Abstract
The bacteria of the Brucella genus are responsible for a worldwide zoonosis called brucellosis. They belong to the α-proteobacteria group, as many other bacteria that live in close association with a eukaryotic host. Importantly, the Brucellae are mainly intracellular pathogens, and the molecular mechanisms of their virulence are still poorly understood. Using the complete genome sequence of Brucella melitensis, we generated a database of protein-coding open reading frames (ORFs) and constructed an ORFeome library of 3091 Gateway Entry clones, each containing a defined ORF. This first version of the Brucella ORFeome (v1.1) provides the coding sequences in a user-friendly format amenable to high-throughput functional genomic and proteomic experiments, as the ORFs are conveniently transferable from the Entry clones to various Expression vectors by recombinational cloning. The cloning of the Brucella ORFeome v1.1 should help to provide a better understanding of the molecular mechanisms of virulence, including the identification of bacterial protein-protein interactions, but also interactions between bacterial effectors and their host's targets.
The α-subdivision of Proteobacteria (or purple bacteria) is of fundamental biological relevance because it contains not only free-living organisms (e.g., Rhodobacter and Caulobacter), but also other members displaying a wide range of associations with eukaryotic organisms. These include (1) Agrobacterium, an extracellular plant pathogen that transforms its host by integrating its T-DNA into the plant's genome, (2) Rhizobium and related species, which are nitrogen-fixing symbionts of legumes, (3) Rickettsia, which live as obligate intracellular pathogens of man, but also insects, (4) Wolbachia, which are intracellular symbionts of insects, and finally, (5) Bartonella and Brucella, which are facultative intracellular pathogens of mammals. Considering their tendency to form close, if not intracellular associations with eukaryotic cells, it is not surprising that these bacteria developed a subtle expertise in manipulating or subverting their host's cellular processes. However, most of the strategies they exploit are still to be discovered at the molecular level. Brucella does not make exception to this rule.
Brucella sp., the etiological agents of brucellosis, have significant impact on both livestock and human health worldwide. In livestock, infection is primarily associated with abortion and infertility, whereas in man it often results in a chronic, debilitating disease known as brucellosis or undulant fever (Acha and Szyfres 1989). Treatment is difficult and prolonged, largely as a result of the intracellular nature of this pathogen (Young 1989). Although live attenuated vaccines played a crucial role in successful eradication programs (B. abortus strain 19 and B. melitensis Rev 1), these vaccines remain virulent for man (Spink et al. 1962). To date, no safe, effective vaccine is available for use in humans. Brucella is also considered as a potential agent of biological warfare or terrorist threat, largely as a result of its chronic debilitating clinical course, problematic treatment, and lack of suitable vaccines (Christopher et al. 1997; Franz et al. 1997).
Chronic intracellular persistence has been postulated as the reason why mammalian hosts can remain infected for life (Enright 1990). The understanding of virulence mechanisms used by Brucella remains sketchy. The chronic nature of brucellosis is likely to be multifactorial, utilizing both the ability of Brucella to evade immune detection and adaptation to intracellular survival inside both phagocytic and nonphagocytic cells. This strategy involves the inhibition of the phagosomal-lysosomal maturation pathway and the deviation of intracellular trafficking, which allows the bacterium to reach its endoplasmic reticulum-derived replicative niche (Celli et al. 2003). The type IV secretion system of Brucella (called VirB) plays a key role in this “vacuolar-jacking” (O'Callaghan et al. 1999; Delrue et al. 2001). Effectors of the VirB system are thought to interfere with presently unidentified eukaryotic targets.
Recently, the genome sequences of two Brucella species (DelVecchio et al. 2002a; Paulsen et al. 2002) and preliminary data from a third one became available. Comparative genomics provide insights into aspects of Brucella virulence that were only suspected before. First, although a few genes are homologous to known bacterial virulence factors that await experimental validation, the absence of genes encoding most classical virulence factors was observed (Moreno and Moriyon 2002). These data explain why Brucella was recently called a “nasty” (Moreno and Moriyon 2002), “furtive” (Letesson et al. 2002), or even a “stealthy” bug (Kohler et al. 2003). More importantly, these genomic sequences revealed the astonishing similarities between Brucella (an intracellular pathogen of mammals) and Mesorhizobium loti (a nitrogen fixing symbiont of plant) or Agrobacterium tumefaciens (an extracellular plant pathogen).
Considering the available genomes of Brucellae, function has been established by phenotypic or biochemical analysis for only a minority (around 200) of the 3198 predicted genes identified on their two chromosomes. Screens for transpositional mutants attenuated in infection models yielded 184 mutants (Delrue et al. 2004), suggesting that these genes have a function in the infection process. Moreover, 688 genes correspond to what appears in GenBank as hypothetical proteins, and most of them are conserved in at least one other α-Proteobacteria. Elucidation of the pathogenic mechanisms used by Brucella and specific host-pathogen interactions requires functional analysis of both Brucella and eukaryotic host gene products. Therefore, our overall objective is the large-scale analysis of protein function in Brucella sp.
