Abstract
Pseudomonas aeruginosa, a common inhabitant of soil and water, is an opportunistic pathogen of growing clinical relevance. Its genome, one of the largest among bacteria [5570 open reading frames (ORFs)] approaches that of simple eukaryotes. We have constructed a comprehensive gene collection for this organism utilizing the annotated genome of P. aeruginosa PA01 and a highly automated and laboratory information management system (LIMS)-supported production line. All the individual ORFs have been successfully PCR-amplified and cloned into a recombination-based cloning system. We have isolated and archived four independent isolates of each individual ORF. Full sequence analysis of the first isolate for one-third of the ORFs in the collection has been completed. We used two sets of genes from this repository for high-throughput expression and purification of recombinant proteins in different systems. The purified proteins have been used to set up biochemical and immunological assays directed towards characterization of histidine kinases and identification of bacterial proteins involved in the immune response of cystic fibrosis patients. This gene repository provides a powerful tool for proteome- and genome-scale research of this organism, and the strategies adopted to generate this repository serve as a model for building clone sets for other bacteria.
The highly adaptable bacterium Pseudomonas aeruginosa inhabits a wide range of environmental niches and causes disease in many species, including humans. This organism leads to a variety of infections in immunocompromised patients as well as destructive chronic respiratory disease in individuals with cystic fibrosis (Ratjen and Doring 2003). Its ability to evade antimicrobial therapy remains a central challenge for clinical infectious disease.
The coding capacity of the relatively large genome of P. aeruginosa, which exceeds 6000 kb for most strains, is thought to contribute to its versatility. P. aeruginosa thus has the potential for expressing a proteome comparable in size and complexity to simple eukaryotes. The genomic repertoire of P. aeruginosa specifies functions that promote survival in diverse environments from the soil and water to respiratory tissue and blood. Relevant to its role as an opportunistic human pathogen are a large number of genes encoding virulence factors, conserved in all strains (Wolfgang et al. 2003). Interestingly, like other bacteria with large genomes, P. aeruginosa displays numerous paralogs for a number of gene families, including regulatory factors, membrane transporters, secretion systems, and adhesins (Stover et al. 2000).
The difficulty in controlling P. aeruginosa infections using conventional antibiotic therapy results in part from an over-abundance of genetic determinants for drug effluxing systems in the genome, such as the presence of 12 members of the resistance-nodulation-cell division (RND) family of multisubunit efflux pumps. The reason for this apparent redundancy of related or identical functions is unclear. One possible explanation for the occurrence of such paralogs holds that they are all differentially regulated enabling survival in various environmental niches. Alternatively, the paralogs may allow for evolutionary fine-tuning of specific but related biochemical activities.
Given its importance as a human pathogen and as an example of a highly adaptable prokaryote, P. aeruginosa exemplifies a useful model organism for the high-throughput (HT) functional analysis of proteomes. The creation and validation of a comprehensive collection of expression-ready genes would provide an important starting point for this approach by enabling the parallel analysis of many or all proteins in a broad variety of experiments. This gene or open reading frame (ORF) collection would allow researchers to: screen for functional activities of interest (Zhu et al. 2001; Phizicky et al. 2003), generate the raw material for various structural analyses, identify immunodominant antigens as well as protective antigens for the development of vaccines (Masignani et al. 2002; McKevitt et al. 2003), and generate network and interaction maps (Uetz et al. 2000; Gavin et al. 2002; Li et al. 2004), and will ultimately serve to annotate the function of the proteome in a manner not apparent from the analysis of gene sequences alone.
An ideal collection of such clones would embody the following virtues: the clone set should be (1) comprehensive; (2) flexible and amenable for many experimental systems—allowing expression from various promoters, on different plasmid backbones, and with peptide tags fused to the ends of the proteins if desired; (3) in protein expression-ready format (i.e., include only coding sequences); (4) catalogued and trackable—allowing for the rapid re-array of relevant subsets of clones as needed; and (5) clonally isolated and sequence verified—allowing the unambiguous assignment of function or property to a specific coding sequence. With such a catalogued clone set, saturating screens that survey the proteome can be performed with efficiency, hits can be identified easily, and, importantly, information regarding all clones (even those with no response) can be extracted and their significance assimilated.
In order to facilitate an understanding of the development of multidrug-resistant P. aeruginosa strains and to aid in the development of a useful vaccine, we have undertaken the development of a gene repository for this organism. This resource would facilitate research in all aspects of basic biology and pathogenesis as well as facilitate the development of novel strategies directed towards the understanding and clinical management of this organism. Here we describe the construction of the gene repository for this pathogen, based on the genome of P. aeruginosa PAO1. The 5570 annotated ORFs were amplified and cloned using a recombinational cloning system (Gateway, Invitrogen). Several expression platforms were tested for two sets of genes from the repository in order to validate the clones and to assess the feasibility of using the repository for HT biochemical and immunological studies.
RESULTS
Project Overview
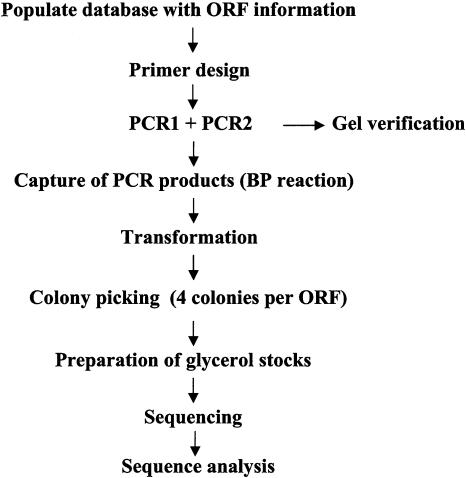
The creation of the P. aeruginosa gene repository occurred in three phases: (1) Preproduction and Development, (2) Production, and (3) Postproduction and Validation. The Preproduction and Development phase focused on several areas including importing and processing the relevant bioinformatics regarding the organism's annotated genome, predicting and organizing a complete set of PCR primers, establishing optimal biochemical conditions and standard operating procedures (SOPs) for the production phase, and settling on a final workflow. Figure 1A shows a schematic representation of the final workflow used in the creation of this repository. During the Production phase, we used our established automated clone production pipeline and laboratory information management system (LIMS), achieving a final throughput of four 96-well plates of genes per week. All processes were carried out in bar-coded containers and tracked by the Oracle-based database at both sample and plate levels along with any relevant process results (gel images, PCR success, colony count, etc.). The Postproduction and Validation phase focused on re-arraying the relevant clones, producing archival sets, and verifying the full-length sequences of the clones.
Figure 1.
Flow diagram of the process of constructing the P. aeruginosa gene repository indicating the major steps involved in the processes of database population, clone production, and validation.
Populating the LIMS with P. aeruginosa Data
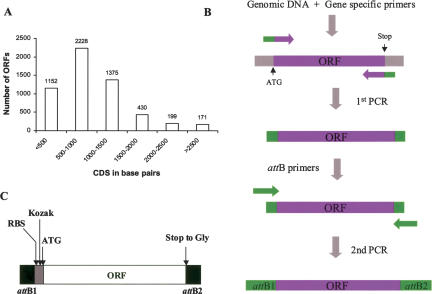
The first step in creating the repository was acquiring the genome sequence information and annotation for the complete set of predicted ORFs for this organism. Complete sequencing and analysis of the P. aeruginosa PA01 genome was completed and published in 2000 (Stover et al. 2000). The annotated genome predicted a coding capacity for 5570 ORFs, of which about half (54.2%) had a function predicted based on sequence; for the rest (45.8%), no function could be assigned (nearly a third of this group having orthologs in other bacteria). We obtained the sequence information for all of the 5570 predicted ORFs from the NCBI FTP site (NC_002516) and the corresponding gene annotation entries from the “Pseudomonas Genome Project” Web page (http://www.pseudomonas.com/) and used them to populate our existing FLEX database. The most relevant features represented in our database and associated with each ORF are CDS sequence (from START to STOP codons), CDS length, G+C content, GI number, and PA number (Pseudomonas unique and specific and alternative gene number). Additionally, a new and unique ID number, which is a unique identifier used by our LIMS, was assigned to each ORF. All these features facilitate the tracking, extraction, analysis, and clustering of data. Two peculiar features of this bacterial genome are the large number of ORFs (see Fig. 2A for ORF size distribution) and its extremely high GC content (as discussed below).
