grow more slowly than the wild type and have the weak, floppy stems seen in knockout mutants of this gene (scale bar, 6 cm). (B) At1g20260 codes for the vacuolar-type H+-ATPase subunit B3. Figure and insets show a representative range of phenotypes in T1 plants 7 wk after germination. In particular, the insets document severe dwarfing, particularly in rosette tissue, whereas flower size and development are not similarly affected (scale bars, 1 cm). (C) At1g20300 codes for a pentatricopeptide repeat protein predicted to be localized in mitochondria. Individual T1 plants (at 7 wk) and duplicate siliques from six independent T1 lines (at 8 wk) illustrate varying degrees of infertility that correlates with phenotype severity (scale bars, 1 cm). Identical symbols indicate the correspondence between a silique pair and its source plant when both are depicted in the panel.
The second example was the effect of silencing At1g20260 (CATMA1a19260), a gene encoding the vacuolar-type H+-ATPase subunit B3 (VHA-B3). V-type H+-ATPases are ubiquitous eukaryotic heteromeric enzyme complexes that acidify endomembrane compartments and are essential for many transport processes involved in development and tolerance to environmental stress (Sze et al. 2002). Classical forward genetics studies in Arabidopsis have characterized a single mutation in V-type ATPases, the det3 mutant (Schumacher et al. 1999). det3 plays an important role in the control of growth and morphogenesis of Arabidopsis seedlings and exhibits severe dwarfing resulting from a reduction in cell expansion. Interestingly, these developmental effects are more pronounced in cells of the hypocotyl, petioles, and inflorescence stems. Despite the severe phenotype, det3 is a weak mutant allele and, consequently, it is likely that lesions in other subunits of the Arabidopsis V-type ATPases have not been described because they lead to lethality, as is the case for null mutations in Neurospora (Ferea and Bowman 1996) and Drosophila (Davies et al. 1996; Guo et al. 1996). The det3 mutant may interfere with signal transduction pathways controlling meristem activity, possibly by regulation through multiple phytohormones (Schumacher et al. 1999). Here, we observed severe dwarfing in the rosette leaves in transgenic plants specifying hpRNA to the H+-ATPase subunit B3 (Fig. 6B, insets). Transgenic plants displayed increased serrations and some leaf and laminar distortions. The majority of the transformants showed some degree of dwarfing that became more pronounced as the plants developed. The range of whole plant phenotypes is shown in Figure 6B. The severity of the symptoms was in agreement with those predicted from the det3 studies and supported the hypothesis that vacuolar ATPases are essential enzymes for maintaining cellular homeostasis. This example demonstrates that GST-induced silencing can be used to obtain mutants deficient in expression of an essential gene, difficult or impossible to obtain by classical insertion mutagenesis.
The third illustration of targeted silencing concerns At1g20300 (CATMA1a19300), encoding a gene product with 11 pentatricopeptide repeats (PPRs) that is predicted to be imported into mitochondria. The function of this gene is completely unknown, but homologs from other plant species are involved in the control of male fertility by regulating expression of mitochondrial “sterility” genes (Bentolila et al. 2002; Brown et al. 2003; Desloire et al. 2003; Kazama and Toriyama 2003; Koizuka et al. 2003; Komori et al. 2004). Eight out of the 10 selected Arabidopsis lines expressing the At1g20300 hpRNA construct developed strong growth phenotypes (representative individuals are shown in Fig. 6C). In general, the suppressed plants were smaller in stature and showed a trend toward early flowering. We observed a marked decrease in fertility correlated with this growth suppression, as illustrated by the reduced or aborted siliques of six individual lines (Fig. 6C, inset). Even in plants with little apparent reduction in phenotype, seed set was markedly reduced relatively to wild-type plants. This example demonstrates that GST-induced silencing can and will give invaluable information on gene function for target genes whose role in plant development is currently unknown.
The length of the GSTs corresponding to the three series of silenced lines described here was 152, 163, and 400 bases. This information and the fact that constructs yielding gross morphological alterations had no obvious size bias (data not shown) suggest that GSTs across the chosen length range may induce gene silencing when expressed as hairpin RNA.
DISCUSSION
From Sequence to Function
Initial analyses of the Arabidopsis genome sequence described fewer than 25,000 genes (The Arabidopsis Genome Initiative 2000). Experimental evidence of gene structure (from full-length cDNAs) and expression was available for only a small proportion of these putative genes and experimental evidence of function available for even fewer. In the past three years, giant strides have been made in compiling cDNA sequences (Haas et al. 2002; Seki et al. 2002; Castelli et al. 2004) and expression data (Schaffer et al. 2000; Kim et al. 2003). Together with improvements in automatic annotation routines and better use of expert manual annotation, these data have enabled us to have a much better view of the coding potential of the Arabidopsis genome (Wortman et al. 2003; http://www.psb.ugent.be/bioinformatics/genomes_ath_index.php). However, these improvements in structural annotation have not been matched by equivalent advances in functional annotation. Functional analysis of Arabidopsis genes or proteins remains a long and painstaking task, but one that can be considerably facilitated by constructing and widely distributing shared functional genomics resources such as collections of insertion mutants (Azpiroz-Leehan and Feldmann 1997; Parinov et al. 1999; Speulman et al. 1999; Sussman et al. 2000; Samson et al. 2002; Sessions et al. 2002; Alonso et al. 2003; Rosso et al. 2003) or cDNA clones (Yamada et al. 2003). The GST collection described here is a novel, versatile resource for functional genomics with applications in expression profiling, reverse genetics, and any other approach that requires gene-specific hybridization.
The GST Collection
The GSTs are formatted to constitute a resource that is durable, versatile, and easy to multiply and distribute. Because of the standard amplicon structure, a set of only 40 universal primers is sufficient for the reamplification of the entire collection, facilitating the iterative production, at limited cost, of the large amount of GST DNAs necessary for the printing of glass microarrays. The extensions that distinguish all rows and all columns help resolve the well-to-well cross-contamination unavoidable during the manipulation of DNA and clone repertoires stored in multiwell plates. As illustrated here with the addition of Gateway attB sites, the extension primers are also useful for adding any sequence to either side of the GSTs simply by PCR amplification, making them compatible with a wide range of cloning techniques and vectors. GST cloning into Gateway entry vectors has established the collection as a set of bacterial clones, which greatly facilitates distribution of the resource.
