Abstract
Intracellular parasites can alter the cellular machinery of host cells to create a safe haven for their survival. In this regard, microsporidia are obligate intracellular fungal parasites with extremely reduced genomes and hence, they are strongly dependent on their host for energy and resources. To date, there are few studies into host cell manipulation by microsporidia, most of which have focused on morphological aspects. The microsporidia Nosema apis and Nosema ceranae are worldwide parasites of honey bees, infecting their ventricular epithelial cells. In this work, quantitative gene expression and histology were studied to investigate how these two parasites manipulate their host’s cells at the molecular level. Both these microsporidia provoke infection-induced regulation of genes involved in apoptosis and the cell cycle. The up-regulation of buffy (which encodes a pro-survival protein) and BIRC5 (belonging to the Inhibitor Apoptosis protein family) was observed after infection, shedding light on the pathways that these pathogens use to inhibit host cell apoptosis. Curiously, different routes related to cell cycle were modified after infection by each microsporidia. In the case of N. apis, cyclin B1, dacapo and E2F2 were up-regulated, whereas only cyclin E was up-regulated by N. ceranae, in both cases promoting the G1/S phase transition. This is the first report describing molecular pathways related to parasite-host interactions that are probably intended to ensure the parasite’s survival within the cell.
Introduction
Parasitism is a type of biological interaction between organisms of different species whereby the parasite benefits at the expense of the host. Host cells have developed a defense machinery to resist pathogen invasion and replication. In order to limit pathogen growth, these systems include mechanism such as the fusion of phagolysosomals, the production of reactive oxygen and reactive nitrogen intermediates, nutrient sequestration or cell suicide (apoptosis) in order to limit pathogen growth [1]. As a counterpoint, obligate parasites have to exploit their host to complete the processes critical for their survival. It is interesting how some intracellular parasites can reprogram their host’s cells to create a safe haven, utilizing the cellular machinery to acquire the necessary resources [2]. The implementation of these defence systems and the ability of successful pathogens to mitigate their effects are ultimately mediated by changes in both the levels and activities of key proteins.
Apoptosis is one mechanism the host uses to control pathogen dissemination, and it is a morphologically and biochemically distinct form of programmed cell death. Apoptosis plays a major role in the removal of damaged or unwanted cells during development, in maintaining tissue homeostasis and it is involved in the aging of multicellular organisms. It is also recognized as an important defense mechanism of defence against viral, bacterial and parasitic pathogens during innate and adaptive immunity [3]. However some intracellular parasites can modulate apoptosis by either inducing or inhibiting the process thereby allowing the parasite to successfully reproduce [3, 4, 5, 6, 7].
Intracellular pathogens may also have other effects on infected cells. For example, Toxoplasma cruzi induces broad modulations of the host’s cellular machinery, altering the expression of up to 353 murine genes. This modification provides insight into how the parasite survives, replicates, and persists in the infected host, and ultimately it defines the clinical outcome of infection [8]. In addition, the host cell’s proteome responds in a dramatic way to Toxoplasma gondii infection, causing changes in protein expression and modifications to key metabolic pathways, including glycolysis, lipid and sterol metabolism, mitosis, apoptosis, and structural-protein expression. These changes suggest that the parasite provokes global reprogramming of the cell’s metabolism, revealing a complex molecular relationship between the host and parasite [1].
Microsporidia are eukaryotes, obligate intracellular fungal parasites that infect a wide range of vertebrates and invertebrates, and cause economically important losses in animal species. Microsporidian genomics and cell biology are the consequence of an extreme reduction driven by an intimate relationship with the host cell. During their life cycle, they have an extracellular spore (called environmental spore) but apart from this form, all the remaining stages develop during the infection of the host, maintaining direct contact with the host throughout the life cycle, from entry to egress.
Although microsporidial infection has often been linked to immunocompromised individuals in vertebrate hosts, there is no such prerequisite for the infection of insects. Indeed, microsporidia can cause serious disease in economically relevant insects like silkworm and crop pollinators. In this regard, Apis mellifera bees are recognized as a model system to study social interactions, immunity and disease in social insects [9]. Specifically, honey bees infection by microsporidia has been associated with significant colony losses [10]. To date, two microsporidian species are known to infect A. mellifera worldwide: Nosema apis [11] and N. ceranae [12]. Both infect the epithelial cells of the honey bee ventriculus (digestive tract) but the pathogenic events in the target tissue have not been described in detail.
Until recently there were only few studies of how microsporidia manipulated host cells and most of these were focused on morphology. In the last few years, it has become clear that some microsporidia produce more specific changes to gene expression in the infected host than other pathogens [13]. Nowadays, this large group of organisms is described as highly exploitative intracellular parasites of the host cell environment [14]. In terms of honey bee microsporidia, previous studies have shown that N. ceranae reduces apoptosis in the bee ventriculi which is the target tissue [15, 16] and induced an effect on genes involved in the homeostasis and renewal of intestinal tissues [17]. However, how this parasite affects other processes in host’s cells that serve to protect them against infection remains unclear.
Here we explore how intracellular parasites manipulate their host cell environment at the molecular level by studying quantitative gene expression in tissues following infection of honey bees with the both Nosema species. Our aim was to study the modifications that infection by these parasites produces in the process of host cell cycle and in the apoptosis by determining the expression of genes involved in the pathways related with these events. In parallel, we used TUNEL (Terminal deoxynucleotide transferase mediated X-dUTP nick end labelling) to assess the rate of apoptosis in these infected tissues after induction.
Materials and methods
Experimental infection
Honey bees used in this work were collected from experimental colonies located at “Centro de Investigación Apícola y Agroambiental- IRIAF”. No permits for the use of bees were required and the field studies did not involve endangered or protected species. A frame with capped brood of A. mellifera iberiensis worker bees was taken from a Nosema-free colony and maintained in an incubator at 35°C until the bees emerged [18, 19]. Newly emerged worker bees were carefully removed from the frame, confined to cages and kept in a different incubator for five days at 33°C. Individual bees were infected as described previously [18], using 2 μl of water containing 100,000 N. ceranae or N. apis spores. Spores were Percoll-purified and confirmed as single species by PCR [20]. Uninfected control bees were fed with 2 μl of water alone. In total, 25 bees were included in each group (N. apis, N. ceranae and Control). Ten days after infection 15 bees per group were treated with 2 μl of 2% cycloheximide solution (Sigma C7698; 94% purity), as an apoptosis inducer by death receptors [21], while the remaining 10 bees for each group were not treated with cycloheximide. The full experimental design is shown in Table 1. In this way, a total of 6 experimental groups were established as follows: A-group experimentally infected with N. apis; AH- group experimentally infected with N. apis and treated with cycloheximide; C-group experimentally infected with N. ceranae; CH-group experimentally infected with N. ceranae and treated with cycloheximide; T- Control, bees not infected with Nosema sp.; TH—Control, bees not infected with Nosema sp. but treated with cycloheximide.
Table 1. Experimental design.
A. mellifera iberiensis bees were infected with N. apis or N. ceranae or non-infected.
|
N. apis N = 25 |
N. ceranae N = 25 |
Uninfected N = 25 |
|
|---|---|---|---|
| Cell cycle, mitochondria & hormone expression | Group A n = 10 |
Group C n = 10 |
Group T n = 10 |
| Apoptosis expression* | Group AH n = 10 |
Group CH n = 10 |
Group TH n = 10 |
| Apoptotic index* | Group AH n = 5 |
Group CH n = 5 |
Group TH n = 5 |
(*) Groups treated with cycloheximide on day 10 post infection.