Figure 2.
Size distribution of the 5570 predicted P. aeruginosa PA01 ORFs (A). Schematic representation of the two-stage PCR reaction strategy used for amplification of the P. aeruginosa ORFs (B). Amplification of an ORF (purple) during the first PCR reaction and generation of the attB recombination sequences (green) during the first and second PCR reactions by use of overlapping primers are indicated. Schematic representation of the configuration of the amplified ORFs in the repository (C) showing attB1 and attB2 Gateway recombination sites, Ribosomal binding site (RBS), Kozak sequence. Normalized initiation codons (ATG) are indicated at the 5′ end of the ORF, and stop-Gly substitution at the 3′ end of the ORFs is also indicated.
Oligonucleotide Design and Gene Amplification
Amplification of the predicted ORFs was carried out by a strategy using two consecutive rounds of PCR amplification (Fig. 2B). The first round of PCR was executed using genomic DNA as template and a pair of matched 5′ and 3′ oligonucleotides containing gene-specific sequences and a short sequence matching part of the attB1 and attB2 sequences, respectively. In the second PCR reaction, the rest of the attB sites were added by using the product of the first PCR reaction as template along with primers containing complete attB1 and attB2 sequences which overlapped the first-round primers. This two-step PCR approach represents an alternative to one-step PCR that efficiently produces entry clones for the Gateway system. By requiring shorter primers for amplification of the target genes it significantly reduces the cost of the primers, which was the single most expensive reagent associated with the construction of this gene repository. More importantly, we found that using shorter oligos reduces the incidence of nucleotide misincorporations and `N - 1' products associated with synthesis of long oligonucleotides. Finally, detailed quality control could be run on a single lot of attB primers, which was then used for all reactions, thus ensuring consistent conditions.
Pairs of oligonucleotides for PCR amplification of each of the 5570 ORFs were automatically calculated by software developed in-house (www.hip.harvard.edu, click on Informatics then Programs) that uses the nearest-neighbor algorithm to create primer pairs with a specified Tm for the gene-specific sequences that are anchored at either end of the coding sequence followed by appending the partial attB tails. Based on the high GC content of this bacterial genome (66% GC-rich) and on preliminary data obtained during PCR optimization studies, the melting temperature for the gene-specific part of the primers was set at 65°C. The forward primers were designed to provide a uniform initiating codon (ATG) for all ORFs. In addition, sequences specifying a ribosome binding site (RBS) and an optimal Kozak sequence were introduced at the appropriate positions between the ATG and the recombination site (see Methods for representative examples of forward and reverse primers). The reverse primers were designed to replace the respective stop codon with the glycine codon (GGA), thus allowing read-through and the addition of C-terminal tags if desired. The primer design thus resulted in the following configuration: attB1-RBS-Kozak-ORF-attB2, with all the initiating codons normalized to ATG and all stop codons converted to GGA (Fig. 2B). A total of 11,140 oligonucleotides (in well-position matched sets of forward and reverse primer plates) were obtained (Operon). Depending on the protein expression vector used, this arrangement allows for the addition of N-terminal tags, C-terminal tags or both. It should be noted that because there is no stop codon, there is a minimum of eight amino acids that will always be appended to the C-terminal end of the protein, which correspond to the amino acids encoded by the recombination site. In addition, when N-terminal tags are desired, additional residues will be added between the end of the tag and the initiation codon of the gene. These residues correspond to the amino acids encoded by the recombinant site and by the sequences of the introduced RBS and Kozak sequences. Despite these anticipated compromises, we felt that this offered the most versatile configuration for this clone set.
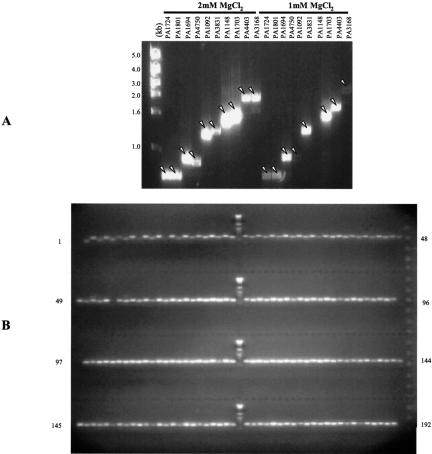
To establish optimal conditions for the PCR amplification step, a set of 10 Pseudomonas genes, representing a range of sizes, were selected and tested using 50 ng of genomic DNA from P. aeruginosa PA01 and DNA polymerases from different suppliers. We obtained the best success rate using the GC-Rich enzyme mix from Roche (data not shown). Additional optimization revealed that increasing the MgCl2 concentration to 2 mM provided better success and yields (see Fig. 3A). In preliminary tests, the nucleotide misincorporation rate for this enzyme at the higher magnesium concentration was equivalent to that obtained under standard conditions (1 mM MgCl2), establishing that the dramatically better PCR success outweighed concerns about reduced fidelity. Next, we validated the PCR protocol by targeting a full plate of genes with a CDS range from 200 to 2700 nucleotides: 100% success rates were obtained for both PCR and BP capture based on gel analysis of the PCR reactions and insert analysis of the clones resulting from the BP reactions, respectively (data not shown).
Figure 3.
PCR amplification of P. aeruginosa ORFs. GelStar-stained agarose gel showing PCR amplification of the test set (10 genes) under two different concentrations of MgCl2 (A). GelStar-stained agarose gel showing the PCR products from two 96-well plates from the production phase (B).
We then used these PCR conditions for the amplification of the 5570 ORFs. We monitored the PCR results for every reaction for the first twelve 96-well plates using an automated agarose gel system developed in the lab (with capacity for 192 Samples). Given the high PCR success rate (99%) and uniformity of the PCR reactions observed (see Fig. 3B), for the remainder of the reactions we used a sampling approach wherein randomly selected samples from each plate were run on diagnostic gels to monitor PCR success.
Cloning of PCR Products
The PCR products were captured in the Gateway entry vector pDONR201 by in vitro recombination between the attB sites flanking the coding sequences and the attP sites of the entry vector. Given the purity of the PCR products (Fig. 3B), an aliquot of the PCR reaction was transferred directly into the recombination reaction without further purification. Capture reaction conditions were identified that yielded 100% success with both the 10- and 94-gene tests, as confirmed by gene-specific PCR analysis of the resulting clones. During the production phase of the project, PCR insert analysis was not performed and the positive capture reactions were defined by the appearance of the expected numbers of transformant colonies (∼10-50). Transformation reactions were done in an automated fashion into T1-resistant DH5α cells and plated onto 48-sector agar plates. These specially designed plates can each culture half of the genes from a 96-well plate, and they are compatible with robotic plating and colony picking. Four colonies were selected for each ORF, and thereafter maintained separately. Selecting multiple independent transformants increases the likelihood of obtaining a mutation-free clone. The first round of cloning was accomplished by a single person in four months and yielded acceptable transformants for 96.7% of the genes (5387 genes; Table 1) and 21,548 glycerol stocks.
Table 1.
Summary of the Results From the P. aeruginosa Gene Repository Clone Production
| Cloning attempt | No. of genes cloned | % of ORFs | Time takeh (weeks) |
|---|---|---|---|
| Phase I | 5387 | 96.7 | 16 |
| Phase II | 5491 | 98.6 | 1 |
| Phase III | 5570 | 100 | 1 |
Number of ORFs that resulted in positive BP transformations, ORFeome coverage, and time involved in the three cloning phases of this project.