Functional genomics projects have to keep up with evolving genome annotation. The flexible GST format makes it possible to add amplicons, plasmids, and expression vectors as additional transcription units are described. Because the GSTs are amplified from genomic DNA templates, any sequence/gene of interest is straightforwardly accessible. Thus, the GST repertoire provides a better genome representation than the best combined collection of cDNA clones, in which for multicellular eukaryotes up to half of the predicted genes may be missing. In that respect, it is interesting to note that transcript profiling data obtained with CATMA v1 microarrays and mutant phenotypes observed in lines expressing GST hpRNA indicate that genes not documented in the latest genome annotation (TIGR release 5.0), but identified in an independent structural annotation effort (http://www.psb.ugent.be/bioinformatics/genomes_ath_index.php), are transcriptionally functional. This observation is in agreement with a study based on a whole-genome oligonucleotide tiling array showing that many transcription units have been overlooked in the annotation process (Yamada et al. 2003). Altogether, the experimental data stress the necessity to integrate the results obtained from independent prediction tools together with information about function, such as expression profiles and mutant phenotypes, to continuously improve the genome annotation.
Much thought went into designing the GST resource to achieve maximum versatility and ease of use. In addition to the applications in transcript profiling and reverse genetics described below, other uses for the resource can be imagined. For example, the amplicons could be synthesized with either 24-column or 16-row biotinylated-extension primers, making it possible to purify the sense or antisense single-strand GST DNAs, respectively. Such single-strand DNAs could be used as probes for microarrays. Alternatively, they may serve as specific probes to capture corresponding clones from cDNA libraries. We envisage using the resource for recovering full-length cDNAs from genes for which these are not currently available.
Application of the GST Collection to Transcript Profiling
We have shown that known transcript profile changes can be reproduced in experiments performed with CATMA microarrays. We also illustrate that the integration of genome-scale functional annotation and transcriptome data obtained with the GST collection yields valuable information for the study of biological mechanisms in Arabidopsis.
Multiple studies, often supported by commercial providers, argue that oligonucleotide arrays offer the best solution for microarray transcript profiling experiments (e.g., Zhu and Wang 2000; Hughes et al. 2001). In our hands, CATMA v1 microarrays perform at least as well in terms of dynamic range, sensitivity, and specificity as oligonucleotide microarrays distributed by commercial providers (J. Allemeersch, S. Durinck, R. Vanderhaeghen, P. Alard, R. Maes, K. Seeuws, T. Bogaert, K. Coddens, K. Deschouwer, P. Van Hummelen, et al., in prep.). Moreover, because the creation of the collection was supported by academic laboratories, all information relative to CATMA arrays, including DNA feature design and sequence, is freely available, and GST resources are distributed for a nominal fee.
The CATMA array format permits applications incompatible with other platforms. The collection is flexible and GST amplicons can be cherry-picked for the production of dedicated arrays or combined with other probe sets. Because they are shared by all DNA features, oligonucleotides carrying the extension sequences and labeled with fluorophores may also constitute calibrated references for the study of mRNA abundance (Dudley et al. 2002).
Application of the GST Collection to Reverse Genetics
Posttranscriptional gene silencing is known to be helpful for the study of gene function in Arabidopsis and in many other plant and metazoan species. Yet, no systematic attempts to carry out gene silencing at the genome scale have yet been completed in plants. With the GST collection described here, the technical procedures have been put in place to generate the tens of thousands of constructs necessary for such an endeavor.
We have shown that the introduction of a transgene expressing homologous GST hairpin RNAs into wild-type plants may result in phenotypes that mimic the effects of known null mutations or that indicate the function of essential or unknown genes. Pursuing this effort with a significant portion of Arabidopsis genes should provide useful information on the overall efficacy of the approach, the phenotypic classes that can be obtained and the specificity and stability of the knockdown effects.
We anticipate that the described RNAi tools will complement the numerous Arabidopsis insertion mutant collections that have been built up over the last decade. Insertional mutants constitute a valuable resource, especially since the systematic determination of the sequences flanking the T-DNA or transposable element inserts and the publication of that information via online databases. Yet, insertional mutagenesis in plants suffers from significant drawbacks inherent to the approach: (1) the introduction of inserts into the genome cannot be directed and hundreds of thousands of transformants must be generated to reach saturation; (2) therefore, only a very few genotypes have been used as recipients in Arabidopsis reverse genetics programs; (3) most informative insertions result in null mutations, and they rarely yield hypomorphic alleles displaying a range of phenotypes; (4) embryo-lethal mutants are difficult to recover and gamete-lethal mutants are lost; and (5) combining multiple insertion mutations is cumbersome. In addition, available flanking sequence tags are often hundreds of base pairs away from the actual insertion site and the majority of phenotypes in T-DNA insertion lines do not result from a mutation tagged with a selectable marker. Consequently, obtaining validated homozygous insertion lines in a locus of interest is not trivial. In contrast, targeted gene silencing only requires a few independent transgenics for the study of any given gene, and the same silencing construct can easily be introduced into multiple genetic backgrounds, including mutants of particular interest to investigate potential interactions involving the target gene. Most knockdown lines are not null mutants and generally show diverse phenotypes typical of allelic series. Finally, essential genes may be studied via partial or conditional silencing.
A genome-scale GST collection is attractive for any species in which RNAi silencing is a practical reverse genetics method. In plants it could form the basis of systematic knockdown programs with a variety of approaches: hpRNA expression as illustrated here, virus-induced gene silencing (Lu et al. 2003) or transitive silencing (Van Houdt et al. 2003). Because the GSTs are available as Gateway entry clones, the same basic resource is useful regardless of the strategy, provided the transgene inducing silencing can be built via recombinational cloning. The enhanced plant silencing vectors that will undoubtedly be developed in the coming years will be compatible with the CATMA collection.
Conclusion
To our knowledge, this is the first attempt to construct a clonable GST resource covering the majority of genes in an organism. Previously constructed sequence collections are either oligonucleotide-based and thus unclonable, or are EST, cDNA, or ORF collections and thus not designed to be gene-specific. This novel and versatile resource has great promise for large-scale transcript profiling and reverse genetics projects, and is being actively used in two major projects of this type, CAGE and AGRIKOLA. However, its uses are not limited to these applications. We hope that this resource will be a cornerstone of other future projects. The principles and technologies described here are in no way specific to Arabidopsis or plants in general. Similar resources should prove equally valuable for functional genomics projects in other organisms.