Gene expression study
The bees studied (Table 1) were killed by freezing in liquid nitrogen on day 11 post infection and they were stored at -80°C until use. The ventriculus (infection target tissue) and rectal ampoule (rectum) were individually separated by dissection of each bee and stored in different tubes. Ventriculi were used to study the gene expression and they were stored in RNAlater (Qiagen 76106). The ampoules were used to confirm Nosema infection in each bee or the uninfected state of the control bees.
The RNA from each ventriculus was extracted individually using the RNeasy Tissue Kit (Qiagen) following the Purification of Total RNA from Animal Tissues protocol with on-column DNase digestion (DNase, Qiagen GmbH). Reverse transcription of RNA to cDNA was performed using the QuantiTect RT Kit (Qiagen GmbH).
The candidate genes related with A. mellifera homeostasis (cell cycle, mitochondria activity, apoptosis and hormones; S1 Table) and the house-keeping (HK) genes were selected from the sequences available at GeneBank (http://www.ncbi.nlm.nih.gov/nuccore). Specific primers and TaqMan® probes were designed using the Primer express (Applied Biosystems) software and the theoretical specificity was checked using the Basic Local Alignment Search Tool (BLAST) at GenBank (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The efficiency of each reaction to study the gene regulation was determined by analyzing serial cDNA dilutions.
Selection of housekeeping genes
Four A. mellifera HK genes (GAPDH, EF, β-Actin and 18S; S1 Table) were evaluated to select the most appropriate ones for analysis [22]. To determine the stability of expression (M) of the selected reference genes in the honeybee tissues studied (ventriculi), the four HK genes were analyzed in duplicate in all the samples and the Ct values were subsequently determined with the geNorm software [23]. The two most stable genes (lowest M value) were EF and β-Actin and they were selected as the HK genes for the subsequent analysis of expression.
The relative expression for each gene was calculated using the Relative Expression Software Tools: REST MCS-version 2 (http://www.gene-quantification.de/download.html) and REST 2009-version 2.0.13 (Qiagen GmbH), which use the pair wise fixed reallocation randomization test [24, 25]. All the groups infected with either N. apis or N. ceranae (irrespective of the treatment with cycloheximide) were compared with the uninfected control groups using this software and the efficiency of every reaction was taken into account as recommended elsewhere [24]. The level of significance for determining the up or down regulation for each gene was also determined using this software.
Real-time quantitative PCR
All real-time quantitative PCR reactions were performed using the 7900 HT Sequence Detection System (Applied Biosystems). To assay expression, each 25 μl reaction contained 0.625 U of Taq (TaqMan Gold/Buffer A Pack, cod. 4304441, Applied Biosystems), 10 μl of diluted cDNA (1/100), 300 nM of each primer and 100 nM of each TaqMan probe. The PCR program involved an initial 2 minute incubation at 50°C, a 10 minute denaturation step at 95°C and 40 cycles of 15 seconds at 95°C and a 1 minute of annealing at 60°C. All the cDNA samples obtained after extraction from each bee were run in duplicate. Negative controls for DNA extraction, reverse-transcription and real-time PCR steps were also included. The Ct values were recorded for each gene studied in every sample and the average of the two replicates was calculated.
Nosema spp. infection checking
The success of Nosema infection was determined using real-time PCR in honey bee rectal ampoules, given that this part of the bee gut stores the microsporidia spores until their exit in the faeces. DNA was extracted from the ampoules that had been separated from the ventriculi using the DNeasy tissue protocol (Qiagen GmbH) and it was collected individually in microcentrifuge tubes (one bee per tube) before analyzing it by real-time PCR and TaqMan® chemistry. The primers and probe sequences are shown in S2 Table.
Virus detection
In order to check the exposure of the bees to other naturally acquired and relevant pathogens, the presence of virus was also analysed in the cDNA (5 μL) from each bee in every group (A, AH, C, CH, T and TH). For each group, 10 μL of the mixture was analysed in duplicate for the presence of Kashmir bee virus (KBV), acute bee paralysis virus (ABPV), black queen cell virus (BQCV), chronic bee paralysis virus (CBPV), Deformed wing virus (DWV) and Israeli acute paralysis virus (IAPV) in duplicate. When a virus was detected in any group, all the bees from that group were individually analysed. The sequences of the primers and probes used are shown in S2 Table.
Histology (TUNEL assay)
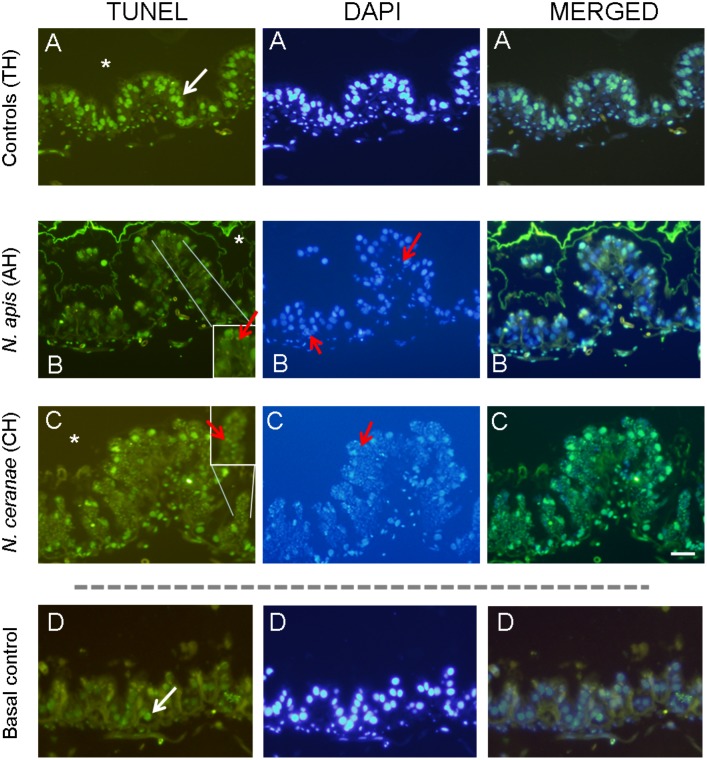
Five Bees from the cycloheximide treated groups (AH, CH and TH) were analysed histologically, to evaluate the effect of microsporidia infection in bees exposed to this strong inducer of apoptosis. The alimentary canal and other tissues (ventriculum with the Malpighian tubules attached, the small intestine and the rectum) were removed from the bees, divided into sections and fixed in 10% buffered formalin before they were paraffin embedded. The TUNEL (Terminal deoxynucleotide transferase mediated X-dUTP nick end labelling) assay was performed on 5 μm thick sections to quantify apoptosis, as described previously [26, 15]. Briefly, tissue sections were deparaffinised and rehydrated through an ethanol series before rinsing in phosphate buffered saline (PBS). The tissues were then treated with Proteinase K (20 μg ml–1, 15 min, room temperature) and then with 0.1% (w:v) Triton X-100 in PBS (10 min). Subsequently, the sections were incubated for 1 h at 37°C in the dark, using a humid chamber and the In situ Cell Death Detection Kit (Roche) according to the manufacturers’ instructions. The sections were then washed with PBS and mounted in Vectashield (Vector Labs) containing DAPI (Sigma) at a final concentration of 1 μg/ml, to label all the cell nuclei (blue). The sections were examined on an Olympus BX61 epifluorescence microscope equipped with filter sets for fluorescence microscopy: ultraviolet (UV, 365 nm, exciting filter UG-1). Photographs were obtained with a digital camera Olympus DP50 and processed using the Adobe PhotoShop 7.0 software (Adobe Systems). Uninfected bees and no treated with cycloheximide were used to determine the basal apoptosis (Basal control) just to show the natural apoptosis in the tissue. Cells undergoing apoptosis were scored (fluorescing green under blue excitation light) and the ventricular cells of three bees per group were counted to determine the number with apoptotic nuclei and the proportion of cells undergoing apoptosis (No. of apoptotic cells x 100 / total No. of cells) as described previously [15]. Approximately 100 cells were examined in representative areas of each ventricular sample (10 areas per sample, approximately 1,000 cells per ventriculum and bee).
