Abstract
We have devised a screen for genes that suppress the loss of contact inhibition elicited by overexpression of the protooncogene MYCN. The initial application of this screen detected nine distinctive suppressors within a representative human cDNA library. One of these genes was ING4, a potential tumor suppressor gene that maps to human chromosome 12p13. Ectopic expression of ING4 suppressed the loss of contact inhibition elicited by either MYCN or MYC but had no direct effect on cellular proliferation. Pursuing the possibility that ING4 might be a tumor suppressor gene, we found inactivating mutations in ING4 transcripts from various human cancer cell lines. In addition, we used comparative genomic hybridization to detect deletion of the ING4 locus in 10-20% of human breast cancer cell lines and primary breast tumors. Ectopic expression of ING4 attenuated the growth of T47D human breast cancer cells in soft agar. We conclude that ING4 is a strong candidate as a tumor suppressor gene.
Keywords: mycn, proliferation, cell transformation, mutation, breast cancer
The ING4 gene belongs to the ING (INhibitor of Growth) tumor suppressor family (1). The five members of the family identified to date all contain a highly conserved plant homeodomain finger motif in the C-terminal end of the proteins. The plant homeodomain finger is found in transcription factors that modulate chromatin structure (2), and the ING gene products interact with protein complexes containing histone acetyltransferase and deacetylase (1). Thus, although the exact functions of ING genes have not been elucidated, the gene products are thought to participate in transcriptional regulation by means of chromatin remodeling (1). The products of ING genes have been shown to interact with and enhance the function of p53 in gene transcription, apoptosis, and DNA repair (1, 3-5). Allelic loss and reduced expression of ING1 and ING3 have been reported in various tumors (1) and in head and neck carcinomas (6), respectively, suggesting roles as tumor suppressors in human cancer.
A recent report demonstrated that ING4 attenuates the growth of human glioblastoma transplants in nude mice by suppressing angiogenesis (7). We have obtained independent evidence that ING4 is a tumor suppressor gene. The evidence emerged from our efforts to identify genes that can suppress the transformation of cells in vitro by MYC family oncogenes. In particular, we screened for genes that could suppress the loss of contact inhibition elicited by ectopic expression of the MYCN protooncogene in Rat1A cells. We designed the screen to select for genes that specifically suppress loss of contact inhibition but do not directly inhibit cellular proliferation. We hypothesized that such genes are likely to encode tumor suppressors whose functions can guard against tumorigenesis.
The screen has identified nine genes to date, among which was ING4. We found that ING4 is frequently mutated in various human cancer cell lines. We also detected frequent deletions of the ING4 locus in specimens from human breast cancers. Ectopic expression of ING4 in a breast cancer cell line attenuated growth of the cells in soft agar. The results sustain the view that ING4 is a tumor suppressor gene and validate the screen as a tool to identify candidate tumor suppressors.
Materials and Methods
Plasmids and Tissue Culture Cells. Rat1A cells that overexpress MYCNER were established by transfecting Rat1A cells with pBabe(puro)/MYCNER (generously provided by W. Weiss, University of California, San Francisco) and selecting for puromycin (Sigma) resistance at 2 μg/ml. The resulting cell line was denoted as Rat1A/MYCNER. To establish Rat1A/MYCNER cells that express the tetracycline-responsive transcription activator (tTA), tTA was cloned into the pBabe(Neo) retroviral vector and used to produce virus in a BOSC23 ecotropic packaging cell line. Rat1A/MYCNER cells were infected with the pBabe(neo)/tTA virus and selected for neomycin resistance at 400 μg/ml Geneticin (GIBCO/BRL). The resulting cell line was denoted as Rat1A/MYCNER/tTA. The pRevTRE retroviral vector and a human brain cDNA expression library in pRevTRE were purchased from Clontech. The Rat1A and Rat1A-derived cell lines were grown in DMEM containing 10% FBS but lacking phenol red. Fifty-millimolar solutions of azidothymidine (AZT) (Sigma) and 5-fluorouracil (5-FU) (Sigma) were prepared in PBS. Doxycycline (Sigma) and 4-hydroxytamoxifen (4-OHT) (Calbiochem) were prepared at 400 μM in water and 100 μM in ethanol, respectively. The human cancer cell lines IMR-32, SK-N-AS, SK-N-BE, H23, H82, N417, T47D, MDAmb231, and ACHN and normal lung fibroblasts, IMR-90, were purchased from American Type Culture Collection and grown according to instructions from the provider.
Northern Blot Analyses. Total cellular RNA was prepared from cells by using the Qiagen RNeasy Midi kit (Valencia, CA) and analyzed by a standard Northern blot method (NorthernMax kit, Ambion, Austin, TX).
RT-PCR/Sequence Analyses. The Titan One Tube RT-PCR system (Roche) was used to amplify the coding sequence of ING4 transcripts from 1 μg of total cellular RNA, with a primer pair of 5′-ATGGCTGCGGGGATGTATTTGGAAC-3′ and 5′-CTATTTCTTCTTCCGTTCTTGGGAGCAG-3′. Subsequently, the amplified PCR fragments were cloned into the pCR2.1 vector by using the Topo-TA cloning kit (Invitrogen), and the individual clones were sequenced.