The first step in that process is the cloning of a (nearly) complete set of protein-encoding open reading frames (ORFs), or “ORFeome” in a reusable and flexible recombinational cloning system (Walhout et al. 2000b). Such an ORFeome should provide a resource for systematic analysis of the molecular mechanisms that enable Brucella to pursue its intracellular lifestyle within its eukaryotic host. Because protein homology searches did yield only a few effector candidates, functional genomic and proteomic approaches should be applied to identify gene products potentially involved in the interaction between the bacterium and its host. The Brucella ORFeome is the platform to achieve this objective. Additionally, the Brucella ORFeome might also contribute to a better understanding of related species such as α-Proteobacteria (LeVier et al. 2000; Briones et al. 2001; Lopez-Goni et al. 2002; Lestrate et al. 2003).
RESULTS
Primer Design
Prior to high-throughput (HT) ORFeome cloning, the overall quality of the B. melitensis genome annotation was validated. In particular, the most likely start codons of the 3198 predicted ORFs were repredicted relative to the original genome annotation (DelVecchio et al. 2002a), as their identification is crucial for the various proteomic uses downstream of the ORFeome project. Starting from the GenBank release of the B. melitensis 16M genome (NC_003317 and NC_003318 accession nos.; DelVecchio et al. 2002a), a consortium of experts manually corrected the predicted start codons of the previously annotated ORFs. A Web site named the “Brucella melitensis database” containing various types of information about the ORFs and their homologs was constructed to allow such annotations (see Web sites references for the URL). Briefly, start codons were corrected to have a predicted ribosome-binding site (RBS), allow typical operonic junction if applicable, and avoid extended overlaps with neighboring ORFs. The close homology with proteins deduced from other α-Proteobacteria did often help to identify the most likely start codon in the B. melitensis genome. Among the 3198 ORFs manually examined, 908 modifications of start codon prediction were made, among which 565 ORFs were shortened, and 334 ORFs were extended. There are 2223 ORFs preceded by a RBS (defined as four bases of the AGGAGGU sequence, of which 1583 are either AGGA, GGAG, GAGG, or AGGU) at positions -1 to -20 relative to the newly predicted start codon. The new annotated sequences are available at the “Brucella melitensis database” (see Web sites references).
To allow ORF cloning using the Gateway recombinational system (Hartley et al. 2000; Walhout et al. 2000a), the attB1 and attB2 sequences must be added during the PCR amplification, at the 5′ and 3′ of each ORF, respectively. The attB1 and attB2 sequences were synthesized at the 5′ end of each forward and reverse primer, respectively. The last nucleotide of the stop codon was not incorporated in the reverse primers, to allow C-terminal fusion proteins to be expressed from Destination vectors. The length of the ORF-specific sequence for each primer, typically between 18 and 28 bases, was automatically determined to obtain similar predicted annealing temperature for each pair of primers. Half of all ORF-specific primer sequences were <21 nucleotides, due to the relatively high GC content of B. melitensis ORFs (57.8% on average).
ORF Amplification
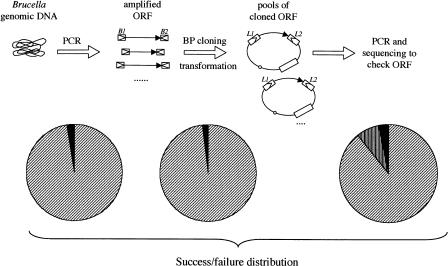
The overall scheme of the B. melitensis ORFeome cloning project was based on the strategy used for the Caenorhabditis elegans ORFeome (Reboul et al. 2003). All PCR amplification and cloning steps were performed in 96-well plates using a “real-estate” that organized ORFs according to increasing size and chromosome location (Fig. 1).
Figure 1.
Overview of the Brucella melitensis ORFeome construction. The main experimental steps of the ORFeome construction, that is, PCR, BP cloning, transformation, and validation of the entry clones, are shown in the top part of the figure. The success/failure distribution is given for the PCR, the presence of transformants after BP reaction, and the validation of the entry clones. The success rates are represented with hatched surfaces, whereas failure rates are shown in black. For the last step, the additional portion (vertically hatched surface) corresponds to entry clones checked by PCR using ORF-specific primers (see text for details). B1 and B2 stand for attB1 and attB2, respectively, and L1 and L2 correspond to attL1 and attL2 sites, respectively.
From 3198 attempted amplifications, 3114 PCR products could be detected by ethidium-bromide staining, suggesting a 97.4% success rate. Only a slight correlation was observed between PCR success rate and ORF size—1.3% of ORFs smaller than 1 kb failed to be amplified, and 4% of ORFs longer than 1 kb were not amplified.