Results
The success of Nosema infection (checked in the bee ampoules) was reflected by the detection of spores in each group. Consequently, all the bees were successfully infected by either N. ceranae or N. apis, while no cross-infection between groups was detected and there was no infection in the uninfected groups. However, one sample from the group A and another bee from the group CH showed a very low level of infection (Ct value > 36 in Nosema infection analysis; see above) and they were not considered for the gene expression analysis.
In none of the samples were CBPV, ABPV, KBV, IAPV or DWV detected. Yet, as all the groups tested positive for BQCV, an additional analysis was carried out to determine the presence of the virus in individual bees. Since more than the 80% of the bees were positive for BQCV in all the groups, viral infection was not considered a significant factor and was not included in the subsequent analyses.
Gene expression
The list of genes selected for study, and the primers and probes used are shown in S3 Table. All the primer pairs tested produced positive amplification with the exception of Cyclin H and BRUCE, thus these genes were not tested further.
Selection of reference HK genes
The ranking of the four candidate reference genes in the honey bee ventriculi according to their average expression stability (M value) was: β-Actin (0.060) <EF (0.066) <GAPDH (0.067) <18S (0.097), from the most stable (lowest M-value) to the least stable (highest M value). All the values were very low and they were much lower than the threshold limit of 1.5 recommended by GeNorm software, reflecting very stable expression. The pairwise variation (V) between the two or three normalization factors was 0 [22], indicating that the use of β-Actin and EF as HK were enough to perform all the subsequent analysis.
Apoptosis related genes
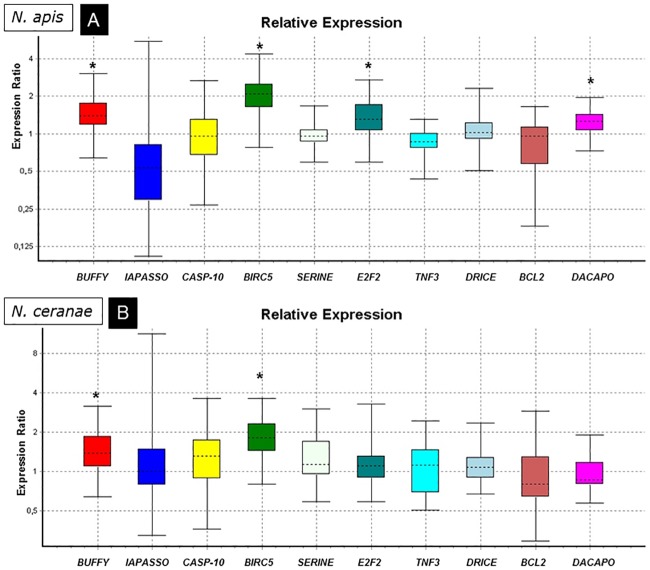
The expression of genes related to apoptosis (Fig 1) was analyzed in the honey bees infected either N. apis (AH; Fig 1A) or N. ceranae (CH, Fig 1B) and it was compared with that in the uninfected control group (TH), all of them treated with cycloheximide. The results showed the up-regulation of Buffy and BIRC5 in both the AH and CH groups for the (P<0.05; Table 2) and the down-regulation of IAPASSO and TNF3 in the AH group alone, that was close to the level of significance (P = 0.055 and P = 0.057, respectively). Finally, the E2F and Dacapo genes were up-regulated in the AH group (P<0.05).
Fig 1. Relative expression ratio plots for apoptosis related genes.
Analysis of groups treated with cycloheximide and infected with N. apis (A) or N. ceranae (B) relative to uninfected bees. (*) significant differences.
Table 2. Expression of studied target genes.
| N. apis | N. ceranae | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene short ID | Reaction Efficiency | Expression | Std. Error | 95% C.I. | P(H1) | Result | Expression | Std. Error | 95% C.I. | P(H1) | Result |
| EF | 0.969 | ||||||||||
| Actin | 0.922 | ||||||||||
| CYTOX | 0.921 | 0.829 | 0.557–1.253 | 0.420–1.828 | 0.174 | 1.208 | 0.905–1.629 | 0.715–2.094 | 0.059 | ||
| TU-MITO | 0.773 | 0.860 | 0.594–1.307 | 0.447–1.878 | 0.262 | 1.096 | 0.788–1.455 | 0.610–1.789 | 0.375 | ||
| S-12 | 1.0 | 0.798 | 0.517–1.224 | 0.382–1.852 | 0.149 | 1.056 | 0.768–1.468 | 0.605–1.832 | 0.582 | ||
| L16 | 0.998 | 1.041 | 0.768–1.494 | 0.565–1.878 | 0.722 | 1.212 | 0.952–1.520 | 0.755–1.833 | 0.022 | UP | |
| LSU | 0.917 | 0.778 | 0.458–1.326 | 0.279–2.860 | 0.219 | 1.145 | 0.878–1.524 | 0.670–1.805 | 0.155 | ||
| M-PHASE | 0.8405 | 0.857 | 0.677–1.023 | 0.467–1.785 | 0.183 | 1.024 | 0.859–1.202 | 0.764–1.513 | 0.708 | ||
| RING | 0.9535 | 0.777 | 0.437–1.349 | 0.358–2.752 | 0.221 | 0.580 | 0.342–1.005 | 0.282–1.803 | 0.007 | DOWN | |
| B1 CYCLIN | 0.8715 | 1.393 | 1.071–1.871 | 0.856–2.324 | 0.001 | UP | 0.900 | 0.684–1.146 | 0.551–1.521 | 0.249 | |
| B3 CYCLIN | 0.9425 | 1.032 | 0.690–1.523 | 0.421–2.422 | 0.820 | 1.043 | 0.697–1.508 | 0.471–1.898 | 0.754 | ||
| K CYCLIN | 0.804 | 0.92 | 0.685–1.230 | 0.541–1.629 | 0.434 | 1.011 | 0.795–1.288 | 0.646–1.595 | 0.896 | ||
| E CYCLIN | 0.977 | 0.831 | 0.651–1.481 | 0.078–2.086 | 0.752 | 1.192 | 0.935–1.502 | 0.718–1.758 | 0.047 | UP | |
| JH | 1.0 | 1.154 | 0.768–1.805 | 0.499–2.580 | 0.325 | 1.112 | 0.756–1.678 | 0.657–2.424 | 0.406 | ||
| VG | 0.905 | 0.611 | 0.249–1.559 | 0.101–2.543 | 0.112 | 1.727 | 0.756–4.300 | 0.355–7.929 | 0.064 | ||
| GAPDH | 0.9855 | 0.795 | 0.550–1.278 | 0.217–1.814 | 0.230 | 0.928 | 0.591–1.382 | 0.302–1.832 | 0.668 | ||
| 18S | 1.0 | 1.147 | 0.688–1.953 | 0.495–3.187 | 0.448 | 0.723 | 0.308–1.276 | 0.220–2.422 | 0.149 | ||
| Buffy | 0.9175 | 1.422 | 1.052–1.945 | 0.783–2.495 | 0.007 | UP | 1.407 | 0.962–2.101 | 0.776–2.701 | 0.034 | UP |
| IAPASSO | 0.977 | 0.529 | 0.230–0.932 | 0.128–3.738 | 0.055 | 1.202 | 0.729–1.939 | 0.398–6.889 | 0.578 | ||
| CASP-10 | 0.994 | 0.951 | 0.572–1.677 | 0.348–2.439 | 0.800 | 1.256 | 0.779–2.109 | 0.434–3.180 | 0.264 | ||
| BIRC5 | 0.99 | 2.022 | 1.423–2.793 | 1.014–3.981 | 0.000 | UP | 1.834 | 1.273–2.704 | 0.842–3.375 | 0.004 | UP |
| SERINE | 0.9575 | 0.953 | 0.817–1.152 | 0.604–1.462 | 0.539 | 1.238 | 0.777–1.901 | 0.599–2.439 | 0.211 | ||
| E2F2 | 0.911 | 1.340 | 0.920–1.944 | 0.730–2.423 | 0.040 | UP | 1.158 | 0.836–1.502 | 0.648–2.823 | 0.337 | |
| TNF3 | 0.867 | 0.840 | 0.648–1.040 | 0.469–1.201 | 0.057 | 1.041 | 0.607–1.667 | 0.522–2.259 | 0.806 | ||
| DRICE | 0.946 | 1.041 | 0.787–1.354 | 0.530–1.867 | 0.721 | 1.102 | 0.804–1.454 | 0.699–2.035 | 0.382 | ||
| BCL2 | 0.988 | 0.790 | 0.471–1.207 | 0.320–1.417 | 0.174 | 0.881 | 0.587–1.522 | 0.324–2.468 | 0.529 | ||
| Dacapo | 0.9545 | 1.229 | 0.976–1.523 | 0.775–1.815 | 0.026 | UP | 0.957 | 0.718–1.380 | 0.604–1.677 | 0.694 | |
EF and βActin were used as reference genes (2000 iterations). UP = up-regulation; DOWN = down-regulation. P(H1) = Probability of an alternate hypothesis that the difference between sample and control groups is due only to chance.