A second round of cloning was then initiated to capture the 183 genes that failed to generate clones during the first round of cloning. To determine whether these genes had failed the PCR amplification step, aliquots of the original PCR reactions were analyzed by agarose electrophoresis. Repeat capture reactions were run for the 60% of these that demonstrated PCR product, and for the remaining clones both PCR and capture reactions were repeated. This yielded 104 additional genes. In the third and final round of cloning, we identified and worked with those genes that failed all previous cloning attempts. This group contained some of the largest genes of this organism (genes larger than 5.5 Kb) and had escaped amplification during the two previous attempts. We successfully amplified them by extending the duration and number of PCR cycles for this group. Despite the PCR success, it was often difficult to capture the genes by recombination because of the decreased efficiency of the Clonase reaction to large inserts. Nevertheless, this third round of cloning eventually resulted in the amplification and capture of all the targeted genes and thus successfully completed the cloning phase of the project (Table 1). Three copies of the complete repository, each containing 22,280 clones, were generated and archived in three different locations, two in Boston and one in Marseille, France. In addition, subsets of genes have been pulled out of the collection at different times, characterized (as shown below) and distributed to various labs in both entry (pDONR201) and bacterial expression vectors for use in different assays.
Sequence Validation of Clones
The strategy to sequence validate the clones was as follows. The clones were re-arrayed to obtain a complete set of one isolate per gene. This left three remaining isolates for each gene in the archive that could be accessed for any failed isolates. The re-arrayed set was submitted for sequencing as pDONR201 entry clones using the Gateway universal sequencing primers (Invitrogen) by Agencourt. The resulting AB1 files were imported and evaluated by Phred software for base calling and quality clipping (Ewing and Green 1998). The sequences were then assembled by Phrap (http://www.phrap.org), and the resulting contigs were then analyzed by in-house software. The clone sequences, including the coding regions and 25 bases flanking each side, were aligned with the reference sequences, and all discrepancies were noted. Penalties were applied to each discrepancy according its consequences at both the nucleotide and polypeptide levels. For example, nonsense and frameshift mutations carry much higher penalties than missense and silent mutations. Discrepancies that occurred in regions of poor sequence quality were marked and further examined. Finally, a summary of all discrepancies for all clones was generated and the clones were categorized as pass, fail, or requires further review. In this manner, we have now analyzed the sequences for over 1900 genes from the repository (a third of the collection) and are currently processing the rest of the genes.
Our criteria for acceptance of clones were as follows. First, the entirety of the gene had to be fully covered by high-quality sequence reads (Phred quality scores ≥20). Second, the sequence had to correspond to the correct reference sequence. Third, the sequence could not contain any insertions, deletions, nonsense, or frameshift mutations. Clones containing discrepancies resulting in two or fewer silent and/or missense mutations were accepted and clearly annotated.
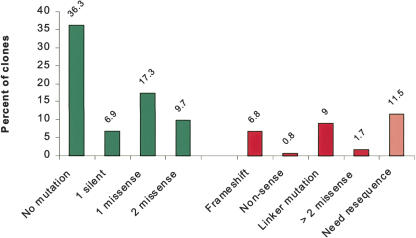
The first objective of the sequence analysis was to identify and retrieve acceptable clones. The second objective was to identify those clones that required additional sequencing, either to fill gaps or to analyze alternate isolates because the first isolate was rejected by the above criteria. Analysis of the sequencing data indicated that 89.0% of the clones were fully covered by high-quality sequence; 6.8% of the clones were only partially sequenced or lacked good-quality reads; and 4.2% of the clones did not yield sequence information due to lack of culture growth. The latter two groups were not considered further in the analysis. Of the clones with good sequence coverage, 99% corresponded to the correct reference sequence, and the remaining 1% all showed 30%-40% homology to the expected gene, but 100% homology to a different gene in the collection.
Of the clones that were fully sequenced, 80% were acceptable by the criteria listed. The results of the sequence analysis are shown in Figure 4. About half of the clones (52%) showed no discrepancies, and the other half (48%) contained discrepancies that result in silent or missense mutations. Unacceptable discrepancies that lead to gross changes in the polypeptide caused 17% of the fully sequenced genes to be rejected. We estimate the error rate of this enzyme to be ∼1 error for every 450 nucleotides. In addition, our analysis also indicated that mutations in the primer region accounted for 9% of the clone mutations. The capture reaction verified that these primer mutations do not affect recombination, and thus these clones were accepted or rejected according to their predicted effects on fusion polypeptides and the criteria above.
Figure 4.
Results of the sequence analysis of 1880 clones from the P. aeruginosa gene collection. Green bars correspond to the different categories of accepted clones. Red bars correspond to the categories of failed clones and pink bars correspond to those that require further sequence. Unless otherwise indicated the mutation analysis refers to the ORF region.
Using this strategy of analyzing one isolate per gene, we were able to accept clones for 70% of the genes submitted for sequencing. Preliminary sequence analysis from a test set of 208 genes (with CDS length up to 2500 bp) for which all four candidate clones were examined revealed a high probability (99%) of identifying an acceptable clone from the archived isolates (data not shown). We are in the process of completing the sequencing of the first candidate for the remaining genes in the collection and the remaining candidates for those genes with unacceptable or uncovered clones from the first round of sequencing.
Protein Expression and Purification
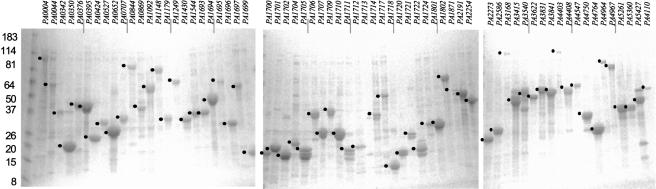
We selected two sets of genes from the repository to be used for the development of HT biochemical and immunogenicity studies. The first, the response regulator set, corresponds to the 71 genes encoding response regulator proteins of the P. aeruginosa two-component signal transduction systems (Supplemental Table I). This group shares similar structural and functional features but encompasses a wide range of molecular weights (14-167 KDa; Rodrigue et al. 2000), and was selected for use in the histidine kinase assays described below. A second set of 89 genes was selected for use in developing an immunogenicity assay. This set included genes encoding substrates and components of the type III secretion system and a number of membrane proteins (Supplemental Table I). The genes in both sets were re-arrayed, sequence verified, and subcloned into bacterial protein expression vectors that add polypeptide tags including N-terminal 6xHis- (pDEST17), C-terminal 6xHis- (pDEST42), and N-terminal GST- (pDEST-GST). The response regulator genes were also cloned in the eukaryotic expression vector pANT7cGST (N. Ramachandran, unpubl.) for use in the rabbit reticulocyte lysates expression system.
Protein expression in bacteria of the response regulator gene set with N-terminal 6xHis- and GST-tags resulted in 90% and 95% success rates, respectively, as judged by SDS-PAGE and Coomassie Blue staining of the total extracts (Fig. 5; Supplemental Table I). Under nondenaturing conditions we found that 75% of the GST-recombinant proteins could be purified from the soluble fraction, whereas only 30% of the 6xHis-tagged proteins were soluble. In contrast, under denaturing conditions 100% of the His-tagged proteins could be recovered and purified (Supplemental Table I). These data are in keeping with earlier results on the relatively poor efficiency of recovering soluble proteins tagged with the His tag compared to those with the GST tag (Braun et al. 2002).
Figure 5.
Expression of the P. aeruginosa response regulator gene set in E. coli. SDS-PAGE and Coomassie Blue staining of total bacterial extracts expressing 6xHis-tagged (A) or GST-tagged (B) response regulator proteins. Identities and characteristics of the proteins for this set are described in Supplemental Table I.