Gateway Cloning
PCR-amplified ORFs were inserted into the Gateway-compatible vector pDONR201 by site-specific recombination. Gateway reactions were performed in 96-well plates, even with the PCR products not visible after gel electrophoresis, as we have previously obtained cloned ORFs from undetected amounts of amplicons (Reboul et al. 2003). The resulting products were then introduced into electrocompetent DH5α-T1r Escherichia coli cells, again using 96-well plates, and the resulting bacterial transformants were selected by growth in liquid medium containing kanamycin. Evaluation of the turbidity of the culture suggested that transformants were absent in 54 wells of the 3198 tested, which yields a 98.3% success rate. Interestingly, transformants were obtained in cases where PCR products were not detected by gel electrophoresis. This confirms that low amounts of PCR fragments, undetectable by ethidium bromide staining, can be sufficient for a successful BP reaction. The expected ORFs were indeed present in 48 Entry clones generated with low amounts of PCR products (see below). The content of wells A12, B12, C12, D12, and E12 of each plate was spread on solid medium in order to estimate the number of transformants by reaction. Using these data, we estimate that ∼80% of the transformant pools contain at least 50 clones, that is, a sample of clones that represent both PCR-mutated and wild-type alleles of each ORF. Liquid cultures of transformant pools were grown to generate glycerol stocks that constitute the B. melitensis ORFeome library, version 1.1.
Validation of the Entry Clones
We verified the identity of the ORF inserted into each Entry clone as follows. Plasmid DNA preparations were performed from the 3198 liquid cultures described in the previous section. These preparations were used as template for a PCR reaction using pDONR201-specific primers. These primers anneal on the pDONR201 vector, in the attL1 and attL2 regions, and with their 3′ end pointing toward the inserted ORF. This allows the amplification of the cloned insert along with ∼200 bp of flanking plasmid vector sequence. The identity of individual ORFs was confirmed by standard dideoxy sequencing reactions using these PCR products as templates to generate ORF sequence tags (OSTs) as described (Reboul et al. 2003). Individual OSTs were compared with ORF sequences predicted from the B. melitensis genome sequences using the BLASTN program (Altschul et al. 1997). Successful sequencing results were obtained for 2852 ORFs.
For the remaining ORFs, we utilized ORF-specific primers for PCR amplification of the same PCR products used as templates for the sequencing reactions. There were 239 amplifications that gave the expected size product, which suggests that the expected ORF is inserted in the corresponding Entry vector. Among the 107 missing ORFs, 36 failed the PCR amplification from genomic DNA. Because all manipulations were done with pools of molecules (Reboul et al. 2003), there may be mixes of clones (including vectors lacking the ORF of interest) for various transformants.
DISCUSSION
ORFeome v1.1 of B. melitensis is composed of 2852 ORFs, each cloned as pools and confirmed by sequencing, and 239 additional entry clones, for which the presence of the expected ORF was determined by PCR. Therefore, 3091 (96.7%) of the 3198 B. melitensis ORFs are included in the ORFeome presented here. This proportion of cloned ORFs is similar to the one reported for the ORF collection reported for Treponema pallidum (McKevitt et al. 2003). This high-cloning efficiency compared with the C. elegans ORFeome v1.1 (Reboul et al. 2003) is primarily due to the fact that genomic DNA, rather than a cDNA library, was used for the PCR amplification step. At this point, we do not know whether all amplified and cloned ORFs are actually expressed by B. melitensis under various conditions, or whether some may actually represent pseudogenes.
Using the “pool” of Entry clones, further versions of the ORFeome can be generated that consist of isolated clones that are free of mutations. Ongoing corrections of the annotations of various Brucella genomes may also yield new ORFs to be included in the B. melitensis ORFeome. Annotation is a real issue for the generation of such ORFeome resources, as a wrong start codon may yield proteins inactive in functional assays as observed for the C. elegans ORFeome version 1.1 (Reboul et al. 2003) and the main issue for generating version 3.1 (Lamesch et al. 2004). Extensive proteomic analyses could also be used to correct the genome annotation, allowing the identification of previously undetected coding sequences. To date, 232 individual proteins were identified using two-dimensional gel electrophoresis and mass spectrometry (DelVecchio et al. 2002b). In addition, sequencing of other strains or species could lead to the discovery of new genes, absent in B. melitensis, that could also be added to the B. melitensis ORFeome to give a multispecies Brucella ORFeome. The Brucella ORFeome should therefore be seen as a dynamic resource, having various versions depending on the amount of ORFs, but also on the presence in the library of clones validated as true wild-type alleles by sequencing. Apart from the normalization of individual clone abundance, the possibility to easily extend the ORFeome library by including ORFs from different strains or species is a major advantage of ORFeome libraries compared with classical genomic libraries.