Southern Blot Analyses. Cells were incubated in a lysis buffer containing 100 mM Tris (pH 8.5), 200 mM NaCl, 5 mM EDTA, 0.2% SDS, and 100 μg/ml proteinase K (Sigma) overnight at 55°C. Genomic DNA from the lysed cells was isolated by a phenol/chloroform/isoamyl alcohol (25/24/1, vol/vol/vol; Invitrogen) extraction and followed by an ethanol precipitation. Enzyme-digested DNA was analyzed by Southern blotting.
Soft Agar Assay. The pMIG retroviral vector (8) or pMIG/ING4 was used to produce virus in a Phoenix amphotropic packaging cell line. The virus was used to infect T47D cells, and the infected cells were plated onto 60-mm dishes in growth medium containing 0.35% soft agar (SeaPlaque, Cambrex, East Rutherford, NJ) between two layers of 0.7% agar-containing media. The plates were photographed on a fluorescent dissection microscope (Leica, Deerfield, IL) after 21 days of incubation in a standard tissue culture incubator.
Array Comparative Genomic Hybridization (CGH) Analysis. Genomewide scanning for copy number assessment by using array-based CGH was performed as described in ref. 9. BAC DNA microarrays were obtained from the core facility at the University of California, San Francisco Comprehensive Cancer Center. Cell line and reference DNA samples were labeled with the fluorescent dye-conjugated nucleotides, Cy3-dUTP and Cy5-dUTP (Amersham Biosciences), respectively. Labeled probes were purified by using MicroSpin G-50 columns (Amersham Biosciences). Approximately 500 ng of each labeled probe was ethanol-precipitated with 65 μg of human cot-1 DNA (Invitrogen) and resuspended in 60 μl of hybridization buffer (50% formamide/10% dextran sulfate/2× SSC/2% SDS/1% yeast tRNA). Probes were denatured at 73°C for 5 min and reannealed at 37°C for 60-90 min before applying them to the slides. A rubber gasket was placed around each array. The reannealed probe mixtures were applied to each array. The slides were placed without cover glass in a humidified chamber (50% formamide/2× SSC) on a rocker at 37°C for 48-72 h. After hybridization, the slides were washed for 15 min at 50°C in 50% formamide/2× SSC (pH 7.0) for 30 min at 50°C in 2× SSC/0.1% SDS and finally for 15 min at room temperature in a buffer containing 0.1 M sodium phosphate and 0.1% Nonidet P-40 (pH 8.0). The slides were mounted with 4′,6-diamidino-2-phenylindole for imaging (1 μM 4′,6-diamidino-2-phenylindole/1× PBS/90% glycerol). TIFF format and 16-bit grayscale images were collected by using a charge-coupled device camera imager and CY3, CY5, and 4′,6-diamidino-2-phenylindole filters. The images were analyzed as described in ref. 9. Data were normalized to the median raw CY3/CY5 ratio and converted to log base 2 to weigh gains and losses equally. The means and standard deviations of the triplicate spots were calculated for the normalized log2 ratios. We considered the copy number increased or decreased if the normalized log2 ratios were >0.3 or less than -0.3 for cell lines, respectively, and >0.2 or less than -0.2 for tumors, because tumors are generally contaminated with normal cells.
Results
A Screen for Genes That Suppress the Loss of Contact Inhibition Induced by MYCN. We sought to identify genes that can suppress oncogenic events without directly affecting normal cellular functions. Loss of contact inhibition is an in vitro property of cells associated with neoplastic transformation. We devised a screen for genes that can suppress loss of contact inhibition but do not directly inhibit cellular proliferation. We used the loss of contact inhibition elicited by MYC family protooncogenes in Rat1A cells, an established line of pseudodiploid rat fibroblasts. We introduced cDNA expression libraries into cells, selected for cells that remained contact-inhibited despite MYC activity, and identified cDNAs that were contained in the selected cells (Fig. 1A, and see below). We designed the screen so that it would be blind to functions that directly inhibit cellular proliferation. This objective was achieved by establishing constitutive expression of the cDNA library before and during the screen. Therefore, cells containing cDNAs that directly inhibited cellular proliferation were eliminated because of their growth disadvantage.
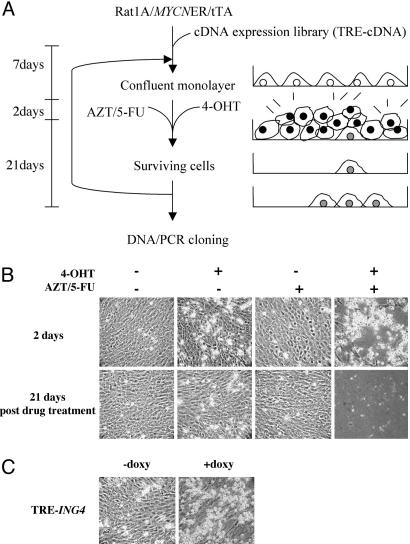
Fig. 1.
A screen for genes that suppress the loss of contact inhibition induced by MYCN. (A) Schematic diagram of the screen. (B) Differential killing of cells with AZT/5-FU in an MYCN-dependent manner. Rat1A/MYCNER cells were grown to a confluent monolayer and incubated for 2 days in the presence or absence of 100 nM 4-OHT and/or 50 μM AZT/50 μM 5-FU. After 2 days, all drugs were withdrawn, and the cells were fed with fresh media every 3 days for 21 days. (C) ING4 suppresses the loss of contact inhibition elicited by MYCN. Rat1A/MYCNER/tTA cells containing pRevTRE-ING4 were grown in the presence or absence of 2 μM doxycycline for the duration of the assay. Confluent monolayers were subjected to 2 days of treatment in the presence or absence of 100 nM 4-OHT and/or 50 μM AZT/50 μM 5-FU.