Cell cycle, mitochondrial activity and hormone related genes
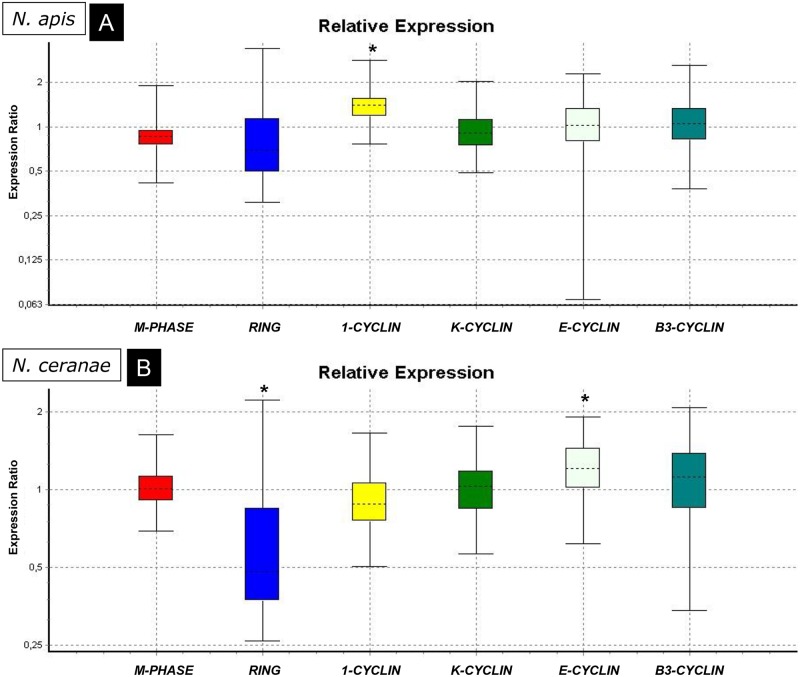
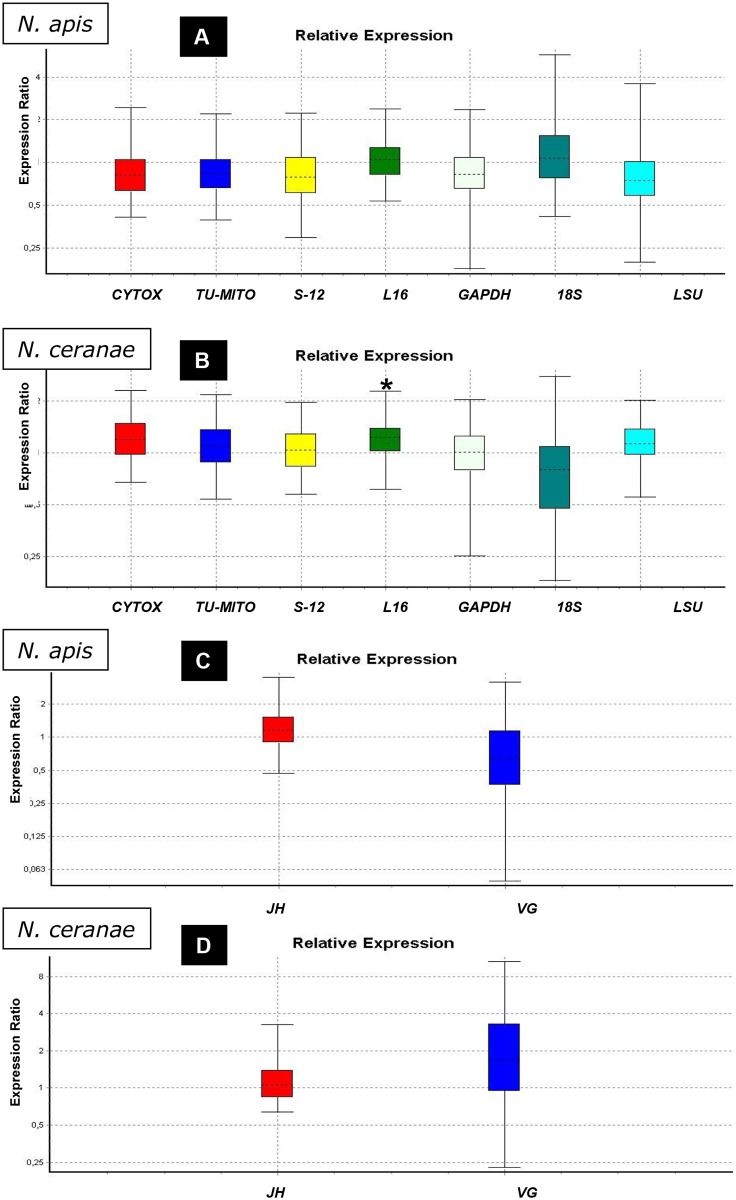
The expression of the genes related to the cell cycle, mitochondrial activity and hormones (Fig 2) was studied in groups A (N. apis infected bees; Fig 2A) and C (N. ceranae infected bees, Fig 2B) and they were compared with those in group T (uninfected bees). Only Cyclin B1 was up-regulated in group A (P<0.05; Table 2) while in the case of group C, one gene related to mitochondrial activity (L16) was up-regulated (P<0.05; Table 2) and another one (CYTOX) was very close to the level of significance for up-regulation (Fig 3A and 3B). Regarding cell cycle related genes (Fig 2), RING was down-regulated in group C while Cyclin E was up-regulated (both P<0.05; Table 2). No significant changes in the expression of VG or JH (Fig 3C and 3D) were observed in any of microsporidia infected bees, although it is important to note the up-regulation for VG close to the level of significance in group C.