In the first step of this ORFeome project, we had to reevaluate the current start codons assignment (DelVecchio et al. 2002a). This allowed new analyses of the B. melitensis genomic sequence, regarding the organization of coding sequences in operons. As reported by Salgado et al. (2000) for E. coli, we observed a high frequency of junctions within directons (i.e., several ORFs pointing in the same orientation) ranging from -10 to +40 nt (data not shown), suggesting that this distance could be used for the prediction of operons. Using this criterion, we found 1392 ORFs organized in 537 operons. This number is probably an underestimation, as long (>40 nt) junctions between ORFs can be observed within operons, for example, between virB1 and virB2 coding sequences, in the B. suis virB operon (Boschiroli et al. 2002). During the correction of genome annotation, we also observed that genomes of Brucella species sequenced to date are closely related, as already suggested by Paulsen et al. (2002). There are only 15 and 28 B. melitensis ORFs having no close homolog in B. abortus and B. suis, respectively. Moreover, within the Rhizobiales, the ORFs of B. melitensis, M. loti, A. tumefaciens, and Sinorhizobium meliloti are closely related. There are 1802 ORFs conserved (E value lower than 10-30 using the tBLASTN program) in all four genomes, whereas 2318 B. melitensis ORFs are conserved in at least one of these bacteria. These data suggest that Brucella could have emerged from a plant pathogen or symbiont, and raise the interesting possibility that some molecular mechanisms involved in the interaction between the bacterium and its host have been conserved (Inon de Iannino et al. 1998; Sola-Landa et al. 1998).
As with the C. elegans ORFeome (Li et al. 2004), the B. melitensis ORFeome is being used for protein-protein interaction mapping studies. We have generated a two-hybrid interaction matrix for the 41 proteins (1681 interactions tested, and 26 proteins found to interact) putatively involved in the two-component systems of Brucella (R. Hallez, M. Wéry, V. Van Mullem, J. Vardenhaute, J.-J. Letesson, and X. DeBolle, unpubl.). Furthermore, in the case of Brucella, it is not only important to identify cognate protein-protein interactions, but also to identify Brucella proteins that specifically interact with proteins from the host. A complete analysis of this aspect of B. melitensis biology awaits the construction of a mammalian ORFeome. It is very likely that Brucella secretes effectors in the cytoplasm (and maybe other subcellular locations) of infected cells, but these effectors and their targets are presently unknown. However, the VirB secretion system is essential for the virulence of this bacterium (O'Callaghan et al. 1999). Thus, one way to identify virulence effectors would be to screen for protein-protein interactions using either VirB proteins or predicted secretion chaperones as baits, with both B. melitensis and human ORFeome clones as preys.
The validation of protein-protein interactions detected using yeast or bacterial two-hybrid methods will also be facilitated using the recombinational cloning from the ORFeome Entry vectors. Vectors compatible with recombinational cloning and allowing expression of ORFs as fusions with tags that can be used for “medium-throughput” coimmunoprecipitation assays (Li et al. 2004) will make confirmation of protein-protein interactions straightforward.
Fusions with tags such as tandem affinity purification tags could also allow the purification of stable complexes to identify the partners involved (Rigaut et al. 1999). Protein tagging with GFP may also be performed to localize protein at cell poles, such as described for proteins involved in the control of cell cycle and morphogenesis in Caulobacter crescentus, another α-Proteobacterium (Jacobs et al. 1999). Of further interest would be the subcellular localization of proteins secreted by Brucella within the host cell, such as with the membrane localization of YpkA from Yersinia pseudotuberculosis (Hakansson et al. 1996) or the nuclear localization of YopM from Y. pestis (Skrzypek et al. 1998). It is expected that Brucella effectors could colocalize with markers of compartments or membranes, such as the endoplasmic reticulum or the endosomes. The introduction of Brucella ORFs into eukaryotic (e.g., yeast) cells to find particular phenotypes associated with their expression (Lesser and Miller 2001) or to identify effector proteins would be relatively easy to set up with an ORFeome resource.
A further application of the ORFeome will be the generation of low-cost DNA microarrays by amplification of the ORFs with a single pair of primers, as all ORFs are inserted in the same vector.
The availability of the ORFeome will also allow high-throughput recombinant protein production, using hosts such as E. coli or yeast. We already know that Brucella ORFs can be expressed in yeast, as two-hybrid interactions may be observed among two-component proteins. We also performed overexpression in E. coli from the ORFeome clones (R. Hallez, unpubl.). These systems may be useful for antigen discovery, as demonstrated for Treponema pallidum (McKevitt et al. 2003). The use of appropriate purification tags would also allow various analyses, including the systematic test of biochemical activities (Martzen et al. 1999) and the detection of proteins able to interact with a given ligand (Zhu et al. 2001). The evaluation of a large number of proteins for their potential effect as protective antigens could also be considered.