We used a Rat1A cell line stably expressing MYCNER, a chimeric molecule between MYCN and the ligand-binding domain of the human estrogen receptor. The fusion renders MYCN activity conditionally dependent on the estrogen analog, 4-hydroxy-tamoxyfen (4-OHT) (10). The use of inducible MYCN avoided potential consequences of prolonged overexpression of MYC protooncogenes, such as genomic instability (11-13). We chose MYCNER rather than MYCER because the cell lines expressing MYCER exhibited loss of contact inhibition even in the absence of 4-OHT (data not shown; see below). In contrast, the Rat1A/MYCNER cell line behaved identically to native Rat1A cells and displayed contact inhibition when grown to a confluent monolayer. When 4-OHT was added to the cell growth media, however, the cells became refractile and overgrew the original monolayer (Fig. 1B).
The cells that lost contact inhibition because of MYCN activity continued to proliferate at confluence and, thus, were killed by addition of the cytotoxic drugs, azido-thymidine (AZT) and 5-fluorouracil (5-FU), to the media. AZT is a thymidine analog that is incorporated into replicating DNA and blocks chain elongation, leading to cell death (14). 5-FU is an inhibitor of de novo thymidylate synthesis and causes the depletion of intracellular thymidine, enhancing the cytotoxic effect of AZT (14, 15). Thus, cells that lost contact inhibition and entered the cell cycle incorporated AZT and died. At 50 μM concentration of AZT/5-FU, the cells were killed effectively only in the presence of 4-OHT (Fig. 1B). A higher concentration than 50 μM or duration longer than 48 h of AZT/5-FU treatment killed the cells even in the absence of 4-OHT (data not shown). The 48-h treatment with 50 μM AZT/5-FU left virtually no surviving cells, with a background frequency of 1 × 10-8. In contrast, Rat1A/MYCER cells at confluence were killed by AZT/5-FU even in the absence of 4-OHT. The cell killings by AZT/5-FU in the presence and absence of 4-OHT differed only by 100-fold, not great enough to permit a screen in the MYCER cell line.
Screen of a cDNA Expression Library and Identification of ING4 as a Suppressor of Loss of Contact Inhibition. To implement the screen, we established a cell line that stably expressed a tetracycline-regulatable transcription factor (tTA) in Rat1A/MYCNER cells, designated Rat1A/MYCNER/tTA. Expression of a human brain cDNA library was driven by the tetracycline response element promoter, such that the cDNA expression could be turned off by adding a tetracycline derivative, doxycycline, to media (16). The conditional expression of cDNAs allowed us to demonstrate dependence of the suppression activity on expression of the cDNAs.
DNA from 1 × 106 independent cDNA clones was used to make retrovirus in an ecotropic virus packaging cell line. The Rat1A/MYCNER/tTA cell line was infected with virus at a multiplicity of infection of 1 to ensure that each infected cell expressed no more than a single cDNA. The cells were grown in the absence of doxycycline before and during the screen so that cDNA expression was constitutive. The cells were then treated with 4-OHT and AZT/5-FU for 48 h. After subsequent feeding of the surviving cells with fresh media every 3 days for 21 days, clonal growth of cells was visible on the plates. The numbers of the colonies varied from 0 to 12 per plate, with an average of 3 per plate and a total yield of ≈150 colonies.
Using an estimate of 30,000 genes in the human genome and assuming 10 as an average copy number for an mRNA, 300,000 clones would represent a screen of approximately one complete genome. We screened 106 cDNA clones, representing a 3.3-fold redundancy. Thus, the 150 clones identified by the screen represented 0.017% of cellular mRNA.
The 150 colonies were divided into 10 groups. Each group of cells was grown to a confluent monolayer, and a second round of screening was performed to eliminate false positives. DNA was isolated from the surviving cells, and the integrated cDNAs were amplified from the DNA by PCR, using primers that flanked the pRevTRE vector cloning site. The PCR amplified bands were cloned individually for sequence analysis. To date, we have cloned and sequenced 47 individual PCR products. There were 16 isolates that encoded apparently full-length cDNAs. Among these were three isolates of cDNAs representing MAD1, a known MYC antagonist (17), and one representing ING4, as well as cDNAs for additional genes.
To confirm that ectopic expression of ING4 suppressed the loss of contact inhibition induced by MYCN, we cloned ING4 into the pRevTRE vector and introduced it into the Rat1A/MYCNER/tTA cell line. When seeded sparsely in tissue culture plates, cells expressing ING4 were killed efficiently by AZT/5-FU (data not shown). This result indicated that ING4 over-expression neither conferred drug resistance nor inhibited DNA replication. At confluence in the presence of 4-OHT, cells overexpressing ING4 survived the AZT/5-FU treatment, whereas the cells were sensitive to the drugs when doxycycline was used to repress the expression of ING4 (Fig. 1C). These results demonstrate that ING4 expression was indeed responsible for suppressing the loss of contact inhibition induced by MYCN.