METHODS
SPADS GST Design
All GSTs were selected with the Specific Primer and Amplicon Design Software (SPADS; Thareau et al. 2003). The database used as the A. thaliana reference nuclear genome contains the five chromosome pseudomolecule sequences (TIGR ASMBL_ID 68170, 51595, 68173, 68164, and 68172) available from TIGR on January 28, 2001 (ftp://ftp.tigr.org/pub/data/a_thaliana/ath1). Parameters for the design of the gene-specific PCR oligonucleotide sequence (without their 5′-extension) were 18 to 25 bases, Tm of 50° to 65°C with a maximum ΔTm of 4°C between paired primers. All other parameters were as described in Results or elsewhere (Thareau et al. 2003). All gene (At) codes listed as GST matches refer to the TIGR release 5.0 genome annotation.
DNA Synthesis
The 40 arbitrary 5′-extensions contain 17 nt, eight to nine being C or G. To avoid hairpin structures, their sequences have no self-complementary repeats of four or more bases separated by three or more bases. They do not contain stretches of more than four contiguous G or C, or any ATG or stop codon in the sense orientation. To avoid PCR primer dimers, the last four bases of an extension (column or row) do not match any sequence in that same extension or in the opposite subset of extensions (rows or columns). All were tested in silico to minimize priming against the BAC plasmid backbone and the E. coli genome.
The primary amplification products were synthesized as follows. A master mix base of 10 μL of 10× Invitrogen PCR buffer (50 mM KCl, 20 mM Tris-HCl at pH 8.3), 3 μL of 50 mM MgCl2 and 75.6 μL of double-distilled filter sterilized water (ddH2O) was made and stored at 4°C. On day of use, 2 μL of a 10 mM dNTP stock and 0.4 μL of Invitrogen Platinum Taq (5 U/μL) were added to the master mix base. Then, 91 μL of the complete master mix was dispensed per well into an ABgene semiskirted PCR plate. GST-specific primers were provided by Sigma-Genosys as mixed pairs in 96-well plate format. These were diluted with 10 mM Tris (pH 8.0) to make a 10 μM working stock, and 4 μL was dispensed into the matching well of the PCR plate containing the complete master mix. Finally, 2.5 ng of a BAC DNA miniprep, containing the gene to which the specific GST primers were designed, was added to the appropriate well. When a BAC clone was not available, 5 μL of a 1 ng/μL Columbia genomic (Col-4) DNA prep was substituted. PCR amplification was carried out using the following program: hold for 5 min at 94°C; 11 cycles of 30 sec at 94°C, 30 sec at 63°C (temperature reduced by 1°C per cycle), and 30 sec at 72°C; 30 cycles of 30 sec at 94°C, 30 sec at 63°C, 30 sec at 72°C; 5 min at 72°C; and 1 min at 4°C for 1 min. To assess quality of product, 1 μL from each well was run on a 2% agarose gel. The reaction setup and cycling conditions were identical for secondary amplification reactions except that the BAC or genomic DNA was replaced with 2 μL of a 100-fold dilution (in 10 mM Tris at pH 8.0, 0.1 mM EDTA) of the primary product plus 2 μL of ddH2O. No purification of the primary product was carried out.
Microarray Spiking Experiment
Arabidopsis Col-4 seeds were sown on plates containing agar-solidified culture medium (1× MS [Duchefa], 0.5 g/L MES at pH 6.0, 1 g/L sucrose, and 0.6% plant tissue culture agar [LabM]). After sowing, plates were cold-stratified for 7 d at 4°C and subsequently transferred to a growth chamber kept at 22°C (24 h photoperiod with 16 h of light at 65 mE/m2 sec PAR). Whole shoots were harvested at growth stage 1.04 (Boyes et al. 2001), frozen immediately in liquid nitrogen, and stored at -70°C until needed. Frozen seedling material was ground, and total RNA was extracted using TRIzol reagent (Invitrogen). The spiking experiment protocols will be detailed further elsewhere with the comparative analysis of results obtained with CATMA microarrays as well as short and long oligonucleotide microarrays distributed by commercial providers (J. Allemeersch, S. Durinck, R. Vanderhaeghen, P. Alard, R. Maes, K. Seeuws, T. Bogaert, K. Coddens, K. Deschouwer, P. Van Hummelen, et al., in prep.). CATMA arrays, or corresponding CATMA-based transcript profiling services are provided by various genomics facilities.
The intensity ratios were calculated as follows. The mean foreground (Fg) intensities for the Cy3 and Cy5 channel measurements were corrected for background (Bg), and the base 2 log ratios were computed from these values. Log ratios were normalized for dye effects by computing Loess regression from MA plots, separately for each print tip and array so that the values were adjusted for dye, print tip, and slide effects (Yang et al. 2002). Log ratios were averaged over each dye-swap pair, and the represented combined ratios were extracted by anti-log conversion of the averaged values.
Microarray Transcript Profile Analysis of Auxin Treatment
To streamline the production of the large DNA fragment collection (CATMA v1), all GST purification steps were automated with the Magnatrix 1200 workstation (Magnetic Biosolutions) using bead technology (V. Wirta, M. Lukacs, A. Holmberg, P. Nilsson, M. Uhlén, R. Bhalerao, and J. Lundeberg, in prep.). Briefly, the GSTs were synthesized by secondary PCR amplification performed with biotinylated “column” extension oligonucleotides (Supplemental Table S1), then bound and washed on streptavidin-coated paramagnetic beads. The PCR products were eluted in deionized water and separated from the beads by magnetic removal. The fully automated protocol is highly efficient for DNA fragments in the GST length range, both in terms of yield and purity. The purified GST fragments were spotted from a 50% dimethyl sulfoxide (DMSO) solution on Ultra-GAPS slides (Corning) with a QArray arrayer (Genetix), then linked to the slide surface by UV treatment (250 mJ/cm2).
The biological samples were prepared as follows. Arabidopsis seeds (Col-0) were germinated and grown in 0.5× MS (Duchefa) liquid medium supplemented with 0.5% sucrose at 22°C (24 h photoperiod with 16 h of light at 75 mE/m2 sec PAR). After 10 d (2 h after dawn), seedlings were treated with 1 μM indole-3-acetic acid for a period of 0, 30, 120, and 240 min. At the end of the treatment, the seedlings were washed once with an excess of MS medium with 0.5% sucrose for 5 min, then immediately frozen in liquid nitrogen and stored at -70°C until needed. Each time-point sample was made of a pool of seedlings grown simultaneously in three independent vials. Frozen seedling material was ground and total RNA was extracted using the QIAGEN RNAeasy kit.