Another possible use for the ORFeome clones is the systematic mutagenesis of genes in the Brucella genome. Usually, gene deletions are performed by a strategy of allelic replacement of the target gene with an antibiotic resistance cassette. Using an antibiotic resistance cassette and a destination vector, each containing attR1 and attR2 sequences in the proper orientation, it should, in principle, be possible to construct plasmids allowing gene replacement by exchanging a given coding sequence with a resistance gene. At the high-throughput level, it should therefore be possible to delete each nonessential gene of the B. melitensis genome. This strategy should also be effective for all bacteria in which transformation and homologous recombination may be applied. This systematic mutagenesis strategy would be interesting, given the high bias found in the large-scale analysis of transposition mutants (Kohler et al. 2002).
In conclusion, the generation of a nearly complete ORFeome for Brucella is a first and important step for undertaking post-genomic approaches that will provide answers to the major questions regarding the molecular mechanisms of virulence, particularly regarding the interactions between these bacteria and their hosts. It is expected that these mechanisms may be conserved in other α-Proteobacteria, as they may adopt similar molecular strategies to interact with a eukaryotic host.
METHODS
Correction of the Annotations of the B. melitensis Genome
The annotated genome files of B. melitensis (named NC_003318.gbk and NC_003317.gbk for the large and small chromosomes, respectively) were downloaded directly from the NCBI ftp site (ftp://ftp.ncbi.nih.gov/genomes/Bacteria/). The Artemis free software, obtained from the Sanger Artemis Web site (http://www.sanger.ac.uk/Software/Artemis/), was used to localize annotations.
For each ORF, we manually checked the predicted start codons using Artemis for sequence examination. The following criteria were used to predict likely start codons. The AUG start codon was preferred to GUG, which itself was preferred to UUG. Typical operonic junctions in which stop codon of the preceding ORF was fused either in the -1 or -4 position (Salgado et al. 2000) to the start codon of the next ORF of an operon were frequently observed and used to locate likely start codons inside operons. The predicted start codons should present an obvious ribosome-binding site (RBS), usually AGGA, GGAG, or GAGG at positions -5 to -20. When close homologs of an ORF were detected, the start codon corresponding to the size of the closest homologs was chosen. If an ORF was shorter than its homologs, and if similarity was detectable by extending the ORF in the same frame, a new start site was searched. In the absence of any homolog, and in the absence of obvious RBS, the longest ORF starting with an AUG or GUG start codon was predicted. The Brucella melitensis database Web site was used to compare the deduced N-terminal peptidic sequence with those of homologous sequences, if any. The comments were incorporated at the Brucella melitensis database Web site and further processed to give a new version of the annotated genome, available in the GenBank format. The set of 3198 protein-coding ORFs was extracted from the corrected version of the annotated genome.
Primer Design
The attB1 segment (5′-GGGGACAAGTTTGTACAAAAAAG CAGGC-3′) was added before the start codon of each forward primer, followed by ORF-specific bases (see below). The attB2 segment (5′-GGGGACCACTTTGTACAAGAAAGCTGGG-3′) was added at the 5′ end of each reverse primer, which was complementary to the end of the ORF, without the last nucleotide of the stop codon, as previously described (Walhout et al. 2000b; Reboul et al. 2003). The size (typically 18 to 28 bases) of the forward and reverse primer parts that are complementary to the ORF were determined to give a similar annealing temperature during the PCR, usually in the range of from 60 to 70°C. The primers were obtained from Illumina Inc. in a 96-well format, and in each plate, the G12 and H12 wells were left empty to incorporate controls (see below).
PCR Amplification of the ORFs
Each PCR reaction was performed in a 50-μL volume, with 1 unit of Platinum HiFi polymerase (Invitrogen), MgSO4 2 mM, dNTP mix (0.2 mM each), forward and reverse primers (0.125 μM each) and genomic DNA (75 ng). The B. melitensis DNA was a kind gift of C. Baldwin laboratory (University of Massachusetts, Amherst). The 25 PCR cycles (94°C for 45 sec, 56°C for 1 min, and 68°C for 1 min/kb) were preceded by heating to 94°C for 2 min, and were followed by a 5-min incubation at 68°C. Confirmation and size of the PCR products was determined by agarose gel electrophoresis using E-gels (Invitrogen).
BP Cloning Reaction of the Amplified ORFs
The BP reactions were performed in 96-well plates using the Invitrogen BP kit, in a final volume of 10 μL containing 2 μL of unpurified PCR product, 75 ng of pDONR201 plasmid (Invitrogen), and 1.5 μL of BP clonase. In each 96-well plate, the G12 well was empty (see below) and the H12 well contained a BP reaction mix without PCR product, to evaluate the background given by the BP reaction. The BP reactions were incubated overnight at 25°C and then stored at -20°C or used directly for bacterial transformation.