We tested whether ING4 expression also suppressed loss of contact inhibition by MYC in a Rat1A/MYCER cell line. ING4 expression did protect the cells from killing by AZT/5-FU at confluence when MYC activity was induced with 4-OHT (data not shown). However, ING4 overexpression did not inhibit or retard the growth of Rat1A cells, primary mouse embryonic fibroblasts, or the ScP2 mouse epithelial cell line (data not shown). We conclude that ING4 does not suppress the role of endogenous MYC in cellular proliferation, but instead, specifically suppresses the loss of contact inhibition elicited by over-expression of MYC.
ING4 Mutations in Human Cancer Cell Lines. Because ING4 was able to suppress loss of contact inhibition induced by MYCN or MYC, we hypothesized that a deficiency of ING4 may play a role in tumorigenesis. To explore this possibility, we first examined whether the expression of ING4 is reduced in various human cancer cell lines (Fig. 2A). The results were inconclusive; in comparison to IMR-90 fibroblasts, modest reductions of expression were observed in some tumor cell lines but not in others.
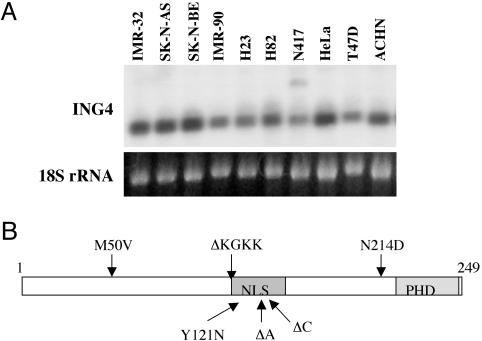
Fig. 2.
ING4 in human cancer cell lines. (A) Expression of ING4. Northern blot analysis was performed on 5 μg of total cellular RNA by using 32P-labeled full-length ING4 cDNA as probe. (B) Schematic diagram of the ING4 protein showing the locations of plant homeodomain (PHD) and nuclear localization signal (NLS). Arrows mark the mutations found in ING4 transcripts from human cancer cell lines.
We then examined ING4 transcripts for mutations that might affect the gene product by cloning the coding sequence of ING4 transcripts with RT-PCR and sequencing at least two independent clones from each cell line. We detected deletions or nucleotide changes in the ING4 transcripts from seven of nine cancer cell lines examined. The results are displayed in Fig. 2B and tabulated in Table 1.
Table 1. Mutations found in ING4 transcripts from human cancer cell lines.
| Cell line | Cancer type | Mutation | Consequence |
|---|---|---|---|
| IMR-32 | Neuroblastoma | Deletion 379-390 | Deletion 4AA* |
| SK-N-AS | Neuroblastoma | Deletion 465C | Frame shift/stop |
| SK-N-BE | Neuroblastoma | Deletion 379-390 | Deletion 4AA* |
| H23 | Lung adenocarcinoma | Missense 361TtoA | Y121N |
| H82 | Small-cell lung carcinoma | Deletion 446A | Frame shift/stop |
| Missense 640AtoG | N214D† | ||
| Deletion 379-390 | Deletion 4AA*† | ||
| N417 | Small-cell lung carcinoma | None | — |
| HeLa | Cervical carcinoma | None | — |
| T47D | Breast ductal cell carcinoma | Deletion 379-390 | Deletion 4AA* |
| ACHN | Colorectal carcinoma | Missense 165AtoG | M50V† |
| Deletion 379-390 | Deletion 4AA† |
Recurrent mutation.
Mutations found in the same allele.
We found a common deletion in the IMR-32 and SK-N-BE (2) neuroblastoma cell lines, the H82 small-cell lung carcinoma cell line, the T47D breast ductal cell carcinoma cell line, and the ACHN colorectal carcinoma cell line. The deletion encompassed 12 nucleotides (379-390), eliminating a sequence of four highly charged amino acid residues (KGKK). These residues are part of a nuclear localization signal in ING4 (5, 7). Thus, the deletion may result in mislocalization of the ING4 protein, thereby impairing its function. Mislocalization to the cytoplasm of the ING1 protein has been reported in brain tumors and invasive breast carcinoma (18, 19). This KGKK amino acid deletion in ING4 appears to arise from mechanisms other than a genomic deletion because the corresponding genomic sequence was intact in T47D cells (data not shown).
Single-nucleotide deletions, 465C and 446A, were found in the SK-N-AS neuroblastoma cell line and H82 lung cancer cell line, respectively. These deletions cause a frameshift in translation and should truncate the gene product, deleting nearly one-half of the molecule, including the highly conserved plant homeodomain finger domain. In a recent report, ING4 proteins with C-terminal truncations have been shown to exert a dominant-negative effect (7). Hence, the 465C and 446A deletion mutations would not only produce nonfunctional proteins but might also inhibit the normal ING4 proteins produced from the wild-type allele.
Two allelic transcripts of ING4 with different mutations were identified in the H82 small-cell lung carcinoma cell line. One contained a single-nucleotide deletion (446A) that truncates the gene product. The other contained the common KGKK deletion and a missense mutation that results in a conservative change in an amino acid N214D (see Table 1). The ACHN colorectal carcinoma cell line also contained two allelic transcripts. Both contained the common 4-aa deletion, but one also contained a missense mutation that results in a conservative change in amino acid sequence M50V (see Table 1).