For each biological sample, 20 μg of total RNA was annealed to 10 μg of anchored oligo(dT) primer (dT20VN; MWG Biotech AG) and reverse-transcribed to cDNA at 42°C during a 105-min reaction. The 30-μL reaction contained 2 mM dNTPs (dTTP:aminoallyl-dUTP in 1:4; unmodified Amersham Biosciences, modified Sigma-Aldrich), first-strand buffer (50 mM Tris-HCl at pH 8.3, 75 mM KCl, 3 mM MgCl2), 10 mM DTT, and 400 U Superscript II (Invitrogen). RNA strand hydrolysis (15 min at 70°C in the presence of 150 mM NaOH) was followed by a purification using MinElute spin columns (QIAGEN) with the provided wash and elution buffers replaced by 80% ethanol and 100 mM NaHCO3 (pH 9.0), respectively. The monofunctional NHS-ester Cy3 or Cy5 fluorophores (Amersham Biosciences) were coupled to the purified cDNA during a 90-min incubation at room temperature. Prior to pooling and spin column purification, ester groups on the unincorporated fluorophores were inactivated using 730 mM hydroxylamine.
Slides were hybridized for 16-18 h using a two-step protocol in the GeneTac hybridization station (Genomic solutions). Prehybridization for 45 min at 42°C (5× SSC, 1% BSA [Sigma-Aldrich], 0.1% SDS, 40 μg of poly(dA) [Sigma-Aldrich], and 20 μg of tRNA [Sigma-Aldrich]) was followed by a hybridization at 42°C with the labeled material in a buffer containing 5× SSC, 25% formamide, 0.1% SDS, 40 μg of poly(dA), and 20 μg of tRNA. Hybridized slides were washed with 2× SSC and 0.1% SDS at 42°C, followed by 0.1× SSC and 0.1% SDS at room temperature, and finally by three washes with 0.1× SSC at room temperature. Slides were scanned at a 10-μm resolution using the G2565BA DNA microarray scanner (Agilent Technologies). The acquired TIFF images were processed using the GenePix 4.1 software (Axon Instruments).
The performed microarray hybridizations constituted a complete experimental loop design. They included eight hybridizations for the comparison of T0, T30, T120, and T240 samples, with reciprocal dye swap, and a T0 self-hybridization (Supplemental Fig. S1). The raw data were preprocessed as follows. Spots corresponding to the intergenic regions were selected, and the median of these control Fg intensities was computed for each slide. Fg intensity measurements below background threshold (i.e., Fg < Bg + 2 × Bg standard deviation) were replaced by the corresponding slide median Fg intergenic control value to reduce the impact of background variability in the statistical analysis of differential expression. The leveled data were Loess-normalized as described above. Mixed ANOVA models, in which some effects are considered fixed and others random, were used to identify genes that were significantly differentially expressed in the experimental time course (Wolfinger et al. 2001). First, array and channel effects were removed from the expression responses by a linear normalization ANOVA model of the form yijkl = μ + Ak + (AD)kl + εijkl, with i (i = 1-21,120, the number of microarray features in the array) indexing the selected GSTs, j (j = 1... 4) indexing the time points (treatments), Ak representing the random array effects (k = 1... 9), (AD)kl representing the random array × dye combinations, or channel effects with k = 1... 9 arrays and l = 1... 2 dyes, and εijkl representing the random error. Second, residuals, denoted as rijkl, and computed by subtracting the fitted values for the effects from the observed values yijkl, were then subjected to 21,120 gene-specific models of the form rijkl = μ + (GD)il + (GT)ij + (GA)ik + γijkl, partitioning gene-specific variation into fixed gene-specific dye effects (GD)il, fixed time-point effects (GT)ij, random spot effects (GA)ik, and random error γijkl. In these gene-specific models, the spot effect serves to account for the spot-to-spot variability inherent in spotted microarray data. To test differences between time points, we used the Wald statistic, which should follow approximate χ2 distribution under the null hypothesis with degrees of freedom equal to 3. To adjust for the multiple testing problem, we set the p-value cutoff at the Bonferroni value of 5% divided by 21,120, or 2.37 × 10-6.
Transcript profiles close to theoretical patterns were identified with the template-matching module of the MultiExperiment Viewer (MeV) software package (Saeed et al. 2003): the four patterns defined up-regulated genes reaching a maximum plateau at 30 and 120 min (up and early) or 120 and 240 min (up and late) or down-regulated genes with a minimum plateau at 30 and 120 min (down and early) or 120 and 240 min (down and late). The biological theme representation analysis performed with the EASE software (Hosack et al. 2003) was based on the TAIR Arabidopsis GO term list (ATH_GO_20040217.txt available at ftp://tairpub:tairpub@ftp.arabidopsis.org/home/tair/Genes/Gene_Ontology/).
Recombinational Cloning and Plant Transformation
For the addition of the attB sites to the GSTs, primary amplicons were used as templates and amplified by Taq polymerase (NEB) with 16 pmoles of the relevant row and column primers (Supplemental Table S1) in 20-μL reactions. This step and all subsequent procedures were carried out in 96-well microtiter plates. The PCR conditions were 1 min at 94°C, followed by 35 cycles of 15 sec at 94°C, 15 sec at 55°C, and 30 sec at 72°C, and terminated by 5 min at 72°C. The PCR reactions contained 2 μL of Red Cresol (Sigma-Aldrich) to facilitate loading on agarose gels for visualization of the products. Figure 3E shows typical results for 48 GSTs listed in Supplemental Table S2 (DNA molecular mass marker: 100-bp DNA Ladder; NEB).
Cloning of the amplified GSTs (Gateway BP reaction) was carried out by adding a 3-μL mix containing 50 ng of pDONR207 vector (Invitrogen), 0.4 μL of BP clonase (Invitrogen), and 1 μL of 5× BP clonase buffer to 2 μL of the amplified GST. The reaction was left overnight at 25°C, then stopped by the addition of 0.5 μL of proteinase K (2 μg/μL) and incubated for 15 min at 37°C. One microliter of the reaction mix was used to transform 10 μL of competent DH5α cells (F′Φ80dlacZ Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17 (rk-, mk+) phoA supE44 λ-thi-1 gyrA96relA1/F′ proAB+ lacIqZΔM15 Tn10(tetr)) prepared as described (Inoue et al. 1990). The cells were incubated with the DNA for 20 min on ice, heat-shocked for 15 sec at 42°C in a waterbath, incubated for 5 min on ice, diluted with 90 μL of SOC medium, and shaken for 1 h at 37°C. The transformation mix was then added to 1 mL of 2× LB medium (containing 15 μg/mL gentamycin to select for transformants) in 2-mL 96-well plates and shaken for 20 h to an OD of 1.4. Presence of the expected GST entry clone in the suspension of transformed cells (0.5 μL) was verified by PCR using 20 pmoles of each of the primers DNR5, 5′-CTGGCAGTTC CCTACTCTCG-3′, and DNR3, 5′-GATGGTCGGAAGAG GCATAA-3′ (Fig. 3F; 100-bp DNA Ladder; NEB). The PCR conditions were as follows: 5 min at 94°C, followed by 35 cycles of 30 sec at 94°C, 30 sec at 55°C, 2 min at 72°C, and terminated by 5 min at 72°C.