Transformation and Plasmid Preparation
In 96-well plates, individual bacterial transformations were carried out using 20 μL of DH5α-T1r Max Efficiency competent cells (Invitrogen), 2 μL of the BP reaction products, and 105 μL of SOC medium. After the transformation reaction, 120 μL of E. coli transformants were transferred to 96-deep well plates (QIAGEN) containing 800 μL of LB with kanamycin (50 μg/mL) using a GenMate robot (Tecan). In each 96-well plate, the G12 well contained a pUC19 vector (2 pg) as positive control of transformation, and the H12 well (containing BP reaction mixes without PCR product) were used as negative control. The transformation mixes of G12 and H12 wells were plated on solid medium supplemented with the appropriated antibiotic (ampicillin, 50 μg/mL for G12 wells, and kanamycin, 50 μg/mL for H12 wells).
After overnight growth at 37°C, 50 μL of each culture was reserved for glycerol stocks and the remaining culture was processed for automated plasmid preparations with a QIAGEN BioRobot 9600.
Test of the Entry Clones
To identify and confirm all ORF entry clones, we performed PCR, followed by sequencing of the PCR product. The PCR was performed as above, except that 2 μL of the plasmid preparation was used as template and universal primers (0.38 μM each) flanking the ORF (5′-TCGCGTTAACGCTAGCATGGATCTC-3′) and (5′-GTAACATCAGAGAGATTTTGAGACAC-3′) were used. The same universal primers were also used for standard dideoxy sequencing reactions as described (Reboul et al. 2003). When sequencing data was not satisfactory, a PCR with ORF-specific primers described above was performed, using the conditions employed for initial amplification of the ORF from genomic DNA.
Acknowledgments
We thank the S. Köhler team for their help with the prediction of the start codons. The European COST845 network is acknowledged for having allowed the consortium to meet and put their efforts together. We thank C. Baldwin and R. Goenka for the generous gift of genomic DNA and the sequencing staff at Agencourt Biosciences for technical assistance. Research in the Universidad de Navarra was supported by the Spanish Ministerio de Ciencia y Tecnología (AGL2000-0305-C02). We also thank Tracey Clingingsmith for superb administrative assistance. Régis Hallez holds a fellowship from the F.R.I.A. (Fonds pour la formation à la Recherche dans l'Industrie et dans l'Agriculture), and Amélie Dricot was supported by a FSR (Fonds Spécial de Recherche) project.
Footnotes
[The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: C. Baldwin and R. Goenka. The Open Biosystems company will act as a distributor of the Brucella ORFeome clones.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2456204.
References
- Acha, P. and Szyfres, B. 1989. La brucellose. In Zoonoses et maladies transmissibles communes à l'homme et aux animaux, pp. 14-35. Office international des épizooties, Paris, France.
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boschiroli, M.L., Ouahrani-Bettache, S., Foulongne, V., Michaux-Charachon, S., Bourg, G., Allardet-Servent, A., Cazevieille, C., Liautard, J.P., Ramuz, M., and O'Callaghan, D. 2002. The Brucella suis virB operon is induced intracellularly in macrophages. Proc. Natl. Acad. Sci. 99: 1544-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briones, G., Inon de Iannino, N., Roset, M., Vigliocco, A., Paulo, P.S., and Ugalde, R.A. 2001. Brucella abortus cyclic β-1,2-glucan mutants have reduced virulence in mice and are defective in intracellular replication in HeLa cells. Infect. Immun. 69: 4528-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli, J., de Chastellier, C., Franchini, D.M., Pizarro-Cerda, J., Moreno, E., and Gorvel, J.P. 2003. Brucella evades macrophage killing via VirB-dependent sustained interactions with the endoplasmic reticulum. J. Exp. Med. 198: 545-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopher, G.W., Cieslak, T.J., Pavlin, J.A., and Eitzen Jr., E.M. 1997. Biological warfare. A historical perspective. JAMA 278: 412-417. [PubMed] [Google Scholar]
- Delrue, R.M., Martinez-Lorenzo, M., Lestrate, P., Danese, I., Bielarz, V., Mertens, P., De Bolle, X., Tibor, A., Gorvel, J.P., and Letesson, J.J. 2001. Identification of Brucella spp. genes involved in intracellular trafficking. Cell Microbiol. 3: 487-497. [DOI] [PubMed] [Google Scholar]
- Delrue, R.M., Lestrate, P., Tibor, A., Letesson, J.J., and De Bolle, X. 2004. Brucella pathogenesis, genes identified from random large-scale screens. FEMS Microbiol. Lett. 231: 1-12. [DOI] [PubMed] [Google Scholar]
- DelVecchio, V.G., Kapatral, V., Redkar, R.J., Patra, G., Mujer, C., Los, T., Ivanova, N., Anderson, I., Bhattacharyya, A., Lykidis, A., et al. 2002a. The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc. Natl. Acad. Sci. 99: 443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DelVecchio, V.G., Wagner, M.A., Eschenbrenner, M., Horn, T.A., Kraycer, J.A., Estock, F., Elzer, P., and Mujer, C.V. 2002b. Brucella proteomes—A review. Vet. Microbiol. 90: 593-603. [DOI] [PubMed] [Google Scholar]
- Enright, F.M. 1990. The pathogenesis and pathobiology of Brucella infection in domestic animals. In Animal brucellosis (eds. K. Nielsen and J. Duncan), pp. 301-320. CRC Press, Boca Raton, FL.