Fig 2. Relative expression ratio plots for genes related to the cell cycle.
Analysis of the groups infected with N. apis (A) or N. ceranae (B) relative to the uninfected bees. (*) significant differences.
Fig 3. Relative expression ratio plots for genes related to mitochondrial and hormone activity.
Analysis of groups infected with N. apis or N. ceranae relative to the uninfected bees. (*) significant differences. A) Mitochondrial activity in N. apis infected bees. B) Mitochondrial activity in N. ceranae infected bees. The expression for GAPDH and 18S (not used as housekeeping genes) is also represented here. C) Hormone activity in N. apis infected bees. D) Hormone activity in N. ceranae infected bees.
Histology (TUNEL assay)
Our study showed that cycloheximide induced apoptosis in bees (Table 3; Fig 4) with the control bees (TH) showing a high level of cell death (69, 95% ± 14.35). Apoptosis appeared to be reduced in the microsporidia infected tissues although this was no significant (ANOVA, F = 1.109; P>0.05). This reduction in apoptosis was higher in N. apis infected ventriculi (27.86% reduction) than in the N. ceranae infected ventriculi (13.82% reduction). Apoptotic TUNEL positive cells (green stained) were detected along the epithelium in the ventriculi (Fig 4A, 4B and 4C). The amount of positive apoptotic cells were higher in the positive controls (TH, uninfected bees treated with cycloheximide; Fig 4A), compared to ventriculi of basal controls (uninfected, no cycloheximide treated bees, Fig 4D). Ventriculi from infected bees either infected by N. apis (Fig 4B) or N. ceranae (Fig 4C) showed a lower amount of TUNEL positive cells compared to ventriculi from TH group bees (Fig 4A).
Table 3. Percentage of ventricular cell nuclei undergoing apoptosis in cycloheximide treated bees.
| Average | Std. deviation | % Reduction in Apoptosis | ||
|---|---|---|---|---|
| Controls (uninfected) | TH | 69.95 | 14.35 | - |
| Infected with N. apis | AH | 50.46 | 11.70 | 27.86% |
| Infected with N. ceranae | CH | 60.27 | 13.72 | 13.82% |
Fig 4. TUNEL assay.
Representative TUNEL / DAPI stained and merged microscopic images of transverse sections from the ventriculum of A: uninfected (controls TH) B: infected with N. apis (AH) or C: N. ceranae (CH) honey bees and treated with cycloheximide. D: Basal apoptosis in uninfected bees and no treated with cycloheximide (Basal control) was very low. The ventriculum cells were counterstained with DAPI (blue). Scale bar = 100 μm. The ventricular lumen is indicated by an asterisk and spores inside ventricular cells are indicated with red arrows.
Discussion
The natural response of an infected cell is to undergo apoptosis in order to prevent the multiplication and dissemination of the invader. Conversely, the invader must find ways to evade this to be able to reproduce [6]. In this work, microsporidia have been shown to successfully manipulate the host cell’s metabolism to progress along the parasite’s life cycle, not only modifying the expression of apoptotic genes but also other important routes implicated in the host’s cell cycle. These responses seem to offer an important advantage, because similar conclusions have been reported for many intracellular parasites [3–7, 14].
To investigate the expression of some genes related to apoptosis, a potent intrinsic inducer of apoptosis was used (cycloheximide, a protein synthesis inhibitor that halts translational elongation [27]). Studies of the apoptotic index derived when using the TUNEL assay and caspase-3 staining suggested that N. ceranae could prevent the epithelial cells of infected honey bee tissue (ventriculi) from undergoing apoptosis [15]. For that reason, in this work a high level of apoptosis was induced in a group of bees in order to determine if N. ceranae or N. apis infection blocked the effect of this strong apoptosis inducer. The tissue of uninfected-bees treated with cycloheximide exhibited more apoptosis than that infected by microsporidia, with a clear reduction in the apoptotic index evident in TUNEL assays. Indeed, this is the first study to confirm this effect in N. apis infected tissues. As such, microsporidia infection clearly inhibits cycloheximide-induced apoptosis (intrinsic route) as described in Leishmania donovani infected macrophages [28].
This inhibition of apoptosis is consistent with the observed modifications in the expression of the apoptosis related genes studied. Infection with both microsporidia species up-regulates buffy and BIRC5, two genes with important function in programmed cell death (Fig 5). The BIRC5 (baculoviral IAP repeat-containing 5; also called Survivin) is a member of the inhibitor of apoptosis (IAP) family and it encodes a protein that inhibits caspase activation, thereby negatively regulating apoptosis [29–31]. On the other hand, buffy was identified in Drosophila melanogaster and it encodes a Bcl-2-like pro-survival protein [32]. The Bcl-2 (B cell lymphoma-2) protein family plays a central role in the intrinsic apoptotic pathway, controlling the integrity of the outer mitochondrial membrane. Consequently, in honey bees the up-regulation of BIRC5 and buffy is associated with a reduction in the number of cells that finally enter apoptosis in the infected tissues. These results agree with a previous work where inhibitors of apoptosis proteins were seen to be up-regulated after N. ceranae infection in sensitive bees while caspase 10 gene expression was not modified [16]. Consequently, IAP family seems to play a key role in the inhibition of these processes. Additionally, this is the first report of such changes in N. apis infected bees, confirming that the inhibition of apoptosis is a common response of the host to the benefit of this group of intracellular parasitic fungi.
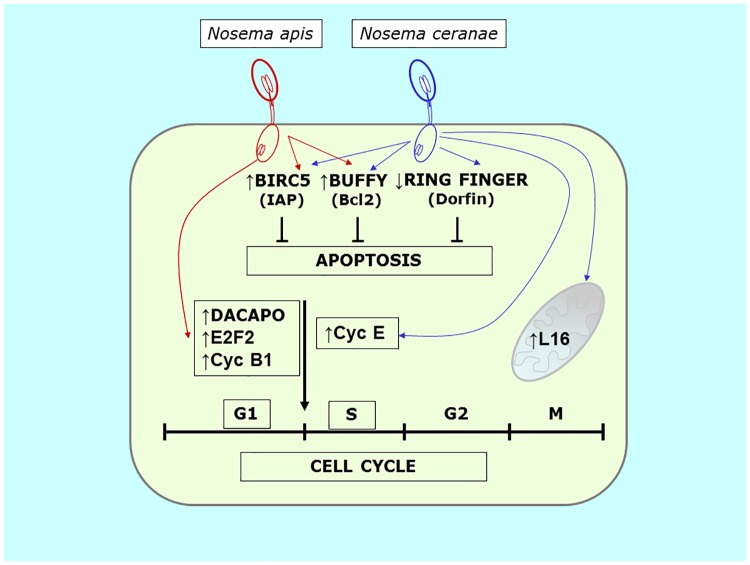
Fig 5. Scheme of the genes altered in N. ceranae and N. apis infection in honey bees and the effects on host’s cell cycle, apoptosis and mitochondrial activity.
We also note that IAPASSO (viral IAP-associated factor; VIAF) and TNF3 (Tumor Necrosis Factor receptor-associated factor 3 interacting protein 1) displayed a tendency towards down-regulation (P = 0.055 and p = 0.057, respectively) in N. apis infected bees. IAPASSO is a conserved inhibitor of apoptosis interacting factor that modulates caspase activation during apoptosis [33] and TNF3 is a member of the TNF superfamily proteins involved in complex pathways that regulate cellular survival, proliferation, differentiation and apoptosis [34, 35]. Consequently, the tendency to down-regulate both these genes in conjunction with buffy and BIRC5 overexpression fits into the theory that apoptosis is inhibited by the microsporidium N. apis.