Subcloning of GSTs from an entry clone into the destination vector pAGRIKOLA (Gateway LR reaction) was carried out by adding a 4-μL mix containing 75 ng of pAGRIKOLA, 1 μL of LR clonase (Invitrogen), and 1 μL of 5× LR clonase buffer to 1 μL (75 ng) of the attL1-GST-attL2 cassette DNA. Cassette DNA was prepared either by alkaline lysis plasmid purification from transformed bacteria (Montage Plasmid Miniprep 96 Kit; Millipore) or by PCR (the products obtained by the validation of the entry clones described above). The reaction conditions and bacterial transformation were as for the BP reactions, except that 50 μg/mL kanamycin was used to select transformed cells. The suspension of transformed cells (0.5 μL) was verified by PCR using 20 pmoles of each of the primers Agri51/Agri56/Agri64/Agri69 (Fig. 5D; Low DNA Mass Ladder; Invitrogen). The PCR conditions were as follows: 5 min at 94°C, followed by 35 cycles of 30 sec at 94°C, 30 sec at 55°C, 2 min at 72°C, and terminated by 5 min at 72°C. Plasmid DNA was prepared from the transformed bacteria by alkaline lysis (Montage Plasmid Miniprep 96 Kit; Millipore).
The A. tumefaciens strain GV3101 (Holsters et al. 1980) carrying helper plasmids pMP90 (Koncz and Schell 1986) and pSOUP (Hellens et al. 2000) was used for Arabidopsis transformation. Agrobacterium cells were transformed with pAGRIKOLA constructs as described using the protocol of An et al. (1988). Clone validation was performed as for E. coli LR clonase products verification (Fig. 5E; Low DNA Mass Ladder; Invitrogen). Arabidopsis plants were transformed using the floral dip method (Clough and Bent 1998).
More detailed cloning and transformation protocols are available online via the AGRIKOLA project Web site (http://www.agrikola.org/).
Acknowledgments
The CATMA Consortium efforts were supported by multiple grants, namely, No-2000-009 from the French Génoplante program to P. Hilson and P. Rouzé, BMBF-312274 to H. Lehrach, BIO2000/3094/E from the Spanish Biotechnology program-MCYT to J. Paz-Ares, grants from the Wallenberg Consortium Nord (RNA platform 1b) to R.P. Bhalerao and J. Lundeberg, from the University of Lausanne (Genomics Project) to P. Reymond and E.E. Farmer, and from the BBSRC as part of the GARNET consortium (UK Investigating Gene Function Initiative) to J. Beynon. The construction of a Compendium of Arabidopsis Gene Expression profiles based on CATMA arrays is supported by the CAGE demonstration project (QLRT-2001-02035). The transfer of the CATMA GSTs into bacterial plasmids and the production of silenced Arabidopsis lines is supported by the AGRIKOLA grant (QLRT-2001-01741). The CAGE and AGRIKOLA projects are funded in the 5th European Framework Programme. We are grateful to Stefanie Albrecht, Caroline Buysschaert, Beate Hoffman, Yolanda Leo del Puerto, Anders Holmberg, Antje Krüger, Morten Lukacs, Ruth Maes, Peter Nilsson, Jennie Rowley, Kurt Seeuws, Rudy Vanderhaeghen, and Raimundo Villarroel for technical assistance in GST synthesis, microarray production, and GST cloning; to Philippe Alard, Eric Bonnet, Björn De Meyer, David Lonsdale, and Martin Trick for assistance with the CATMA database; to Philippe Grevet for assistance with the AGRIKOLA database and Web site; to Mark A. Bennett for establishing the plant transformation database; to Wendy Byrne, Bertrand Dubreucq, Mathilde Grelon, Melanie Lück, Raphael Mercier, Hakim Mireau, Jean-Christophe Palauqui, and Matthieu Simon for assistance with production and analysis of transgenic plants; and to Uli Schneider and Gerrit Beemster for helpful discussions. We thank Peter Waterhouse and Chris Helliwell for sharing with us materials prior to publication and for very useful advice on the use of plant hpRNA vectors and Martine De Cock for help in preparing the manuscript. The TAMU BAC clones were graciously provided by Marcel Salanoubat (Génoscope). Y. Moreau contributed to this work while on leave at the Center for Biological Sequence Analysis, Danish Technical University, Lyngby, Denmark.
Footnotes
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2544504.
[Supplemental material is available online at www.genome.org. The pAgrikola sequence data have been submitted to GenBank under accession no. (AY568055) and the transcript profiling data to Array Express under accession nos. (E-MEXP-30 and E-MEXP-65). The GST amplicons and CATMA spotted microarrays are available from the Nottingham Arabidopsis Stock Centre (NASC; http://nasc.nott.ac.uk). NASC will also distribute the cloned GST repertoires. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: C. Helliwell, M. Salanoubat, and P. Waterhouse.]