A single-nucleotide missense mutation that changes a tyrosine to asparagine (Y121N) was detected in the H23 lung adenocarcinoma cell line. Because the structure and charge of these two amino acids differ greatly, this missense mutation may have a functional consequence for ING4.
We also observed a deletion of three nucleotides (392-395) that creates an in-frame replacement of adjacent lysine and glutamine residues (KG) with a single serine (S) in the H23, N417, and HeLa cell lines. This sequence variation has been reported in GenBank in different mRNA isolates from different tissue sources. Additional searches of GenBank revealed that the mouse ING4 homolog also exists in two forms (KG or S). Given this cross-species conservation, it seems unlikely that the deletion affects ING4 function. It is not clear whether the deletion represents a polymorphism or is the result of alternative splicing. No additional alterations were found in ING4 transcripts from N417 lung cell carcinoma cells or the HeLa cervical cancer cell line. ING4 transcripts from IMR-90 normal lung fibroblasts did not contain any sequence variations.
The ING4 Locus Is Deleted in Human Breast Cancer Cell Lines. We performed Southern blot analysis to search for deletions of ING4 in cancer cells. The ING4 hybridization signal obtained with DNA isolated from the T47D human breast ductal carcinoma cell was substantially reduced in comparison with that obtained with DNA from the H23, H82, and HeLa cancer cell lines (Fig. 3A). This result raised the possibility of a genomic deletion of the ING4 gene in T47D, in addition to the mutation we detected in ING4 transcripts (see Table 1 and above).
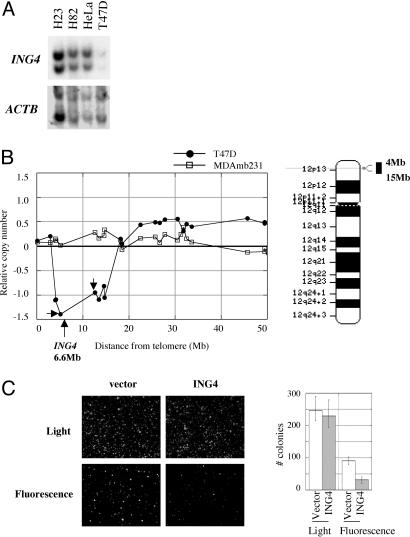
Fig. 3.
Ectopic reconstitution of ING4 in T47D breast cancer cells. (A) Southern blot. Genomic DNA was digested with BamHI and analyzed by Southern blotting with 32P-labeled full length cDNA of ING4 or the actin gene, ACTB,as hybridization probes. (B) CGH analysis of T47D (filled circles) and MDAmb231 (open squares) cell lines. Loss and gain of DNA are reflected in the relative copy number score on the y axis of the graph. Relative copy numbers were determined by the ratio of the test DNA to the reference DNA. Therefore, a -0.5 score indicates that the test DNA contained half the copy number compared with the reference DNA, reflecting the loss of one copy in the test DNA. As such, loss of two copies would score -1.0. Small arrows mark the location of BAC probes, RP11-272L6 at 5.1 Mb and RP11-59H1 at 12.6 Mb from telomere. A cytological banding of chromosome 12, the location of ING4, and the extent of deletion in T47D are shown. (C) Suppression of growth in soft agar. T47D cells infected with the retroviral constructs, ING4-IRES-GFP and IRES-GFP, were plated in soft agar and then incubated for 21 days. Colonies were counted with the aid of either light or fluorescent microscopy.
The National Institutes of Health human genome sequence data set maps ING4 to chromosome 12p13.31, 6.6 Mb from the telomere of the short arm of chromosome 12. We assessed the extent of the deletion surrounding the ING4 gene in T47D cells by comparative genomic hybridization (CGH). CGH had been performed previously on a set of 55 breast cancer cell lines, which included T47D (unpublished work of K.C. and J.W.G.). The CGH data showed an 11-Mb deletion in 12p13 of T47D cells (Fig. 3B). In comparison, no deletion was detected in this region in another breast cancer cell line, MDAmb231 (Fig. 3B). T47D DNA scored -1.39 and -0.95 with probes located 5.1 Mb (BAC RP11-272L6) and 12.6 Mb (BAC RP11-59H1) from the telomere, respectively (Fig. 3B). These scores indicate loss of at least two genomic copies. T47D cells are hypotriploid (www.atcc.org). Therefore, it appears that T47D cells contain a deletion of at least two ING4 copies.
Analysis of the CGH data for the complete set of 55 breast cancer cell lines revealed that two BAC probes that flank ING4, BAC RP11-272L6 and BAC RP11-59H1, were lost in 13 (23.6%) and in 6 (11.1%) cell lines, respectively. Given the frequency of deletion for these BAC probes, we conclude that the frequency of ING4 loss in human breast cancer cell lines is between 10% and 20%.
Ectopic Expression of ING4 Attenuates Colony Formation by T47D Cells in Soft Agar. We tested whether ectopic expression of wild-type ING4 might affect the behavior of T47D cells. We used a retroviral vector to express ING4 in T47D cells. The construct coexpressed GFP via an internal ribosomal entry site (IRES) linked to ING4. The efficiency of infection with viruses carrying either ING4-IRES-GFP or GFP alone was 40% at the time of plating.