- Franz, D.R., Jahrling, P.B., Friedlander, A.M., McClain, D.J., Hoover, D.L., Bryne, W.R., Pavlin, J.A., Christopher, G.W., and Eitzen Jr., E.M. 1997. Clinical recognition and management of patients exposed to biological warfare agents. JAMA 278: 399-411. [DOI] [PubMed] [Google Scholar]
- Hakansson, S., Galyov, E.E., Rosqvist, R., and Wolf-Watz, H. 1996. The Yersinia YpkA Ser/Thr kinase is translocated and subsequently targeted to the inner surface of the HeLa cell plasma membrane. Mol. Microbiol. 20: 593-603. [DOI] [PubMed] [Google Scholar]
- Hartley, J.L., Temple, G.F., and Brasch, M.A. 2000. DNA cloning using in vitro site-specific recombination. Genome Res. 10: 1788-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inon de Iannino, N., Briones, G., Tolmasky, M., and Ugalde, R.A. 1998. Molecular cloning and characterization of cgs, the Brucella abortus cyclic β(1-2) glucan synthetase gene: Genetic complementation of Rhizobium meliloti ndvB and Agrobacterium tumefaciens chvB mutants. J. Bacteriol. 180: 4392-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, C., Domian, I.J., Maddock, J.R., and Shapiro, L. 1999. Cell cycle-dependent polar localization of an essential bacterial histidine kinase that controls DNA replication and cell division. Cell 97: 111-120. [DOI] [PubMed] [Google Scholar]
- Kohler, S., Foulongne, V., Ouahrani-Bettache, S., Bourg, G., Teyssier, J., Ramuz, M., and Liautard, J.P. 2002. The analysis of the intramacrophagic virulome of Brucella suis deciphers the environment encountered by the pathogen inside the macrophage host cell. Proc. Natl. Acad. Sci. 99: 15711-15716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler, S., Michaux-Charachon, S., Porte, F., Ramuz, M., and Liautard, J.P. 2003. What is the nature of the replicative niche of a stealthy bug named Brucella? Trends Microbiol. 11: 215-219. [DOI] [PubMed] [Google Scholar]
- Lamesch, P., Milstein, S., Hao, T., Rosenberg, J., Li, N., Sequerra, R., Bosak, S., Doucette-Stamm, L., Vandenhaute, J., Hill, D.E., et al. 2004. C. elegans ORFeome Version 3.1: Increasing the coverage of ORFeome resources with improved gene predictions. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- Lesser, C.F. and Miller, S.I. 2001. Expression of microbial virulence proteins in Saccharomyces cerevisiae models mammalian infection. EMBO J. 20: 1840-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lestrate, P., Dricot, A., Delrue, R.M., Lambert, C., Martinelli, V., De Bolle, X., Letesson, J.J., and Tibor, A. 2003. Attenuated signature-tagged mutagenesis mutants of Brucella melitensis identified during the acute phase of infection in mice. Infect. Immun. 71: 7053-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letesson, J.J., Lestrate, P., Delrue, R.M., Danese, I., Bellefontaine, F., Fretin, D., Taminiau, B., Tibor, A., Dricot, A., Deschamps, C., et al. 2002. Fun stories about Brucella: The “furtive nasty bug”. Vet. Microbiol. 90: 317-328. [DOI] [PubMed] [Google Scholar]
- LeVier, K., Phillips, R.W., Grippe, V.K., Roop II, R.M., and Walker, G.C. 2000. Similar requirements of a plant symbiont and a mammalian pathogen for prolonged intracellular survival. Science 287: 2492-2493. [DOI] [PubMed] [Google Scholar]
- Li, S., Armstrong, C.M., Bertin, N., Ge, H., Milstein, S., Boxem, M., Vidalain, P.O., Han, J.D., Chesneau, A., Hao, T., et al. 2004. A map of the interactome network of the metazoan C. elegans. Science 303: 540-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Goni, I., Guzman-Verri, C., Manterola, L., Sola-Landa, A., Moriyon, I., and Moreno, E. 2002. Regulation of Brucella virulence by the two-component system BvrR/BvrS. Vet. Microbiol. 90: 329-339. [DOI] [PubMed] [Google Scholar]