The use of cell-free protein expression systems offers an appealing alternative to in vivo expression, particularly in the HT context. As cell-free systems use pre-made and characterized extracts, they avoid the need to transform, grow, induce, and lyse cells. To determine the applicability of this technology for characterization of our clones, we have successfully adopted Yokoyama's protocol (Kigawa et al. 1995) for the production of bacterial S30 extracts and their use for protein expression (Murthy et al. 2004). We used a subset of 24 clones from the response regulator gene set (in pDEST17) to evaluate a bacterial cell-free expression system. Using 50-μL reactions in pre-aliquoted 96-well plates, we obtained equivalent results, in terms of success of expression, to those obtained in vivo (Murthy et al. 2004). The yields of protein from the cell-free reaction, as well as those recovered after performing one-step affinity purification, were much lower for the cell-free system. However, the use of the cell-free system for analytical purposes facilitates the protein expression and purification operation and significantly increases the throughput of the process.
The 89 genes in the second gene set (Supplemental Table II) were subcloned and expressed in BL21DE3 cells as N-terminal 6xHis-fusions. The relative success rates for expression of nonmembrane- and membrane-associated proteins were 91% and 59%, respectively (Supplemental Table II). Furthermore, the 90% success rate for expression of soluble proteins agrees with the rate obtained for the response regulator gene set described above (which is composed of soluble proteins). Figure 6 shows the IMAC purification results for the 67 expressed proteins that were employed in the ELISA assay described below.
Figure 6.
Proteins used in the ELISA assays. SDS-PAGE and Coomassie Blue staining of proteins expressed in bacteria as 6xHis-tagged recombinant proteins and subjected to Ni2+NTA purification. Identities and characteristics of the proteins for this set are described in Supplemental Table II.
Assays Using Recombinant Proteins
Developing a Histidine Kinase Substrate Assay
Bacteria use two-component systems to sense and respond to changes in their environments. In P. aeruginosa, a number of two-component systems have been shown to be involved in signal transduction, regulating the expression of diverse genes, particularly those encoding virulence factors. These include control of expression of fimbrial and flagellar components (Ishimoto and Lory 1992; Hobbs et al. 1993; Ritchings et al. 1995) and production of the alginate capsule (Deretic et al. 1989; Wozniak and Ohman 1991; Goldberg and Dahnke 1992). Several mechanisms of intrinsic antimicrobial drug resistance are also under the control of two-component systems (Macfarlane et al. 1999; Drenkard and Ausubel 2002; McPhee et al. 2003). Screens of P. aeruginosa mutants for attenuation of virulence in model systems of infection also identified two-component systems as regulators of unknown virulence traits (Tan et al. 1999; Potvin et al. 2003).
Prototypically, the two-component system is composed of two proteins, a response regulator and a histidine kinase (usually membrane-associated). Upon stimulation, the histidine kinase undergoes autophosphorylation at a histidine residue, followed by transfer of the phosphate to an aspartic acid on the response regulator. The phosphorylated response regulator stimulates the transcription of target genes by binding to their regulatory sequences (Stock et al. 2000). Genes encoding histidine kinases are commonly genetically linked to those of their cognate response regulators in defined operons. However, it is not known to what degree the histidine kinases can also phosphorylate alternative response regulators. There are 62 two-component systems predicted in the P. aeruginosa genome (Rodrigue et al. 2000), more than in any other characterized prokaryote. In addition to the operon-linked histidine kinase/response regulators, several unlinked “orphan” response regulators and one “orphan” histidine kinase have been identified. The creation of a comprehensive set of protein-encoding clones for this organism provided a unique opportunity to explore the specificity and activities of these important proteins.
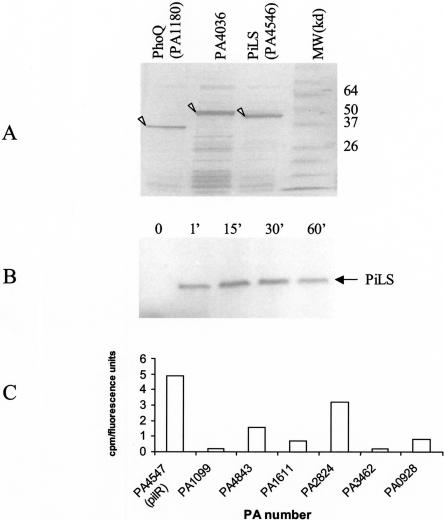
We sought to develop an in vitro system to screen for substrates of the response regulator kinases. In order to establish the conditions for detecting kinase activity, we chose three kinases: two well characterized kinases, PhoQ (PA1180) and PilS (PA4546), and the putative “orphan” kinase PA4036. Guided by prediction algorithms of secondary structures and transmembrane domains and experience with PilS (Boyd and Lory 1996), the genes for the three kinases were truncated by removing the transmembrane segments in order to enhance the chances of producing soluble proteins. All three kinases expressed well as 6xHis-N-terminal fusion proteins but remained in the insoluble fraction, requiring purification from inclusion bodies and re-folding (Fig. 7A). Autophosphorylation assays with these proteins demonstrated enzymatic activity for PilS (Fig. 7B) but not for PhoQ or PA4036 (data not shown).
Figure 7.
Histidine kinase assays. Coomassie Blue staining of the recombinant proteins refolded from inclusion bodies (A). Autoradiogram of PilS after undergoing autophosphorylation and SDS-PAGE (B). Phosphorylation of a subset of response regulators by PilS (C) expressed as cpm incorporated by the substrates (corrected by the amount of substrate in each reaction).
In order to generate substrates and conditions to measure PilS histidine kinase activity, we isolated and subcloned all of the 71 genes encoding for the predicted P. aeruginosa response regulators, including the bona fide substrate of this enzyme, PilR (Supplemental Table II; Fig. 5). For the kinase assays described here, all 71 response regulators were subcloned into a C-terminal GST-fusion vector with a T7 promoter (pANT7cGST) and were expressed using the rabbit reticulocyte system (Promega). Protein expression levels were assessed by western blot using anti-GST antibodies (Supplemental Fig. 1). In our initial experiments we immobilized a set of seven proteins to 96-well anti-GST plates to measure their ability to act as substrates of the PilS kinase. Figure 7C shows the relative levels of phosphorylation of the bona fide substrate of this enzyme, PilR, as well as that of the other response regulators after correcting for the amount of substrate used in the assay. As expected, our positive control, PilR, demonstrated a strong signal. Interestingly, another response regulator, PA2824, was phosphorylated nearly as strongly, and a third, PA4843, showed above-background incorporation of phosphate. These findings suggest that PilS (and potentially other histidine kinases) can phosphorylate alternative substrates at rates similar to that of their bona fide substrates. The physiological relevance of the phosphorylation of PA2824 by PilS is currently being evaluated at the genetic level.
Developing an Immune Response Assay
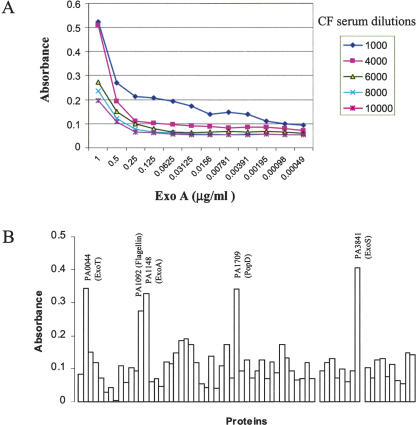
Given Pseudomonas' ability to evade antimicrobial therapy, significant emphasis has been placed on identifying an effective vaccine. Unfortunately, the identification of appropriate antigens has remained elusive. The assembly of this collection of clones offers the opportunity to screen for antigens at the proteome level. To establish conditions for this analysis we selected ExoA (PA1148), one of the proteins in this set previously identified to elicit humoral immune response in cystic fibrosis (CF) patients colonized by P. aeruginosa (Brauner et al. 1993; Moss et al. 2001; Banwart et al. 2002). We used this protein and sera from CF patients and noninfected control individuals to identify conditions that specifically detect this reactivity. Figure 8A shows the titration curves of one of the CF sera against different amounts of recombinant ExoA (PA1148). Based on these results, we tested the reactivity of sera of several CF patients and that of a normal control at 1:4000 dilutions against all of the 73 proteins shown in Figure 6. Figure 8B shows the relative reactivity of the CF17 serum against these proteins after subtracting the reactivity obtained with the normal control sera. Not only ExoA (PA1148) showed significant levels of reactivity with the patient sera; we were also able to identify other apparent immunodominant antigens such as ExoT (PA0044), ExoS (PA3841), flagellin (PA1092), and PopD (PA1709). Notably, the pattern of reactivity was similar among the three patient sera analyzed, with the latter four proteins being identified by all three sera (data not shown). All of these proteins are either surface localized or secreted, further confirming that extracellular location of proteins is a key factor in immune recognition of bacterial antigens by the infected host. ExoA (PA1148) is a toxin delivered by the Type II secretion system, ExoS (PA3841) and ExoT are effectors of the type III secretion system, PopD (PA1709) belongs to the type III delivery system, and flagellin (PA1092) is a flagellar filament protein.