Regarding the cell cycle, it has been suggested that the G1 phase is vital to decide if a cell will go on to differentiate, multiply, apoptose or become quiescent or senescent [36]. A balance between cell proliferation and apoptosis is essential for the development of the multicellular organism [32]. In our study, a modification of cell cycle related genes was observed in an epithelium (bee ventriculi) considered to be non-replicative beyond its basal germinative layer.
Curiously, while both microsporidia inhibit apoptosis through the same pathways (Buffy and BIRC5), the modifications to cell cycle related genes differ. However, both Nosema species caused the up-regulation of some cyclins in the infected cells: over expression of cyclin B1 (cycB1) by N. apis and cyclin E (cycE) by N. ceranae (Fig 5).
N. apis infected tissues also over expressed dacapo and E2F2, both of them closely related with the progression of the G1-S phase. Dacapo is a cyclin dependent kinase inhibitor that coordinates the rates of G1-S and G2-M progression, maintaining normal rates of proliferation when cell cycle controls are perturbed [37]. Also, E2F2 (E2F Transcription Factor 2) is a transcription factor maximally expressed late in G1 that plays a pivotal role in G1/S transition [38, 39]. Over expression of both these genes suggest that the infected cells undergo the G1-S phase progression. However, the up-regulation of cyclin B1 (G2/Mitotic-specific Cyclin-B1) was initially unexpected since it codes for a regulatory protein involved in mitosis and it contributes to all or none switch-like behavior of the cell in deciding to commit to mitosis. E2F2 was overexpressed in rabbit corneal endothelial cells (arrested in G1-phase) and it coexisted with high levels of cyclin B1 [40] and there was strong evidence of progression from the G1-phase arrested state to at least the G2-phase in these tissues. Our data suggest a progression from G1 to S phase is being promoted in N. apis infected tissues (Fig 5).
In N. ceranae infected bees, significant changes were observed in the expression of cyclin E (CycE) and RING (RING-finger protein 19 or Dorfin). A previous study of the transcriptome of A. mellifera infected by N. ceranae identified mRNA for a number of cyclins (as cyclin E) and other proteins closely related to cell cycle [41] although no data on expression were available. Our results show an up-regulation of CycE, which appears to be the most important cyclin for the G1 to S transition [42, 43]. Consequently, and as in N. apis infection, G1-S phase progression is apparently being stimulated in N. ceranae infected cells, albeit through this different pathway.
Additionally, down regulation of the RING gene (RING-finger protein 19 or Dorfin) was observed after N. ceranae infection. This gene codes for the E3 ubiquitin-protein ligase RNF19A which in Caenorhabditis elegans has been described as one of the three core components of a complex that target toxins and intracellular pathogen proteins for degradation [44]. In general these RING finger E3 proteins can influence the balance between proliferation and apoptosis. In response to apoptotic stimuli the E3 activity of IAPs leads to their auto-ubiquitination, degradation, and progression toward cell death [45]. Ubiquitin pathways have been described as a specific mechanism of host defence against microsporidia infection [44]. Although RING was studied in bees from Group C here (cell cycle assay) its down-regulation provides more information about apoptosis and corroborates the results obtained in group CH (treated with cycloheximide). In fact, these data suggest another pathway to inhibit apoptosis that may be activated by N. ceranae infection.
Microsporidia are obligate intracellular parasites with extremely reduced genomes and a dependence on host-derived ATP. Honey bee cells infected with N. ceranae were enlarged and the cytoplasm contained a larger number of host mitochondria and free ribosomes. Several mitochondria were close to and surrounded the plasmalemma of meronts [18], similar to sporulating Buxtehucdea scaniae [46], suggesting a energy supply external to the parasite. For this reason, the regulation of five host mitochondrial related genes was also included in this study. However only one of them (MRP L16) was seen to be up-regulated and only in N. ceranae infected bees (Fig 5). MRP L16 is a nuclear gene that encodes for the mitochondrial ribosomal protein L16. This polypeptide plays an important role in the assembly and structure of the peptidyl transferase centre of the ribosome, and it is crucial for the correct behavior of mito-ribosomes in yeast [47]. MRP over expression depends mainly on glucose repression and de-repression, which decreases or elevates MRP mRNA and protein levels, respectively [40]. It is important to note that N. ceranae infection has been reported to produce a nutritional and energetic stress in bees that leads to a major ingestion of food that is rich in glucose [48–50]. On the other hand, the stable accumulation of L16 depends on the presence of mitochondrial rRNA [47], so changes in the integrity of mitochondrial activity directly affect the levels of this protein too. The related genes studied in this work, CYTOX, TU-MITO, S12 and LSU, suggest normal mitochondrial activity that would allow the accumulation of L16 protein inside this organelle. In fact, the expression of Cytochrome C oxidase subunit VI gene is remarkable high, at levels that are almost significant(p = 0.059), which would fit into the theory of the mitochondria overworking during N. ceranae infection. Mitochondrial protein modulation by intracellular parasites has been reported previously in Toxoplasma gondii infection [1]. The direct or indirect perturbation of mitochondria dynamics may play a crucial role in the sustained viability of host cells preserving the pathogen’s replication niche or alternatively triggering the apoptotic process to circumvent immune effector cells. It will be crucial to identify the host’s molecular mechanisms and the pathogenic factors involved in such control [7]. Finally, no significant results were observed in terms of mitochondrial activity in bees infected by N. apis. This again indicates that both microsporidia affect the same host in a different manner.
Finally, the expression of vitellogenin and juvenile hormone were also assessed here. These molecules have been related to the bees’ immune response and to other physiological activities such as in behavioural development [51, 52], and some modifications to their expression were previously reported in bees infected by Nosema spp. [53–55]. However, no significant modification was detected here for either hormone, probably because the effects of infection were only studied in the target tissue (ventricular cells) and the hormone secreting organs were not assessed.
In multicellular organisms, cell proliferation and death must be regulated to maintain tissue homeostasis. Many observations suggest that this regulation may be at least partially achieved by coupling the process of cell cycle progression and programmed cell death through a set of common factors. Evidence is accumulating that manipulation of the cell cycle may either prevent or induce an apoptotic response arguing in favor of a link between the cell cycle and apoptosis [56]. For bee microsporidia, this is the first report describing molecular pathways related to parasite-host interactions that probably serve to promote their own survival within the cell. New routes of apoptosis involving Buffy and BIRC5 were seen to be modified by both microsporidia, while the effects of the microsporidia on the expression of genes related with the cell cycle indicate that both Nosema species apparently promote the G1/S phase through different pathways (cyclin B1/ dacapo/ E2F2 by N. apis, cyclin E byN. ceranae). Lastly, the changes to mitochondrial markers appear to further support a distinct response to the two Nosema species studied as only N. ceranae infection leads to higher mitochondrial activity. All these evidences demonstrate the strong interaction between the host cells and this group of parasites. These results increase further our knowledge on the specific pathways that these microsporidia uses to survive and multiply in the host, shedding light on the pathogenic mechanism of A. mellifera microsporidia, and hence on the relationship between these insects and their intracellular parasites.
Supporting information
Genes related to cell cycle, mitochondrial activity, apoptosis, hormone activity and housekeeping genes.