As anticipated from the design of the original screen, we observed that ectopic expression of ING4 had no effect on the proliferation of T47D cells (data not shown). Moreover, confluent T47D cells displayed contact inhibition despite their apparent deficiency in ING4 (data not shown), suggesting that functional ING4 may not be required for contact inhibition. We then extended the analysis to an additional in vitro correlate of neoplastic transformation, anchorage-independent propagation in soft agar. GFP or ING4-IRES-GFP-containing cells were visualized by fluorescent microscopy, whereas plating efficiency was assessed by light microscopy. Ectopic expression of ING4 in T47D cells attenuated growth in soft agar by 3-fold compared with the cells that contained vector alone (Fig. 3C). In contrast, ING4 expression did not affect colony formation by the MDAmb231 cell line (data not shown), which contains no deletions in the ING4 locus detectable by CGH (Fig. 3B). These results authenticate the ability of ING4 to suppress aspects of neoplastic transformation and suggest that T47D cells are indeed deficient for ING4.
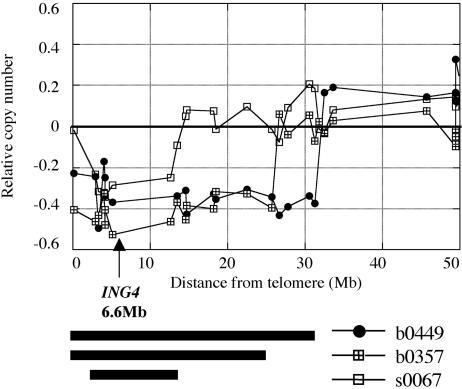
The ING4 Locus Is Deleted in 10-20% of Human Primary Breast Tumors. To extend our study to specimens from human tumors, we analyzed data from a CGH analysis of 146 breast cancers (K.C., J.W.G., S. DeVries, and F. M. Waldman, unpublished work). We found that the DNA represented by the two BAC probes flanking ING4, RP11-272L6, and RP11-59H1, was deleted at a frequency of 9.9% and 20.3%, respectively. The CGH scores for the probes ranged between -0.26 and -0.57, suggesting deletion of one copy. The data for three representative tumors are shown in Fig. 4. Tumor C contained a 10-Mb deletion around the ING4 locus, whereas tumors A and B had larger deletions of 31 Mb and 26 Mb, respectively. The deletion frequency of 10-20% in primary breast tumors is comparable to that estimated for the cell lines derived from breast tumors. These data sustain the idea that a deficiency of ING4 may play a role in the genesis of at least some breast cancer.
Fig. 4.
Genomic deletion of the ING4 locus in primary breast tumors. CGH analysis of three tumors, A (filled circles), B (sectored squares), and C (open squares), are shown in a graph. The extent of each deletion is represented with bars.
Discussion
A Screen for Genes That Suppress the Loss of Contact Inhibition Induced by MYCN. We have devised a screen to identify genes that can suppress the loss of contact inhibition elicited by overexpression of the protooncogene MYCN. The screen was designed to identify genes that have no direct effect on cell proliferation. Accordingly, tumor suppressors such as p53 and RB, and cell cycle inhibitors such as p21, were not expected to be identified by the screen, because their overexpression blocks cell proliferation. By the same reasoning, we did not expect to identify direct inhibitors of MYC, because deletion of endogenous MYC severely retards cell proliferation (20, 21).
Nine genes have been identified by the screen to date, including ING4 and MAD1. MAD1 is a known antagonist of MYC (17), whose overexpression has been shown to inhibit cellular proliferation (22, 23). However, the repeated detection of MAD1 by our screen should not be due to a direct effect on cellular proliferation (see above). We investigated whether this discrepancy was due to the expression level of MAD1. Indeed, when we infected Rat1A cells with the virus containing a MAD1 expression construct at a high multiplicity of infection, proliferation of the cells was severely retarded (S.K., unpublished result). We conclude that the screen selected cells that were expressing MAD1 below the threshold required for direct inhibition of proliferation but above a threshold required for an effect on contact inhibition.
We do not yet know how ING4 suppresses the loss of contact inhibition induced by MYC-family oncogenes. It is possible, for example, that ING4 inhibits the function of MYC protein directly. In preliminary studies using a luciferase reporter, ING4 did not inhibit transcriptional activation by MYC (S.K., unpublished result). A more comprehensive examination by microarray analyses may provide insights into this issue. Alternatively, ING4 may be working downstream of transcriptional control by MYC or MYCN.
ING4 Mutations in Human Cancer Cell Lines. We found only nominal reductions of ING4 expression in a number of human cancer cell lines. Instead, mutations in ING4 transcripts were prevalent; seven of nine cancer cell lines contained mutant transcripts. Several cell lines contained a mutation that deletes part of a nuclear localization signal (KGKK). This recurrence suggests a selective advantage for cells that acquire the mutation. It remains to be seen whether this mutation also recurs in primary tumors. Other cell lines contained single-nucleotide deletions that result in the C-terminal truncation of the ING4 proteins. Such truncations have a dominant-negative effect (7), which has the potential to create a null state in even those cells that are heterozygous for the wild-type ING4 gene. The KGKK deletion, C-terminal truncation mutation (Δ446A), and an amino acid substitution mutation (Y121N) were functionally inactive in suppressing loss of contact inhibition (S.K., unpublished result). Wild-type ING4 transcripts were also found in three of the cell lines that contained mutant transcripts, including T47D breast cancer cells. Our studies showed that the hypotriploid T47D cells have lost at least two copies of ING4 and contain mutant transcripts (KGKK deletion). Ectopic expression of ING4 in T47D cells attenuated their growth in soft agar, indicating that T47D cells are impaired for ING4 function despite the presence of wild-type ING4 transcripts. These findings suggest that haploinsufficiency of ING4 may play a role in tumorigenesis.