References
- Alonso, J.M., Stepanova, A.N., Leisse, T.J., Kim, C.J., Chen, H., Shinn, P., Stevenson, D.K., Zimmerman, J., Barajas, P., Cheuk, R., et al. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653-657. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An, G., Ebert, P.R., Mitra, A., and Ha, S.B. 1988. Binary vectors. In Plant molecular biology manual (eds. S.B. Gelvin et al.), pp. 1-19. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- The Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796-815. [DOI] [PubMed] [Google Scholar]
- Azpiroz-Leehan, R. and Feldmann, K.A. 1997. T-DNA insertion mutagenesis in Arabidopsis: Going back and forth. Trends Genet. 13: 152-156. [DOI] [PubMed] [Google Scholar]
- Bentolila, S., Alfonso, A.A., and Hanson, M.R. 2002. A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proc. Natl. Acad. Sci. 99: 10887-10892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes, D.C., Zayed, A.M., Ascenzi, R., McCaskill, A.J., Hoffman, N.E., Davis, K.R., and Görlach, J. 2001. Growth stage-based phenotypic analysis of Arabidopsis: A model for high throughput functional genomics in plants. Plant Cell 13: 1499-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, G.G., Formanová, N., Jin, H., Wargachuk, R., Dendy, C., Patil, P., Laforest, M., Zhang, J., Cheung, W.Y., and Landry, B.S. 2003. The radish Rfo restorer gene of Ogura cytoplasmic male sterility encodes a protein with multiple pentatricopeptide repeats. Plant J. 35: 262-272. [DOI] [PubMed] [Google Scholar]
- Castelli, V., Aury, J.-M., Jaillon, O., Wincker, P., Clepet, C., Menard, M., Cruaud, C., Quétier, F., Scarpelli, C., Schächter, V., et al. 2004. Whole genome sequence comparisons and “full-length” cDNA sequences: A combined approach to evaluate and improve Arabidopsis genome annotation. Genome Res. 14: 406-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H. and Sharp, B.M. 2002. Oliz, a suite of Perl scripts that assist in the design of microarrays using 50mer oligonucleotides from the 3′ untranslated region. BMC Bioinformatics 3: 27.1-27.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, S., Creelman, R.A., Mullet, J.E., and Wing, R.A. 1995. Construction and characterization of a bacterial artificial chromosome library of Arabidopsis thaliana. Plant Mol. Biol. Rep. 13: 124-128. [Google Scholar]
- Clough, S.J. and Bent, A.F. 1998. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735-743. [DOI] [PubMed] [Google Scholar]
- Crowe, M.L., Serizet, C., Thareau, V., Aubourg, S., Rouzé, P., Hilson, P., Beynon, J., Weisbeek, P., Van Hummelen, P., Reymond, P., et al. 2003. CATMA: A complete Arabidopsis GST database. Nucleic Acids Res. 31: 156-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, S.A., Goodwin, S.F., Kelly, D.C., Wang, Z., Sözen, M.A., Kaiser, K., and Dow, J.A.T. 1996. Analysis and inactivation of vha55, a gene encoding the vacuolar ATPase B-subunit in Drosophila melanogaster reveals a larval lethal phenotype. J. Biol. Chem. 271: 30677-30684. [DOI] [PubMed] [Google Scholar]
- Desloire, S., Gherbi, H., Laloui, W., Marhadour, S., Clouet, V., Cattolico, L., Falentin, C., Giancola, S., Renard, M., Budar, F., et al. 2003. Identification of the fertility restoration locus, Rfo, in radish, as a member of the pentatricopeptide-repeat protein family. EMBO Rep. 4: 588-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley, A.M., Aach, J., Steffen, M.A., and Church, G.M. 2002. Measuring absolute expression with microarrays with a calibrated reference sample and an extended signal intensity range. Proc. Natl. Acad. Sci. 99: 7554-7559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferea, T.L. and Bowman, B.J. 1996. The vacuolar ATPase of Neurospora crassa is indispensable: Inactivation of the vma-1 gene in repeat-induced point mutation. Genetics 143: 147-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girke, T., Todd, J., Ruuska, S., White, J., Benning, C., and Ohlrogge, J. 2000. Microarray analysis of developing Arabidopsis seeds. Plant Physiol. 124: 1570-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Y., Kaiser, K., Wieczorek, H., and Dow, J.A.T. 1996. The Drosophila melanogaster gene vha14 encoding a 14-kDa F-subunit of the vacuolar ATPase. Gene 172: 239-243. [DOI] [PubMed] [Google Scholar]
- Haas, B.J., Volfovsky, N., Town, C.D., Troukhan, M., Alexandrov, N., Feldmann, K.A., Flavell, R.B., White, O., and Salzberg, S.L. 2002. Full-length messenger RNA sequences greatly improve genome annotation. Genome Biol. 3: research0029.1-0029.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen, G. and Guilfoyle, T. 2002. Auxin-responsive gene expression: Genes, promoters and regulatory factors. Plant Mol. Biol. 49: 373-385. [PubMed] [Google Scholar]
- Hannon, G.J. 2002. RNA interference. Nature 418: 244-251. [DOI] [PubMed] [Google Scholar]
- Hartley, J.L., Temple, G.F., and Brasch, M.A. 2000. DNA cloning using in vitro site-specific recombination. Genome Res. 10: 1788-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens, R.P., Edwards, E.A., Leyland, N.R., Bean, S., and Mullineaux, P.M. 2000. pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42: 819-832. [DOI] [PubMed] [Google Scholar]
- Helliwell, C. and Waterhouse, P. 2003. Constructs and methods for high-throughput gene silencing in plants. Methods 30: 289-295. [DOI] [PubMed] [Google Scholar]
- Hilson, P., Small, I., and Kuiper, M.T.R. 2003. European consortia building integrated resources for Arabidopsis functional genomics. Curr. Opin. Plant Biol. 6: 426-429. [DOI] [PubMed] [Google Scholar]
- Himanen, K., Vuylsteke, M., Vanneste, S., Alard, P., Boucheron, E., Vercruysse, S., Chriqui, D., Van Montagu, M., Inzé, D., and Beeckman, T. 2003. Transcript profiling of early lateral root initiation. Proc. Natl. Acad. Sci. 101: 5146-5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway, A.J., van Laar, R.K., Tothill, R.W., and Bowtell, D.D.L. 2002. Options available—from start to finish—for obtaining data from DNA microarrays II. Nat. Genet. Suppl. 32: 481-489. [DOI] [PubMed] [Google Scholar]