Figure 8.
ELISA assay using P. aeruginosa recombinant. Titration curves using serum from a CF patient and various amounts of ExoA (A). Results from the ELISA of 73 P. aeruginosa proteins using serum from a CF patient and normalized by the results obtained with normal serum (B).
DISCUSSION
Genome sequencing projects have produced an immense amount of information regarding the organization, evolution, and coding capacity of the various genomes covered. All this has propelled a trend in biology to address scientific questions on a whole-cell or system-wide basis, in contrast to the classical reductionistic approaches. There is an ever-increasing need to develop tools and resources that can build on the genome information and facilitate the understanding of the biological function of the thousands of uncharacterized gene products that have been identified through sequencing. Technologies such as DNA microarrays have been developed and successfully applied to analysis of gene expression at the RNA level (Brown and Botstein 1999). However, progress in more ambitious areas, such as comprehensive in vivo and in vitro analysis of protein function, protein-protein interactions, or dissection complex signaling or developmental pathways, has been until very recently limited by the availability of state-of-the-art resources that make use of the recently gained genome information and novel cloning technologies (Uetz et al. 2000; Brizuela et al. 2001; Gavin et al. 2002; Reboul et al. 2003; Li et al. 2004).
The use of bacteria as model systems to begin developing HT tools for studying protein function offers a number of advantages. More than 600 bacterial genomes have been sequenced or are near completion (http://wit.integratedgenomics.com/GOLD), leading to excellent tools and predictive methods for identifying protein-encoding ORFs. The genetic tractability and manageable genome size of bacteria facilitate experimentation and hypothesis testing. Moreover, the study of Pseudomonas in particular offers the opportunity to understand its virulence, its ability to evade antimicrobials, and its interaction with the host response. Nearly half of its predicted ORFs have no assigned function, and novel tools are needed to identify both vaccine candidates and new targets for therapeutic intervention. The creation of a repository of well characterized clones provides an opportunity to develop these tools and employ them in discovery.
We used the annotated P. aeruginosa PA01 genome sequence (Stover et al. 2000) and an automated and LIMS-supported production line to construct a gene repository for this human pathogen. The ORFs in this repository are cloned in a recombination-based cloning system (Gateway) to allow HT manipulation of the ORFs and expression of proteins with alternative tagging schemes. After three successive rounds of amplification and capture, transformants were obtained for all 5570 predicted ORFs. Sequence analysis of a single isolate for each of 1900 ORFs (one-third of the total) yielded an overall success rate of ∼70% of all tested isolates. The 30% failure rate illustrates the importance of selecting multiple isolates for each attempted ORF and the need for careful quality control to ensure that acceptable clones are identified and that all clones are annotated. To estimate the overall likelihood of obtaining a good clone for each ORF, we can consider that for each failed ORF, there are three remaining isolates. If we assume that the remaining isolates also have an overall failure rate of ∼30%, then sequencing the three remaining isolates for each failed ORFs should yield an acceptable clone 97% of the time (1 - 0.303 = 0.97). This is in good agreement with the results obtained from 208 genes where all four isolates were analyzed and for which we obtained an acceptable clone for 206 of the genes (99%).
The mutation rate of 1 error every 450 bases observed for this project is notably higher than we have observed in other cloning projects in our lab. This high mutation rate is a result of the necessity to use a polymerase and PCR conditions that allowed the amplification of the high-GC-content ORFs of this organism. On the other hand, the intrinsic organization of the P. aeruginosa genome (as well as those of other prokaryote organisms) facilitated several of the steps involved in the construction of the repository. The high density of coding regions and the absence of introns in the bacterial genomes allowed the use of genomic DNA preparations as template for the PCR amplification of genes. Genomic DNA is an ideal PCR template because it is perfectly normalized and easy to prepare in high quantities. Thus, ample template can be added to all reactions allowing for a reduction in amplification cycles, which in turn reduces the frequency of mutations. PCR conditions were identified that gave a very high hit rate, ∼99%, and for which there were virtually no aberrant products (Fig. 3). This may have been due to the relatively high melting temperature used for the hybridization cycle due to the high GC content. The high specificity of the PCR obviated the need to purify the PCR products before the capture reaction, thus increasing the throughput considerably. Finally, the average ORF size for bacterial genomes is smaller than that of eukaryote organisms, facilitating the amplification of the ORFs and reducing the error rate.
The transfer of over 150 genes of interest to several protein expression vectors demonstrates the ease with which these clones can be used to subclone into virtually any vector. Expression levels for these recombinant proteins as hybrid proteins fused at their N-termini to 6x-His or GST are comparable to those observed in the lab for clones from other collections produced in the Gateway system such as our human clones (Braun et al. 2002).
Furthermore, we have demonstrated the feasibility of using these expressed proteins in two important aspects of therapeutic discovery regarding Pseudomonas. In the first, an HT format for immunodetection assays was used to probe the sera of patients with CF for the presence of antibodies to P. aeruginosa proteins. Sera from CF patients displayed immunoreactivity against a number of the tested P. aeruginosa proteins that were not detected in noninfected individuals. These included reactivity to several components of the type III secretion apparatus located on the outer surface of bacteria, as well as secreted proteins such as ExoS and ExoA. Experiments are currently underway to expand the scale of this approach in order to examine the proteome of Pseudomonas for potential immundominant antigens. Correlating the immune response to specific components of the bacterial proteome with the clinical condition of individual patients may serve as a useful guide in selecting proteins suitable for vaccine development.
In a second approach, we used active purified PilS histidine kinase to probe a collection of response regulators of the two-component signal transduction system to identify potential substrates. In addition to its bona fide substrate PilR, two other putative substrates were identified. Based on the existing annotation for these genes, one of the phosphorylated proteins (PA 2824) is a probable sensor/response regulator hybrid, and the second one (PA 4843) is a probable two-component response regulator. Further disruptions of these genes and analysis of the mutants in the context of PilS stimulation is underway to validate these findings.
It is apparent that added power can be brought to the body of information generated by various genome-sequencing projects by using this information to create tangible tools to study the functions of the proteome. With a resource that enables the expression of all proteins in the proteome, it will be possible to execute experiments that probe the myriad of protein activities extant in the cell, providing an opportunity to functionally annotate many of the predicted ORFs for which there is no assigned biochemical function. Here we report the creation of one such resource, a gene repository for P. aeruginosa, an opportunistic human pathogen. This type of repository will greatly accelerate research in various areas of functional proteomics. Moreover, this resource can provide important tools for investigations in more applied fields of vaccines and anti-infective agents. This type of gene repository will also facilitate the identification of protective antigens by “reverse vaccinology” and “genetic vaccination” approaches (Masignani et al. 2002). Similarly, expressed proteins can be used in antibiotic discovery research by identifying small molecules that interact with specific components of the pathogen's proteome, a subset of which could result in inhibitors of essential functions. It is very likely that we will see in the near future an outpouring of similar repositories for other bacterial pathogens, providing essential components of the systems biologist's toolbox.