ING4 displays parallel effects in Rat1A fibroblasts and T47D breast tumor cells. In both settings, the gene appears to have no direct effect on the cell division cycle, yet it can suppress the loss of contact inhibition elicited by MYC genes in Rat1A cells and inhibit the growth of T47D cells in soft agar. Loss of contact inhibition and growth in soft agar are generally coordinate manifestations in vitro of neoplastic transformation, but it is not known whether these two complex phenotypes are linked mechanistically. We conclude that ING4 is capable of suppressing aspects of cellular behavior that are thought to play a role in tumorigenesis. To date, we have attempted to test the effect of ING4 on tumorigenesis proper only with T47D cells, and that test failed because the cells did not form tumors in nude mice.
The ING4 protein has been shown to interact directly with p53 and enhance its activity as a transcription factor (5). T47D cells contain a single copy of p53 with a missense mutation that renders the protein nonfunctional (24). Therefore, the ING4 effect on colony formation in soft agar by T47D cells appears to be independent of p53 function.
ING4 Maps to a Region Frequently Deleted in Human Cancer. Loss of heterozygosity in 12p13 that includes the ING4 gene has been reported in 5-26% of childhood acute lymphoblastic leukemia (25-27), 12-43% of prostate cancer (28, 29), and 26% of ovarian cancer (30). This chromosomal region also carries two other candidate tumor suppressors, TEL/ETV6 and p27KIP, at 11.8 and 12.7 Mb from the telomere, respectively. We cannot exclude the possibility that the loss of these two genes plays a role in tumorigenesis. Our analysis of CGH data revealed that 10-20% of primary breast tumors also have deletions in 12p13. The deletions appear to affect only one copy of the gene; no genomic mutations were found in the remaining allele of ING4 in the three tumors presented in Fig. 4. Reduction in the ING4 transcript level due to a single copy deletion may contribute to tumorigenesis, and/or ING4 transcripts from the remaining allele may contain inactivating alterations such as the KGKK deletion mutation.
The work reported here demonstrates that ING4 can suppress two in vitro parameters of neoplastic transformation: loss of contact inhibition and growth in soft agar. In addition, we have found that ING4 is frequently affected by mutations or deletions in cell lines derived from human neuroblastomas and from cancers of the breast, colon, and lung, as well as in primary specimens of human breast cancer. The gene has also been implicated in the control of angiogenesis during tumor formation by transplanted glioblastoma cells (7). We conclude that ING4 is a strong candidate as a tumor suppressor gene in humans.
Acknowledgments
We thank Drs. Michelle Le Beau and William Weiss for critical review of our manuscript and Chris Yoo, Alana Welm, and members of the Bishop laboratory for helpful discussions. This work was supported by National Institutes of Health Grant CA44338 (to J.M.B.); the G. W. Hooper Research Foundation; U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research Contract DE-AC03-76SF00098 (to J.W.G.); National Institutes of Health, National Cancer Institute Grant P50 CA58207 (to J.W.G.); and the Avon Foundation.
Author contributions: S.K. designed and performed research and wrote the paper; S.K., K.C., and J.W.G. contributed new reagents/analytic tools; S.K. and K.C. analyzed data; and J.M.B. supervised research.
Abbreviations: AZT, azido-thymidine; 5-FU, 5-fluorouracil; 4-OHT, 4-hydroxy-tamoxifen; CGH, comparative genomic hybridization; IRES, internal ribosome entry site; tTA, tetracycline-responsive transcription factor.
References
- 1.Feng, X., Hara, Y. & Riabowol, K. (2002) Trends Cell Biol. 12, 532-538. [DOI] [PubMed] [Google Scholar]
- 2.Aasland, R., Gibson, T. J. & Stewart, A. F. (1995) Trends Biochem. Sci. 20, 56-59. [DOI] [PubMed] [Google Scholar]
- 3.Nagashima, M., Shiseki, M., Miura, K., Hagiwara, K., Linke, S. P., Pedeux, R., Wang, X. W., Yokota, J., Riabowol, K. & Harris, C. C. (2001) Proc. Natl. Acad. Sci. USA 98, 9671-9676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagashima, M., Shiseki, M., Pedeux, R. M., Okamura, S., Kitahama-Shiseki, M., Miura, K., Yokota, J. & Harris, C. C. (2003) Oncogene 22, 343-350. [DOI] [PubMed] [Google Scholar]