- Holsters, M., Silva, B., Van Vliet, F., Genetello, C., De Block, M., Dhaese, P., Depicker, A., Inzé, D., Engler, G., Villarroel, R., et al. 1980. The functional organization of the nopaline A. tumefaciens plasmid pTiC58. Plasmid 3: 212-230. [DOI] [PubMed] [Google Scholar]
- Hosack, D.A., Dennis Jr., G., Sherman, B.T., Lane, H.C., and Lempicki, R.A. 2003. Identifying biological themes within lists of genes with EASE. Genome Biol. 4: R70.1-R70.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, T.R., Mao, M., Jones, A.R., Burchard, J., Marton, M.J., Shannon, K.W., Lefkowitz, S.M., Ziman, M., Schelter, J.M., Meyer, M.R., et al. 2001. Expression profiling using microarrays, fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol. 19: 342-347. [DOI] [PubMed] [Google Scholar]
- Inoue, H., Nojima, H., and Okayama, H. 1990. High efficiency transformation of Escherichia coli with plasmids. Gene 96: 23-28. [DOI] [PubMed] [Google Scholar]
- Kazama, T. and Toriyama, K. 2003. A pentatricopeptide repeat-containing gene that promotes the processing of aberrant atp6 RNA of cytoplasmic male-sterile rice. FEBS Lett. 544: 99-102. [DOI] [PubMed] [Google Scholar]
- Kim, H., Snesrud, E.C., Haas, B., Cheung, F., Town, C.D., and Quackenbush, J. 2003. Gene expression analyses of Arabidopsis chromosome 2 using a genomic DNA amplicon microarray. Genome Res. 13: 327-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight, J. 2001. When the chips are down. Nature 410: 860-861. [DOI] [PubMed] [Google Scholar]
- Koizuka, N., Imai, R., Fujimoto, H., Hayakawa, T., Kimura, Y., Kohno-Murase, J., Sakai, T., Kawasaki, S., and Imamura, J. 2003. Genetic characterization of a pentatricopeptide repeat protein gene, orf687, that restores fertility in the cytoplasmic male-sterile Kosena radish. Plant J. 34: 407-415. [DOI] [PubMed] [Google Scholar]
- Komori, T., Ohta, S., Murai, N., Takakura, Y., Kuraya, Y., Suzuki, S., Hiei, Y., Imaseki, H., and Nitta, N. 2004. Map-based cloning of a fertility restorer gene, Rf-1, in rice (Oryza sativa L.). Plant J. 37: 315-325. [DOI] [PubMed] [Google Scholar]
- Koncz, C. and Schell, J. 1986. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204: 383-396. [Google Scholar]
- Li, F. and Stormo, G.D. 2001. Selection of optimal DNA oligos for gene expression arrays. Bioinformatics 17: 1067-1076. [DOI] [PubMed] [Google Scholar]
- Liscum, E. and Reed, J.W. 2002. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 49: 387-400. [PubMed] [Google Scholar]
- Lu, R., Malcuit, I., Moffett, P., Ruiz, M.T., Peart, J., Wu, A.-J., Rathjen, J.P., Bendahmane, A., Day, L., and Baulcombe, D.C. 2003. High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J. 22: 5690-5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozo, T., Dewar, K., Dunn, P., Ecker, J.R., Fischer, S., Kloska, S., Lehrach, H., Marra, M., Martienssen, R., Meier-Ewert, S., et al. 1999. A complete BAC-based physical map of the Arabidopsis thaliana genome. Nat. Genet. 22: 271-275. [DOI] [PubMed] [Google Scholar]
- Nielsen, H.B., Wernersson, R., and Knudsen, S. 2003. Design of oligonucleotides for microarrays and perspectives for design of multi-transcriptome arrays. Nucleic Acids Res. 31: 3491-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parinov, S., Sevugan, M., Ye, D., Yang, W.-C., Kumaran, M., and Sundaresan, V. 1999. Analysis of flanking sequences from Dissociation insertion lines: A database for reverse genetics in Arabidopsis. Plant Cell 11: 2263-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee, S.Y., Beavis, W., Berardini, T.Z., Chen, G., Dixon, D., Doyle, A., Garcia-Hernandez, M., Huala, E., Lander, G., Montoya, M., et al. 2003. The Arabidopsis Information Resource (TAIR): A model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 31: 224-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond, C.S., Glasner, J.D., Mau, R., Jin, H., and Blattner, F.R. 1999. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 27: 3821-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso, M.G., Li, Y., Strizhov, N., Reiss, B., Dekker, K., and Weisshaar, B. 2003. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53: 247-259. [DOI] [PubMed] [Google Scholar]
- Rouillard, J.-M., Herbert, C.J., and Zuker, M. 2002. OligoArray: Genome-scale oligonucleotide design for microarrays. Bioinformatics 18: 486-487. [DOI] [PubMed] [Google Scholar]
- Rozen, S. and Skaletsky, H. 2000. Primer3 on the WWW for general users and for biologist programmers. In Bioinformatics methods and protocols, Methods in Molecular Biology, Vol. 132 (eds. S. Misener and S.A. Krawetz), pp. 365-386. Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- Saeed, A.I., Sharov, V., White, J., Li, J., Liang, W., Bhagabati, N., Braisted, J., Klapa, M., Currier, T., Thiagarajan, M., et al. 2003. TM4: A free, open-source system for microarray data management and analysis. BioTechniques 34: 374-378. [DOI] [PubMed] [Google Scholar]
- Samson, F., Brunaud, V., Balzergue, S., Dubreucq, B., Lepiniec, L., Pelletier, G., Caboche, M., and Lecharny, A. 2002. FLAGdb/FST: A database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res. 30: 94-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer, R., Landgraf, J., Pérez-Amador, M., and Wisman, E. 2000. Monitoring genome-wide expression in plants. Curr. Opin. Biotechnol. 11: 162-167. [DOI] [PubMed] [Google Scholar]
- Schiex, T., Moisan, A., and Rouzé, P. 2001. EuGène: An eukaryotic gene finder that combines several sources of evidence. Lect. Notes Comput. Sci. 2066: 111-125. [Google Scholar]
- Schumacher, K., Vafeados, D., McCarthy, M., Sze, H., Wilkins, T., and Chory, J. 1999. The Arabidopsis det3 mutant reveals a central role for the vacuolar H+-ATPase in plant growth and development. Genes & Dev. 13: 3259-3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki, M., Narusaka, M., Kamiya, A., Ishida, J., Satou, M., Sakurai, T., Nakajima, M., Enju, A., Akiyama, K., Oono, Y., et al. 2002. Functional annotation of a full-length Arabidopsis cDNA collection. Science 296: 141-145. [DOI] [PubMed] [Google Scholar]