METHODS
Preparation of Genomic DNA from Pseudomonas aeruginosa
Pseudomonas aeruginosa PA01 was streak-purified in an LB/agar plate, and a single colony was isolated and grown overnight at 37° C in 15 mL of LB. The cells were harvested by centrifugation, and the resulting pellet was resuspended and washed twice with TE (pH 8.0). The cells were then resuspended in 5 mL of TE and lysed by adding 50 μL of 20% SDS solution and gently inverting the tubes a few times. Proteins in the lysate were digested by adding 50 μL of 20 mg/mL proteinase K solution, followed by incubation at 37° C for 1 h. The lysate was extracted with phenol, phenol/chloroform/iso-amyl alcohol, and chloroform. Genomic DNA was precipitated by gentle adding and mixing 2 volumes of ethanol (EtOH). Strings of genomic DNA were harvested with a glass pipette and then extensively washed with a 70% ethanol solution. Finally, the DNA was air dried and dissolved in TE. The concentration of DNA was determined by OD at 260 nm.
Oligonucleotide Design and Synthesis
Gene-specific primers with matching Tms (65°C), for the PCR1 reaction, were calculated for each targeted ORF in a batch mode using a modified nearest-neighbor algorithm (Sugimoto et al. 1995). attB sequences were automatically appended to the gene-specific part of the oligonucleotides. This oligonucleotide calculator is integrated into our in-house laboratory information management system (LIMS) but can also be used as a stand-alone program (which is available at www.hip.harvard.edu, click on Informatics then Programs).
An example of a typical set of gene-specific primers is given below. The gene-specific portion of the oligos is underlined. The rest of the sequence corresponds to the partial attB sequences. Initiation (ATG) codon and complementary sequence for Gly codon (substituting the natural STOP codon) are indicated in bold:
Forward primer: 5′-AAAGCAGGCTCCGAAGGAGATAC CATGAGCGAAACGCCCTCCCCCGC-3′
Reverse primer: 5′-AGAAAGCTGGGTCTCCGCCGCACAGATAGGTACCGGTAC-3′
The primer sequences were arranged automatically on corresponding plates, one 5′ set and one 3′ set, with each primer sequence pair on the same position for each plate. Next, 50 nmol scale synthesis of the oligonucleotides was carried out by OPERON, and oligos were delivered in normalized 50 μM solutions in bar-coded 96-well plates.
A unique set of primers was used for all of the PCR 2 reactions in order to reconstruct the attB sites. Regions of overlap with the PCR 1 primers are underlined.
attB1primer: 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCCGAAGGAGAT-3′
attB2 primer: 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTCC-3′
Amplification of ORFs
PCR amplification reactions were performed in sets of four 96-well plates at a time, using a DNA Engine tetrad (MJ Research). The first PCR reaction was carried out in 30 μL volumes containing 100 ng of chromosomal DNA, 0.5 μM forward and reverse primers, 1 Unit of GC-Rich enzyme (Roche), 200 μM of each dNTP, 1 × GC-Rich Buffer #2, 0.5 M GC-Rich Buffer #3, and MgCl2 adjusted to 2 mM. An initial denaturation step at 94°C for 4 min was followed by 15 cycles of denaturation at 94°C for 1 min, annealing at 65°C for 45 seconds and elongation at 72°C for 5 min. An additional incubation at 72°C for 10 min was then added.
The second PCR reaction was carried out in a 50 μL volume containing 3 μL from the first PCR reaction (as template), 0.5 μM attB primers, 1 U GC-Rich Enzyme (GC-RICH PCR System from Roche), 200 μM of each dNTP, 1 × GC-Rich Buffer #2, 0.5 × GC-Rich Buffer #3, and MgCl2 adjusted to 2 mM. Thermocycling was performed using an initial denaturation step at 94°C for 4 min, followed by 15 cycles of denaturation at 94°C for 1 min, annealing at 65°C for 45 sec, and elongation at 72°C for 5 min. A final incubation at 72°C for 10 min was then added.
Capture of Amplified ORFs
Capture of the amplified ORFs into the entry vector pDONR201 was performed by carrying out 10 μL BP reactions in 96-well plates overnight at 25°C. Composition of the LR reactions was as follows: 4 μL of the second PCR reaction, 100 ng of pDONR 201, 1 μL of BP reaction buffer, and 2 μL BP Clonase (Invitrogen).
Preparation of E. coli-Competent Cells
DH5α T1-resistant cells (Invitrogen) cells were grown to mid-log phase in TB at 37°C, and the resulting cell pellet was resuspended with 1/20th of the original culture volume of ice-cold LB transformation mix (LB pH 6.1 containing 10% (w/v) PEG3350 (Sigma), 5 % (v/v) DMSO (Sigma), 10 mM MgCl2, 10 mM MgSO4, 14 % (v/v) glycerol. The cell suspension was then aliquoted into Eppendorf tubes, snap frozen in dry ice ethanol bath, and stored at -80°C.
Transformation of PEG/DMSO-Competent Cells and Isolation of Clones
Thirteen μL of freshly thawed competent cells were added into the wells of 96-well plates (PCR microplate from Axygen) containing 10 μL of KCM buffer (100 mM KCL, 30 mM CaCl2, and 50 mM MgCl2) and 2.5 μL BP reactions. Plates were incubated on ice for 20 min and then at room temperature for an additional 10 min; 100 μL of TB was then added to each well and the plates were incubated at 37°C for 30 min with agitation (375 rpm). Fifty μL of the transformation mix was then aliquoted onto 25 cm × 25 cm LB plates that had been sectioned into 48 grids. LB-agar in the plates contained 50 μg/mL kanamycin. Transformation plates were loaded onto the MegaPix (Genetix) colony picker. Four independent colonies from each of the 48 LR reactions on a given plate were automatically picked and inoculated into 96-well plates with LB supplemented with 50 μg/mL kanamycin.
Subcloning of ORFs Into Expression (Destination) Vectors
LR reactions were performed in 10 μL volume reactions containing 150 ng destination vector, 1 × LR Clonase buffer, 1 μL LR Clonase (Invitrogen), 0.25 μL topoisomerase, and 1.0 μL of the corresponding entry clone DNA miniprep (QIAGEN). Reactions were incubated 1-3hat25°C, and 2.5 μL of the reaction mix was transformed onto DH5α-competent cells. The cells were selected for the antibiotic resistance marker present in the destination vector (usually ampicillin).
Bacterial Expression and Purification of Proteins
Cell growth, transformation, and protein purification for in vivo production of His- and GST-tagged proteins was performed according to the protocols developed in our laboratory and described elsewhere (Braun et al. 2002). Briefly, purification of His-tagged proteins under denaturing conditions was carried out by growing BL21 star (DE3) pLysS transformants at 37°C as 1-mL cultures in 96-well blocks (Marsh Biomedical), to an O.D600 of ∼0.7 and induced with 1 mM IPTG. After allowing a postinduction growth of 4 h, the cells were harvested, lysed with the lysis buffer (100 mM NaH2PO4, 10 mM Tris-HCl, 6 M guanidinium-HCl, 10% glycerol, 10 mM 2-mercaptoethanol, pH 8.0) and allowed to bind to Ni2+ NTA resin prewashed with lysis buffer. The resin with the bound protein was washed twice with wash buffer (100 mM NaH2PO4, 10 mM Tris-HCl, 8M urea, pH 8.0). The bound protein was eluted in 80 μL of elution buffer (wash buffer containing 500 mM imidazole, pH 8.0). Next, 12.5 μL of the eluate was treated with 12.5 μL of 2 × SDS sample buffer and subjected to SDS-PAGE. The protein bands were visualized by staining with Coomassie Blue.
Western Blot Analysis
Two μL of total cell-free reactions after protein synthesis was subjected to SDS-PAGE, followed by western blot analysis. Briefly, the proteins were transferred onto a PVDF membrane, and the membrane was blocked with 3% BSA for 1 h. The membrane was then probed with anti-GST antibodies (1:2000, Amersham Pharmacia) for 2 h. The blot was washed three times with PBST (PBS buffer plus 0.05% Tween 20) and incubated with HRP-linked anti-mouse secondary antibodies (1:5000, Amersham Pharmacia) for 1 h. After three washes with PBST, the protein bands were detected by chemiluminescence.