- 5.Shiseki, M., Nagashima, M., Pedeux, R. M., Kitahama-Shiseki, M., Miura, K., Okamura, S., Onogi, H., Higashimoto, Y., Appella, E., Yokota, J. & Harris, C. C. (2003) Cancer Res. 63, 2373-2378. [PubMed] [Google Scholar]
- 6.Gunduz, M., Ouchida, M., Fukushima, L., Ito, S., Jitsumori, Y., Nakashima, T., Nagai, N., Nishizaki, K. & Shimizu, K. (2002) Oncogene 21, 4462-4470. [DOI] [PubMed] [Google Scholar]
- 7.Garkavtsev, I., Kozin, S. V., Chernova, O., Xu, L., Winkler, F., Brown, E., Barnett, G. H. & Jain, R. K. (2004) Nature 428, 328-332. [DOI] [PubMed] [Google Scholar]
- 8.Van Parijs, L., Refaeli, Y., Abbas, A. K. & Baltimore, D. (1999) Immunity 11, 763-770. [DOI] [PubMed] [Google Scholar]
- 9.Pinkel, D., Segraves, R., Sudar, D., Clark, S., Poole, I., Kowbel, D., Collins, C., Kuo, W. L., Chen, C., Zhai, Y., et. al. (1998) Nat. Genet. 20, 207-211. [DOI] [PubMed] [Google Scholar]
- 10.Eilers, M., Picard, D., Yamamoto, K. R. & Bishop, J. M. (1989) Nature 340, 66-68. [DOI] [PubMed] [Google Scholar]
- 11.Mai, S., Fluri, M., Siwarski, D. & Huppi, K. (1996) Chromosome Res. 4, 365-371. [DOI] [PubMed] [Google Scholar]
- 12.Felsher, D. W. & Bishop, J. M. (1999) Proc. Natl. Acad. Sci. USA 96, 3940-3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mai, S. & Mushinski, J. F. (2003) J. Environ. Pathol. Toxicol. Oncol. 22, 179-199. [DOI] [PubMed] [Google Scholar]
- 14.Andreuccetti, M., Allegrini, G., Antonuzzo, A., Malvaldi, G., Conte, P. F., Danesi, R., Del Tacca, M. & Falcone, A. (1996) Eur. J. Cancer 32A, 1219-1226. [DOI] [PubMed] [Google Scholar]
- 15.Brunetti, I., Falcone, A., Calabresi, P., Goulette, F. A. & Darnowski, J. W. (1990) Cancer Res. 50, 4026-4031. [PubMed] [Google Scholar]
- 16.Gossen, M. & Bujard, H. (1992) Proc. Natl. Acad. Sci. USA 89, 5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grandori, C., Cowley, S. M., James, L. P. & Eisenman, R. N. (2000) Annu. Rev. Cell Dev. Biol. 16, 653-699. [DOI] [PubMed] [Google Scholar]
- 18.Vieyra, D., Senger, D. L., Toyam, T., Muzik, H., Brasher, P. M., Johnston, R. N., Riabowol, K. & Forsyth, P. A. (2003) Clin. Cancer Res. 9, 5952-5961. [PubMed] [Google Scholar]
- 19.Nouman, G. S., Anderson, J. J., Crosier, S., Shrimankar, J., Lunec, J. & Angus, B. (2003) J. Clin. Pathol. 56, 507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trumpp, A., Refaeli, Y., Oskarsson, T., Gasser, S., Murphy, M., Martin, G. R. & Bishop, J. M. (2001) Nature 414, 768-773. [DOI] [PubMed] [Google Scholar]
- 21.Mateyak, M. K., Obaya, A. J., Adachi, S. & Sedivy, J. M. (1997) Cell Growth Differ. 8, 1039-1048. [PubMed] [Google Scholar]
- 22.Roussel, M. F., Ashmun, R. A., Sherr, C. J., Eisenman, R. N. & Ayer, D. E. (1996) Mol. Cell. Biol. 16, 2796-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vastrik, I., Kaipainen, A., Penttila, T. L., Lymboussakis, A., Alitalo, R., Parvinen, M. & Alitalo, K. (1995) J. Cell Biol. 128, 1197-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Connor, P. M., Jackman, J., Bae, I., Myers, T. G., Fan, S., Mutoh, M., Scudiero, D. A., Monks, A., Sausville, E. A., Weinstein, J. N., et al. (1997) Cancer Res. 57, 4285-4300. [PubMed] [Google Scholar]
- 25.Stegmaier, K., Pendse, S., Barker, G. F., Bray-Ward, P., Ward, D. C., Montgomery, K. T., Krauter, K. S., Reynolds, C., Sklar, J., Donnelly, M., et al. (1995) Blood 86, 38-44. [PubMed] [Google Scholar]
- 26.Cave, H., Gerard, B., Martin, E., Guidal, C., Devaux, I., Weissenbach, J., Elion, J., Vilmer, E. & Grandchamp, B. (1995) Blood 86, 3869-3875. [PubMed] [Google Scholar]
- 27.Takeuchi, S., Bartram, C. R., Miller, C. W., Reiter, A., Seriu, T., Zimmerann, M., Schrappe, M., Mori, N., Slater, J., Miyoshi, I. & Koeffler, H. P. (1996) Blood 87, 3368-3374. [PubMed] [Google Scholar]
- 28.Fromont, G., Joulin, V., Chantrel-Groussard, K., Vallancien, G., Guillonneau, B., Validire, P., Latil, A. & Cussenot, O. (2003) J. Urol. 170, 1394-1397. [DOI] [PubMed] [Google Scholar]
- 29.Kawana, Y., Ichikawa, T., Suzuki, H., Ueda, T., Komiya, A., Ichikawa, Y., Furuya, Y., Akakura, K., Igarashi, T. & Ito, H. (2002) Prostate 53, 60-44. [DOI] [PubMed] [Google Scholar]
- 30.Hatta, Y., Takeuchi, S., Yokota, J. & Koeffler, H. P. (1997) Br. J. Cancer 75, 1256-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]