- Martzen, M.R., McCraith, S.M., Spinelli, S.L., Torres, F.M., Fields, S., Grayhack, E.J., and Phizicky, E.M. 1999. A biochemical genomics approach for identifying genes by the activity of their products. Science 286: 1153-1155. [DOI] [PubMed] [Google Scholar]
- McKevitt, M., Patel, K., Smajs, D., Marsh, M., McLoughlin, M., Norris, S.J., Weinstock, G.M., and Palzkill, T. 2003. Systematic cloning of Treponema pallidum open reading frames for protein expression and antigen discovery. Genome Res. 13: 1665-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, E. and Moriyon, I. 2002. Brucella melitensis: A nasty bug with hidden credentials for virulence. Proc. Natl. Acad. Sci. 99: 1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Callaghan, D., Cazevieille, C., Allardet-Servent, A., Boschiroli, M.L., Bourg, G., Foulongne, V., Frutos, P., Kulakov, Y., and Ramuz, M. 1999. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol. Microbiol. 33: 1210-1220. [DOI] [PubMed] [Google Scholar]
- Paulsen, I.T., Seshadri, R., Nelson, K.E., Eisen, J.A., Heidelberg, J.F., Read, T.D., Dodson, R.J., Umayam, L., Brinkac, L.M., Beanan, M.J., et al. 2002. The Brucella suis genome reveals fundamental similarities between animal and plant pathogens and symbionts. Proc. Natl. Acad. Sci. 99: 13148-13153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboul, J., Vaglio, P., Rual, J.F., Lamesch, P., Martinez, M., Armstrong, C.M., Li, S., Jacotot, L., Bertin, N., Janky, R., et al. 2003. C. elegans ORFeome version 1.1: Experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat. Genet. 34: 35-41. [DOI] [PubMed] [Google Scholar]
- Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., and Seraphin, B. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17: 1030-1032. [DOI] [PubMed] [Google Scholar]
- Salgado, H., Moreno-Hagelsieb, G., Smith, T.F., and Collado-Vides, J. 2000. Operons in Escherichia coli: Genomic analyses and predictions. Proc. Natl. Acad. Sci. 97: 6652-6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrzypek, E., Cowan, C., and Straley, S.C. 1998. Targeting of the Yersinia pestis YopM protein into HeLa cells and intracellular trafficking to the nucleus. Mol. Microbiol. 30: 1051-1065. [DOI] [PubMed] [Google Scholar]
- Sola-Landa, A., Pizarro-Cerda, J., Grillo, M.J., Moreno, E., Moriyon, I., Blasco, J.M., Gorvel, J.P., and Lopez-Goni, I. 1998. A two-component regulatory system playing a critical role in plant pathogens and endosymbionts is present in Brucella abortus and controls cell invasion and virulence. Mol. Microbiol. 29: 125-138. [DOI] [PubMed] [Google Scholar]
- Spink, W., Hall, J., Finstad, J., and Mallet, E. 1962. Immunization with viable Brucella organisms. Bull. World Health Organ. 26: 409-419. [PMC free article] [PubMed] [Google Scholar]
- Walhout, A.J., Sordella, R., Lu, X., Hartley, J.L., Temple, G.F., Brasch, M.A., Thierry-Mieg, N., and Vidal, M. 2000a. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science 287: 116-122. [DOI] [PubMed] [Google Scholar]
- Walhout, A.J., Temple, G.F., Brasch, M.A., Hartley, J.L., Lorson, M.A., van den Heuvel, S., and Vidal, M. 2000b. Gateway recombinational cloning: Application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 328: 575-592. [DOI] [PubMed] [Google Scholar]
- Young, E. 1989. Treatment of brucellosis in humans. In Brucellosis: Clinical and laboratory aspects (eds. E. Young and M. Corbel), pp. 127-141. CRC Press, Boca Raton, FL.
- Zhu, H., Bilgin, M., Bangham, R., Hall, D., Casamayor, A., Bertone, P., Lan, N., Jansen, R., Bidlingmaier, S., Houfek, T., et al. 2001. Global analysis of protein activities using proteome chips. Science 293: 2101-2105. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- ftp://ftp.ncbi.nih.gov/genomes/Bacteria/; NCBI FTP Web site.
- http://www.sanger.ac.uk/Software/Artemis/; Sanger Artemis Web site.
- http://serine.urbm.fundp.ac.be/~seqbruce/GENOMES/Brucella_melitensis/; Brucella melitensis database Web site.