Construction and Production of Histidine Kinase Deletion Mutants
Truncated variants of PilS, PhoQ, and PA4036 were constructed using the same general amplification and cloning strategies used for cloning genes in the repository. Regions of the genes encoding predicted transmembrane helices were identified based on our previous results (Boyd and Lory 1996) and/or by using various resources to predict secondary structures and transmembrane domains (http://bioweb.pasteur.fr/seqanal/interfaces/toppred and http://cubic.bioc.columbia.edu/predictprotein). Deletion mutants coding for protein lacking amino acids 1-205 (PilS), 1-190 (PhoQ), and 1-377 (PA4036) were cloned in pDEST17. Recombinant proteins were expressed and recovered from inclusion bodies as described for other histidine kinases (Uhl and Miller 1996). Briefly, BL21-DE3 (LysS) transformants were grown to mid-log phase (OD600 of 0.6-0.8), and expression was induced with 1 mM IPTG. Cultures were grown for an additional 3 h, pelleted, and washed with wash buffer #1 (20 mM HEPES, pH 7.4, 50 mM KCl, 10% glycerol). The cells were resuspended in 0.02 vol of wash buffer #1 and lysed by sonication. The lysate was centrifuged, and the pellet was washed with wash buffer #2 (10 mM HEPES, pH7.2, 50 mM KCl, 0.1% Triton X-100), resuspended in denaturation buffer (6 M guanidine-HCl, 50 mM Na2HPO4, pH7.2), and dialyzed against 200 × volume of renaturation buffer [(20 mM HEPES, pH 7.4, 50 mM KCl, 10 mM MgCl2,5mM dithiothreitol (DTT), 50% glycerol] for 18 h without stirring. The renaturation buffer was changed and dialysis continued for another 18 h with stirring. The resulting fraction of soluble protein was then recovered and stored at -80°C in small aliquots for future use.
Kinase Assays
Autophosphorylation reactions were performed by incubating the enzyme preparations in 30 μL volumes containing 30 mM Tris, pH 7.5, 10 mM MgCl2, 1 mM DTT, 10 mM ATP, and 30 μCi [γ-33 P] ATP (5000 Ci/mmol, ICN). At the indicated time intervals, 5 μL of the reaction were removed and the reaction was stopped by addition of Laemmly sample buffer (LSB) containing 0.05 M EDTA. Samples were then separated by SDS-PAGE. Gels were fixed in a cold solution composed of 50% methanol, 10% acetic acid, then rinsed briefly in cold water and dry gel on 3 MM paper (Whatman), followed by autoradiography.
Phosphorylation of the response regulator proteins was carried out by incubating PilS with GST-tagged response regulator proteins immobilized in the wells of 96-well plates.
Response regulators in a Gateway converted-pCITE-GST were expressed using a rabbit reticulocyte system (Promega). Then, 15 μL of the extracts were transferred to individual wells of a 96-well plate coated with anti-GST antibodies (Amersham) and incubated at 15°C for 2 h. Plates were then washed with PBS and incubated with blocking buffer (5% nonfat dry milk, 0.2% Tween 20 in PBS). Amounts of proteins bound to the wells were estimated by incubating the wells with monoclonal anti-GST antibodies (Cell Signalling), followed by detection with anti-mouse HRP-conjugated IgG antibodies. (Amersham) using pico ELISA ECL reagents (Pierce).
Phosphorylation of the immobilized proteins was carried out by adding 50 μL of autophosphorylated PilS to each well and allowing the reaction to proceed for 30 min. Conditions of the assay were exactly as described above for autophosphorylation reactions. Phosphorylation was terminated by removing the kinase solution and washing the wells with PBS/0.05% Tween 20. The incorporated radioactivity was then determined by scintillation counting and corrected by the amount of protein present in each well (as determined by the anti-GST determination described above).
ELISA Assays
Ninety-six-well (Nunc-Maxisorp) plates were coated with the indicated amounts of recombinant proteins in 100 μL of PBS buffer (pH 7.4) and incubated overnight at 4°C. The plates were washed three times with PBST (PBS buffer plus 0.05% Tween 20) and then incubated with blocking buffer BB (PBS + 3% BSA + 0.05% Tween 20) for 1 h at room temperature. The plates were washed three times with wash buffer (WB; PBS + 1% BSA + 0.05% Tween 20). The wells were incubated with human sera at 1:4000 dilution in PBS (unless otherwise indicated) for 1 h and washed three times with WB. The plates were then incubated with an HRP-conjugated secondary antibody solution (goat anti-human IgGHRP at 1:2000 dilution in PBS) for 1 h and washed five times with WB and once with PBS. HRP activity was developed with Super Signal ELISA development reagent (Pierce) for 20 min at room temperature, and the ODs at 425 nm were read and plotted.
LIMS
The production LIMS has a three-tier architecture: data storage, business logic, and presentation layers. The database on the backend is Oracle 8i, and stores information such as sequences, clone samples, plates, oligos, processes, protocols, projects, workflows, process results, users, user requests, etc. The middle tier of the system holds the business logic. It encapsulates business rules within an object-oriented model. It is implemented using Java, Java Servlets, Java Server Pages (JSP), the Struts framework, Tomcat, etc., and is J2EE compliant. The database core, application programs, and Web server are housed in a Sun E450 enterprise class server. In addition to the standard Oracle rollback capability, the database is backed up nightly to both hard disk and tape. Because the presentation layer is HTML, users can access the application from anywhere with an internet connection.
Acknowledgments
We thank all past and present members of the Institute of Proteomics who have contributed to the development of the techniques that made this work possible. Special thanks to Dr. Gerald Marsischky for helpful discussions and advice all along this project. We thank Andreas Rolfs for organizing the rearray and submission of clones for sequencing. This work has been supported by grant LORY00V0 from the Cystic Fibrosis Foundation and by funds from the Harvard Medical School.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2482804.