(DOCX)
Sequences of primers and probes used in Real-Time PCR for the detection of pathogens in individual bee samples and PCR conditions were obtained from previously published works.
(DOCX)
Sequences designed for Real-Time PCR to study the cell cycle, mitochondrial activity, apoptosis, hormone activity and housekeeping genes. All of them were designed in this study except 18S and EF that have been published previously.
(DOCX)
Acknowledgments
The authors wish to thank V. Albendea, C. Rogerio, T. Corrales and M. Gajero for their technical support.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by INIA (FEDER) PROJECT RTA2012-00072-C02-01. The funding organization did not play a role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript and only provided financial support in the form of research materials. In fact, this organization only provided funds for acquisition of research materials and they provided no salaries for any author.
References
- 1.Nelson MM, Jones AR., Carmen JC, Sinai AP, Burchmore R, Wastling JM. Modulation of the Host Cell Proteome by the Intracellular Apicomplexan Parasite Toxoplasma gondii. Infect Immun. 2008;76: 828–844. 10.1128/IAI.01115-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta S. Parasite immune escape: new views into host–parasite interactions. Curr Opin Microbiol. 2005;8: 428–433. 10.1016/j.mib.2005.06.011 [DOI] [PubMed] [Google Scholar]
- 3.Lüder CGK, Gross UWE, Lopes MF. Intracellular protozoan parasites and apoptosis: diverse strategies to modulate parasite-host interactions. Trends Parasitol. 2001;17: 480–486. [DOI] [PubMed] [Google Scholar]
- 4.Bruchhaus I, Roeder T, Rennenberg A, Heussler VT. Protozoa parasites: programmed cell death as mechanism of parasitism. Trends Parasitol. 2007; 23: 376–383. 10.1016/j.pt.2007.06.004 [DOI] [PubMed] [Google Scholar]
- 5.Heussler VT, Küenxi P, Rottenberg S. Inhibition of apoptosis by intracellular porotzoan parasites. Int J Parasitol. 2001;31: 1166–1176. [DOI] [PubMed] [Google Scholar]
- 6.James ER, Green DR. Manipulation of apoptosis in the host-parasite interaction. Trends Parasitol. 2004;20: 280–287. 10.1016/j.pt.2004.04.004 [DOI] [PubMed] [Google Scholar]
- 7.Rodrigues V, Cordeiro-da-Silva A, Laforge M, Ouaissi A, Silvestre R, Estaquier J. Modulation of Mammalian Apoptotic Pathways by Intracellular Protozoan Parasites. Cell Microbiol. 2012;14(3): 325–333. 10.1111/j.1462-5822.2011.01737.x [DOI] [PubMed] [Google Scholar]
- 8.Manque PA, Probst C, Pereira MCS, Rampazzo RCP, Ozaki LS, Pavoni DP, et al. Trypanosoma cruzi Infection Induces a Global Host Cell Response in Cardiomyocytes. Infect Immunity 2011;79: 1855–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anonymous. Insights into social insects from the genome of the honeybee Apis mellifera. Nature 2006;443: 931–949. 10.1038/nature05260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Higes M, Martín-Hernández R, Meana A. Nosema ceranae in Europe: an emergent type C nosemosis. Apidologie 2010;41: 375–392. [Google Scholar]
- 11.Zander E. Tierische Parasiten als Krankheitserreger bei der Biene. Leipziger Bienenztg 1909;24: 147–150, 164–166. [Google Scholar]
- 12.Fries I, Feng F, da Silva A, Slemenda SB, Pieniazek NJ. Nosema ceranae n.sp. (Microspora, Nosematidae), morphologycal and molecular characterization of a Microsporidian parasite of the Asian honey bee Apis cerana (Hymenoptera, Apidae). Eurp J Protistol. 1996;32: 356–365. [Google Scholar]
- 13.Szumowski SC, Troemel ER. Microsporidia-host interactions. Curr Opin Microbiol. 2015;26: 10–16. 10.1016/j.mib.2015.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams BAP. Unique physiology of host-parasite interactions in microsporidia infections. Cell Microbiol. 2009;11(11): 1551–1560. 10.1111/j.1462-5822.2009.01362.x [DOI] [PubMed] [Google Scholar]
- 15.Higes M, Juarranz A, Dias-Almeida J, Lucena S, Botías C, Meana A, et al. Apoptosis in the pathogenesis of Nosema ceranae (Microsporidia: Nosematidae) in honey bees (Apis mellifera). Environ Microbiol Rep. 2013;5: 530–536. 10.1111/1758-2229.12059 [DOI] [PubMed] [Google Scholar]
- 16.Kurze C, Le Conte Y, Dussaubat C, Erler S, Kryger P, Lewkowski O, et al. Nosema tolerant honeybees (Apis mellifera) escape parasitic manipulation of apoptosis. PLoS ONE 2015; 10(10): e0140174 10.1371/journal.pone.0140174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dussaubat C, Brunet JL, Higes M, Colbourne JK, Lopez J, Choi JH, et al. Gut pathology and responses to the microsporidium Nosema ceranae in the honey bee Apis mellifera. PLoS ONE 2012;7(5): e37017 10.1371/journal.pone.0037017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higes M, García-Palencia P, Martín-Hernández R, Meana A. Experimental infection of Apis mellifera with Nosema ceranae (Microsporidia) J Invertebr Pathol. 2007;94: 211–217. 10.1016/j.jip.2006.11.001 [DOI] [PubMed] [Google Scholar]
- 19.Martín-Hernández R, Meana A, García-Palencia P, Marín P, Botías C, Garrido-Bailón E, et al. Effect of Temperature on the biotic potential of Honeybee Microsporidia. App Environ Microbiol. 2009;75(8): 2554–2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martín-Hernández R, Botías C, Bailón EG, Martínez-Salvador A, Prieto L, Meana A, et al. Microsporidia infecting Apis mellifera: coexistence or competition. Is Nosema ceranae replacing Nosema apis? Environ Microbiol 2012;14: 2127–2138. 10.1111/j.1462-2920.2011.02645.x [DOI] [PubMed] [Google Scholar]
- 21.Scanlon M, Leitch GJ, Shaw AP, Moura H, Visvesvara GS. Susceptibility to apoptosis is reduced in the microsporidia infected host. J Eukaryot Microbiol. 1999; 46: 34S–35S. [PubMed] [Google Scholar]
- 22.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3: research0034.1–research0034.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scharlaken B, de Graaf DC, Goossens K, Brunain M, Peelman LJ, Jacobs FJ. Reference gene selection for insect expression studies using quantitative real-time PCR: The honeybee, Apis mellifera, head after a bacterial challenge. J Insect Sci. 2008;8: 33. [Google Scholar]
- 24.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30(9):e36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Botías C, Martín-Hernández R, Días J, García-Palencia P, Matabuena M, Juarranz A., et al. The effect of induced queen replacement on Nosema spp. infection in honey bee (Apis mellifera iberiensis) colonies. Environ Microbiol. 2012;14: 845–859. 10.1111/j.1462-2920.2011.02647.x [DOI] [PubMed] [Google Scholar]
- 27.Martin SJ, Highfield AC, Brettell L, Villalobos EM, Budge GE, Powell M, et al. Early redistribution of plasma membrane phosphatidylserine is a general feature of apoptosis regardless of the initiating stimulus: inhibition by overexpression of Bcl-2 and Abl. J Exp Med. 1995;182: 1545–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Donovan MJ, Maciuba BZ, Mahan CE, McDowell MA. Leishmania infection inhibits cycloheximide-induced macrophage apoptosis in a strain dependent manner. Exp Parasitol. 2009;123, 58–64. 10.1016/j.exppara.2009.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altieri DC. Molecular cloning of effector cell protease receptor-1, a novel cell surface receptor for the protease factor Xa. J Biol Chem. 1994;269: 3139–3142. [PubMed] [Google Scholar]