- Sessions, A., Burke, E., Presting, G., Aux, G., McElver, J., Patton, D., Dietrich, B., Ho, P., Bacwaden, J., Ko, C., et al. 2002. A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simillion, C., Vandepoele, K., Van Montagu, M., Zabeau, M., and Van de Peer, Y. 2002. The hidden duplication past of Arabidopsis thaliana. Proc. Natl. Acad. Sci. 99: 13627-13632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, N.A., Singh, S.P., Wang, M.-B., Stoutjesdijk, P.A., Green, A.G., and Waterhouse, P.M. 2000. Total silencing by intron-spliced hairpin RNAs. Nature 407: 319-320. [DOI] [PubMed] [Google Scholar]
- Speulman, E., Metz, P.L.J., van Arkel, G., te Lintel Hekkert, B., Stiekema, W.J., and Pereira, A. 1999. A two-component Enhancer-Inhibitor transposon mutagenesis system for functional analysis of the Arabidopsis genome. Plant Cell 11: 1853-1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman, M.R., Amasion, R.M., Young, J.C., Krysan, P.J., and Austin-Phillips, S. 2000. The Arabidopsis knockout facility at the University of Wisconsin-Madison. Plant Physiol. 124: 1465-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze, H., Schumacher, K., Müller, M.L., Padmanaban, S., and Taiz, L. 2002. A simple nomenclature for a complex proton pump: VHA genes encode the vacuolar H+-ATPase. Trends Plant Sci. 7: 157-161. [DOI] [PubMed] [Google Scholar]
- Taylor, N.G., Scheible, W.-R., Cutler, S., Somerville, C.R., and Turner, S.R. 1999. The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11: 769-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thareau, V., Déhais, P., Serizet, C., Hilson, P., Rouzé, P., and Aubourg, S. 2003. Automatic design of gene-specific sequence tags for genome-wide functional studies. Bioinformatics 19: 2191-2198. [DOI] [PubMed] [Google Scholar]
- Turner, S.R. and Somerville, C.R. 1997. Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 9: 689-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houdt, H., Bleys, A., and Depicker, A. 2003. RNA target sequences promote spreading of RNA silencing. Plant Physiol. 131: 245-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varotto, C., Richly, E., Salamini, F., and Leister, D. 2001. GST-PRIME: A genome-wide primer design software for the generation of gene sequence tags. Nucleic Acids Res. 29: 4373-4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhout, A.J.M., Sordella, R., Lu, X., Hartley, J.L., Temple, G.F., Brasch, M.A., Thierry-Mieg, N., and Vidal, M. 2000. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science 287: 116-122. [DOI] [PubMed] [Google Scholar]
- Waterhouse, P.M. and Helliwell, C.A. 2003. Exploring plant genomes by RNA-induced gene silencing. Nat. Rev. Genet. 4: 29-38. [DOI] [PubMed] [Google Scholar]
- Wesley, S.V., Helliwell, C.A., Smith, N.A., Wang, M., Rouse, D.T., Liu, Q., Gooding, P.S., Singh, S.P., Abbott, D., Stoutjesdijk, P.A., et al. 2001. Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27: 581-590. [DOI] [PubMed] [Google Scholar]
- Wolfinger, R.D., Gibson, G., Wolfinger, E.D., Bennett, L., Hamadeh, H., Bushel, P., Afshari, C., and Paules, R.S. 2001. Assessing gene significance from cDNA microarray expression data via mixed models. J. Comput. Biol. 8: 625-637. [DOI] [PubMed] [Google Scholar]
- Wortman, J.R., Haas, B.J., Hannick, L.I., Smith Jr., R.K., Maiti, R., Ronning, C.M., Chan, A.P., Yu, C., Ayele, M., Whitelaw, C.A., et al. 2003. Annotation of the Arabidopsis genome. Plant Physiol.. 132: 461-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, D., Li, G., Wu, L., Zhou, J., and Xu, Y. 2002. PRIMEGENS: Robust and efficient design of gene-specific probes for microarray analysis. Bioinformatics 18: 1432-1437. [DOI] [PubMed] [Google Scholar]
- Yamada, K., Lim, J., Dale, J.M., Chen, H., Shinn, P., Palm, C.J., Southwick, A.M., Wu, H.C., Kim, C., Nguyen, M., et al. 2003. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302: 842-846. [DOI] [PubMed] [Google Scholar]
- Yang, Y.H., Dudoit, S., Luu, P., Lin, D.M., Peng, V., Ngai, J., and Speed, T.P. 2002. Normalization for cDNA microarray data: A robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, T. and Wang, X. 2000. Large-scale profiling of the Arabidopsis transcriptome. Plant Physiol. 124: 1472-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://genomics.bio.uu.nl; the department of Molecular Genetics at the Utrecht University provides information about the microarray printing facility and databases of the CATMA and TFC microarrays that are produced in-house.
- http://genoplante-info.infobiogen.fr/FLAGdb/; the FLAGdb Arabidopsis genome annotation database with flexible graphical display of structural and functional data.
- http://nasc.nott.ac.uk/; the Nottingham Arabidopsis Stock Centre (NASC) distributes CATMA arrays and CATMA GST PCR amplicons, and will provide E. coli strains carrying GST entry or expression Gateway clones from the AGRIKOLA project.
- http://www.agrikola.org/; the Arabidopsis Genomic RNAi Knock-Out Line Analysis (AGRIKOLA) project.
- http://www.arabidopsis.org/; the Arabidopsis Information Resource (TAIR).
- http://www.arabidopsis.org/info/2010_projects/2003_Report.pdf; the multinational coordinated Arabidopsis thaliana functional genomics project, annual report 2003, edited by the Multinational Arabidopsis Steering Committee (MASC).
- http://www.catma.org/; the Arabidopsis GST database.
- http://www.microarrays.be/; the MicroArray Facility (MAF) of the Flanders Interuniversity Institute for Biotechnology (VIB) provides hybridization services based on CATMA microarrays.
- http://www.psb.rug.ac.be/CAGE/; the Compendium of Arabidopsis Gene Expression (CAGE) project.
- http://www.psb.ugent.be/bioinformatics/genomes_ath_index.php; Arabidopsis genome annotation project at the Bioinformatics and Evolutionary Genomics group, (VIB—Ghent University).
- http://www.unil.ch/ibpv/microarrays.htm; the functional genomics project of the Department of Plant Molecular Biology of the University of Lausanne presents expression data for plant defense-related research.
- ftp://ftp.tigr.org/pub/data/a_thaliana/ath1; The Institute for Genomic Research (TIGR) Arabidopsis genome annotation data, release 5.0.
- ftp://tairpub:tairpub@ftp.arabidopsis.org/home/tair/Genes/Gene_Ontology/; Gene Ontology (GO) annotations for Arabidopsis genes annotated by TAIR and TIGR according to the GO Consortium controlled vocabularies.