References
- Banwart, B., Splaingard, M.L., Farrell, P.M., Rock, M.J., Havens, P.L., Moss, J., Ehrmantraut, M.E., Frank, D.W., and Barbieri, J.T. 2002. Children with cystic fibrosis produce an immune response against exoenzyme S, a type III cytotoxin of Pseudomonas aeruginosa. J. Infect. Dis. 185: 269-270. [DOI] [PubMed] [Google Scholar]
- Boyd, J.M. and Lory, S. 1996. Dual function of PilS during transcriptional activation of the Pseudomonas aeruginosa pilin subunit gene. J. Bacteriol. 178: 831-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, P., Hu, Y., Halleck, A., Shen, B., Harlow, E., and LaBaer, J. 2002. High-throughput purification of human proteins from bacteria. Proc. Natl. Acad. Sci. 99: 2654-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauner, A., Cryz, S.J., Granstrom, M., Hanson, H.S., Lofstrand, L., Strandvik, B., and Wretlind, B. 1993. Immunoglobulin G antibodies to Pseudomonas aeruginosa lipopolysaccharides and exotoxin A in patients with cystic fibrosis or bacteremia. Eur. J. Clin. Microbiol. Infect. Dis. 12: 430-436. [DOI] [PubMed] [Google Scholar]
- Brizuela, L., Braun, P., and LaBaer, J. 2001. FLEXGene Repository: From sequenced genes to gene repositories for high-throughput functional biology and proteomics. Mol. Biochem. Parasitol. 118: 155-165. [DOI] [PubMed] [Google Scholar]
- Brown, P.O. and Botstein, D. 1999. Exploring the new world of the genome with DNA microarrays. Nat. Genet. 21: 33-37. [DOI] [PubMed] [Google Scholar]
- Deretic, V., Dikshit, R., Konyecsni, W.M., Chakrabarty, A.M., and Misra, T.K. 1989. The algR gene, which regulates mucoidy in Pseudomonas aeruginosa, belongs to a class of environmentally responsive genes. J. Bacteriol. 171: 1278-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drenkard, E. and Ausubel, F.M. 2002. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416: 740-743. [DOI] [PubMed] [Google Scholar]
- Ewing, B. and Green, P. 1998. Base-calling of automated sequencer traces using phred.II. Error probabilities. Genome Res. 8: 186-194. [PubMed] [Google Scholar]
- Gavin, A.C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J., Rick, J.M., Michon, A.M., and Cruciat, C.M., et al. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141-147. [DOI] [PubMed] [Google Scholar]
- Goldberg, J.B. and Dahnke, T. 1992. Pseudomonas aeruginosa AlgB, which modulates the expression of alginate, is a member of the NtrC subclass of prokaryotic regulators. Mol. Microbiol. 6: 59-66. [DOI] [PubMed] [Google Scholar]
- Hobbs, M., Collie, E.S., Free, P.D., Livingston, S.P., and Mattick, J.S. 1993. PilS and PilR, a two-component transcriptional regulatory system controlling expression of type 4 fimbriae in Pseudomonas aeruginosa. Mol. Microbiol. 7: 669-682. [DOI] [PubMed] [Google Scholar]
- Ishimoto, K.S. and Lory, S. 1992. Identification of pilR, which encodes a transcriptional activator of the Pseudomonas aeruginosa pilin gene. J. Bacteriol. 174: 3514-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kigawa, T., Muto, Y., and Yokoyama, S. 1995. Cell-free synthesis and amino acid-selective stable isotope labeling of proteins for NMR analysis. J. Biomol. NMR. 6: 129-134. [DOI] [PubMed] [Google Scholar]
- Li, S., Armstrong, C.M., Bertin, N., Ge, H., Milstein, S., Boxem, M., Vidalain, P.O., Han, J.D., Chesneau, A., Hao, T., et al. 2004. A map of the interactome network of the metazoan C. elegans. Science 303: 540-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane, E.L., Kwasnicka, A., Ochs, M.M., and Hancock, R.E. 1999. PhoP-PhoQ homologues in Pseudomonas aeruginosa regulate expression of the outer-membrane protein OprH and polymyxin B resistance. Mol. Microbiol. 34: 305-316. [DOI] [PubMed] [Google Scholar]
- Masignani, V., Rappuoli, R., and Pizza, M. 2002. Reverse vaccinology: A genome-based approach for vaccine development. Expert Opin. Biol. Ther. 2: 895-905. [DOI] [PubMed] [Google Scholar]
- McKevitt, M., Patel, K., Smajs, D., Marsh, M., McLoughlin, M., Norris, S.J., Weinstock, G.M., and Palzkill, T. 2003. Systematic cloning of Treponema pallidum open reading frames for protein expression and antigen discovery. Genome Res. 13: 1665-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhee, J.B., Lewenza, S., and Hancock, R.E. 2003.Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol. Microbiol. 50: 205-217. [DOI] [PubMed] [Google Scholar]
- Moss, J., Ehrmantraut, M.E., Banwart, B.D., Frank, D.W., and Barbieri, J.T. 2001. Sera from adult patients with cystic fibrosis contain antibodies to Pseudomonas aeruginosa type III apparatus. Infect. Immun. 69: 1185-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy, T.V.S., Wu, W., Qiu, Q.Q., Shi, S., LaBaer, J., and Brizuela, L. 2004. Bacterial cell-free system for high-throughput protein expression and a comparative analysis of E. coli cell-free and whole cell expression systems. Prot. Exp. Purif. 36: 217-225. [DOI] [PubMed] [Google Scholar]
- Phizicky, E., Bastiaens, P.I., Zhu, H., Snyder, M., and Fields, S. 2003. Protein analysis on a proteomic scale. Nature 422: 208-215. [DOI] [PubMed] [Google Scholar]
- Potvin, E., Lehoux, D.E., Kukavica-Ibrulj, I., Richard, K.L., Sanschagrin, F., Lau, G.W., and Levesque, R.C. 2003. In vivo functional genomics of Pseudomonas aeruginosa for high-throughput screening of new virulence factors and antibacterial targets. Environ. Microbiol. 5: 1294-1308. [DOI] [PubMed] [Google Scholar]
- Ratjen, F. and Doring, G. 2003. Cystic fibrosis. Lancet 361: 681-369. [DOI] [PubMed] [Google Scholar]
- Reboul, J., Vaglio, P., Rual, J.F., Lamesch, P., Martinez, M., Armstrong, C.M., Li, S., Jacotot, L., Bertin, N., and Janky, R., et al. 2003. C. elegans ORFeome version 1.1: Experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat. Genet. 34: 35-41. [DOI] [PubMed] [Google Scholar]
- Ritchings, B.W., Almira, E.C., Lory, S., and Ramphal, R. 1995. Cloning and phenotypic characterization of fleS and fleR, new response regulators of Pseudomonas aeruginosa which regulate motility and adhesion to mucin. Infect. Immun. 63: 4868-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigue, A., Quentin, Y., Lazdunski, A., Mejean, V., and Foglino, M. 2000. Two-component systems in Pseudomonas aeruginosa: Why so many? Trends Microbiol. 8: 498-504. [DOI] [PubMed] [Google Scholar]
- Stock, A.M., Robinson, V.L., and Goudreau, P.N. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69: 183-215. [DOI] [PubMed] [Google Scholar]
- Stover, C.K., Pham, X.Q., Erwin, A.L., Mizoguchi, S.D., Warrener, P., Hickey, M.J., Brinkman, F.S., Hufnagle, W.O., Kowalik, D.J., Lagrou, M. et al. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406: 959-964. [DOI] [PubMed] [Google Scholar]
- Sugimoto, N., Nakano, S., Katoh, M., Matsumura, A., Nakamuta, H., Ohmichi, T., Yoneyama, M., and Sasaki, M. 1995. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry 34: 11211-11216. [DOI] [PubMed] [Google Scholar]
- Tan, M.W., Rahme, L.G., Sternberg, J.A., Tompkins, R.G., and Ausubel, F.M. 1999. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc. Natl. Acad. Sci. 96: 2408-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhl, M.A. and Miller, J.F. 1996. Integration of multiple domains in a two componenet sensor protein: The Bordetella pertussis BvgAS phosphorelay. EMBO J. 15: 1028-1036. [PMC free article] [PubMed] [Google Scholar]
- Uetz, P., Giot, L., Cagney, G., Mansfield, T.A., Judson, R.S., Knight, J.R., Lockshon, D., Narayan, V., Srinivasan, M., and Pochart, P. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403: 623-627. [DOI] [PubMed] [Google Scholar]
- Wolfgang, M.C., Kulasekara, B.R., Liang, X., Boyd, D., Wu, K., Yang, Q., Miyada, C.G., and Lory, S. 2003. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. 100: 8484-8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak, D.J. and Ohman, D.E. 1991. P. aeruginosa AlgB, a two-component response regulator of the NtrC family, is required for algD transcription. J. Bacteriol. 173: 1406-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, H., Bilgin, M., Bangham, R., Hall, D., Casamayor, A., Bertone, P., Lan, N., Jansen, R., Bidlingmaier, S., and Houfek, T. 2001. Global analysis of protein activities using proteome chips. Science 293: 2101-2105. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.ncbi.nlm.nih.gov/genomes/altik.cgi?gi=163&db=Genome; NCBI, Microbial genomes.
- http://www.pseudomonas.com; Pseudomonas Genome project.
- http://wit.integratedgenomics.com/GOLD; Genomes online database.
- http://bioweb.pasteur.fr/seqanal/interfaces/toppred.html; TopPred: Topology prediction of membrane proteins.
- http://cubic.bioc.columbia.edu/predictprotein; The PredictProtein server.
- http://www.phrap.org; The Phred/Phrap/Consed System home page.