- 30.Altieri DC. Splicing of effector cell protease receptor-1 mRNA is modulated by an unusual retained intron. Biochem. 1994;33: 13848–13855. [DOI] [PubMed] [Google Scholar]
- 31.Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nature Med. 1997;3: 917–921. [DOI] [PubMed] [Google Scholar]
- 32.Quinn L, Coombe M, Mills K, Daish T, Colussi P, Kumar S, et al. Buffy, a Drosophila Bcl-2 protein, has anti-apoptotic and cell cycle inhibitory functions. Eur Mol Biol Organ J. 2003;22: 3568–3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilkinson JC, Richter BWM, Wilkinson AS, Burstein E, Rumble JM, Balliu B, et al. VIAF, a Conserved Inhibitor of Apoptosis (IAP)-interacting Factor That Modulates Caspase Activation. J Biol Chem. 2004; 279: 51091–51099. 10.1074/jbc.M409623200 [DOI] [PubMed] [Google Scholar]
- 34.Locksley RM, Killeen N, Lenardo MJ. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell 2001;104: 487–501. [DOI] [PubMed] [Google Scholar]
- 35.Bodmer J, Schneider P, Tschopp J. The molecular architecture of the TNF superfamily. Trends Biochem Sci. 2002;27(1): 19–26. [DOI] [PubMed] [Google Scholar]
- 36.Neganova I, Lako M. G1 to S phase cell cycle transition in somatic and embryonic stem cells. J Anatomy 2008;213: 30–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reis T, Edgar BA. Negative regulation of dE2F1 by cyclin-dependent kinases controls cell cycle timing. Cell 2004;117: 253–264. [DOI] [PubMed] [Google Scholar]
- 38.Royzman I, Whittaker AJ, Orr-Weaver TL. Mutations in Drosophila DP and E2F distinguish G1-S progression from an associated transcriptional program. Genes Dev. 1997;11: 1999–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duronio RJ, Bonnette PC, O'Farrell PH. Mutations of the Drosophila dDP, dE2F, and cyclin E genes reveal distinct roles for the E2F-DP transcription factor and cyclin E during the G1-S transition. Mol Cell Biol. 1998;18:141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Joyce NC, Harris DL, Mc Alister JC, Ali RR, Larkin DF. Effect of overexpressing the transcription factor E2F2 on cell cycle progression in rabbit corneal endothelial cells. Invest Ophthalmol Vis Sci. 2004;45: 1340–1348. [DOI] [PubMed] [Google Scholar]
- 41.Aufauvre J, Misme-Aucouturier B, Viguès B, Texier C, Delbac F, et al. (2014) Transcriptome Analyses of the Honeybee Response to Nosema ceranae and Insecticides. PLoS ONE 9(3): e91686 10.1371/journal.pone.0091686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knoblich JA, Sauer K, Jones L, Richardson H, Saint R, Lehner CF. et al. Cyclin E controls S phase progression and its down-regulation during Drosophila embryogenesis is required for the arrest of cell proliferation. Cell 1994;77: 107–120. [DOI] [PubMed] [Google Scholar]
- 43.Richardson H, O’Keefe LV, Marty T, Saint R. Ectopic cyclin E expression induces premature entry into S phase and disrupts pattern formation in the Drosophila eye imaginal disc. Development 1995;121:3371–3379. [DOI] [PubMed] [Google Scholar]
- 44.Bakowski MA, Desjardins CA, Smelkinson MG, Dunbar TA, Lopez-Moyado IF, Rifkin SA, et al. Ubiquitin- mediated response to microsporidia and virus infection in C. elegans. PLoS Pathog. 2014;10:e1004200 10.1371/journal.ppat.1004200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang Y, Fang S, Jensen JP, Weissman AM, Ashwell JD. Ubiquitin Protein Ligase Activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science 2000;288: 874–877. [DOI] [PubMed] [Google Scholar]
- 46.Vavra J, Larsson JIR. Structure of the Microsporidia In: Murray W., Louis M.W., editors. The Microsporidia and Microsporidiosis. Washington DC: American Society of Microbiology;1999. pp. 7–84. [Google Scholar]
- 47.Pan C, Mason TL. Identification of the yeast nuclear gene for the mitochondrial homologue of bacterial ribosomal protein L16. Nucleic Acids Res. 1995;23: 3673–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mayack C, Naug D. Energetic stress in the honeybee Apis mellifera from Nosema ceranae infection. J Invertebr Pathol. 2009;100: 185–188. 10.1016/j.jip.2008.12.001 [DOI] [PubMed] [Google Scholar]
- 49.Aliferis KA, Copley T, Jabaji S. Gas chromatography-mass spectrometry metabolite profiling of worker honey bee (Apis mellifera L.) hemolymph for the study of Nosema ceranae infection. J Insect Physiol. 2012;58: 1349–1359. 10.1016/j.jinsphys.2012.07.010 [DOI] [PubMed] [Google Scholar]
- 50.Vidau C, Panek J, Texier C, Biron DG, Belzunces lP, Le Gall M, et al. Differential proteomic analysis of midguts from Nosema ceranae-infected honeybees reveals manipulation of key host functions. J Invertebr Pathol. 2014;121: 89–96. 10.1016/j.jip.2014.07.002 [DOI] [PubMed] [Google Scholar]
- 51.Robinson GE, Vargo EL. Juvenile hormone in adult eusocial Hymenoptera: gonadotropin and behavioral pacemaker. Arch Insect Biochem Physiol. 1997;35(4): 559–583. [DOI] [PubMed] [Google Scholar]
- 52.Nelson CM, Ihle KE, Fondrk MK, Page RE, Amdam GV. The gene vitellogenin has multiple coordinating effects on social organization. PLoS Biology. 2007;5(3):e62 10.1371/journal.pbio.0050062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goblirsch M, Huang ZY, Spivak M. Physiological and behavioral changes in honey bees (Apis mellifera) induced by Nosema ceranae infection. PLoS ONE 2013;8(3): e58165 10.1371/journal.pone.0058165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ares AM, Nozal MJ, Bernal JL, Martin-Hernandez R, Higes M, Bernal J. Liquid chromatography coupled to ion trap-tandem mass spectrometry to evaluate juvenile hormone III levels in bee hemolymph from Nosema spp. infected colonies. J Chromatogr B 2012;899: 146–153. [DOI] [PubMed] [Google Scholar]
- 55.Ma Z, Li C, Pan G, Li Z, Han B, Xu J, et al. Genome-Wide Transcriptional Response of Silkworm (Bombyx mori) to Infection by the Microsporidian Nosema bombycis. PLoS ONE 2013;8(12): e84137 10.1371/journal.pone.0084137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pucci B, Kasten M, Giordano A. Cell Cycle and Apoptosis. Neoplasia 2000;2: 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes related to cell cycle, mitochondrial activity, apoptosis, hormone activity and housekeeping genes.
(DOCX)
Sequences of primers and probes used in Real-Time PCR for the detection of pathogens in individual bee samples and PCR conditions were obtained from previously published works.
(DOCX)
Sequences designed for Real-Time PCR to study the cell cycle, mitochondrial activity, apoptosis, hormone activity and housekeeping genes. All of them were designed in this study except 18S and EF that have been published previously.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.