Abstract
Genetic association studies have identified 215 risk loci for inflammatory bowel disease 1–8, which have revealed fundamental aspects of its molecular biology. We performed a genome-wide association study of 25,305 individuals, and meta-analyzed with published summary statistics, yielding a total sample size of 59,957 subjects. We identified 25 new loci, three of which contain integrin genes that encode proteins in pathways identified as important therapeutic targets in inflammatory bowel disease. The associated variants are correlated with expression changes in response to immune stimulus at two of these genes (ITGA4, ITGB8) and at previously implicated loci (ITGAL, ICAM1). In all four cases, the expression increasing allele also increases disease risk. We also identified likely causal missense variants in the primary immune deficiency gene PLCG2 and a negative regulator of inflammation, SLAMF8. Our results demonstrate that new common variant associations continue to identify genes relevant to therapeutic target identification and prioritization.
Inflammatory bowel disease (IBD) is a chronic, debilitating, disorder of the gastrointestinal tract that includes two common disease subtypes, Crohn’s disease and ulcerative colitis. Disease pathogenesis is poorly understood but is likely driven by a dysregulated immune response to unknown environmental triggers in genetically susceptible individuals. Treatment regimes often use potent immunomodulators to achieve and maintain remission of symptoms. However, patients commonly experience side effects, lose response to treatment, or develop complications of IBD, with many ultimately requiring major abdominal surgery. Previous genome-wide association studies (GWAS) and targeted follow-up using the Immunochip have been very successful at identifying genetic risk loci for IBD, but increased biological understanding has not yet had a significant impact on therapy for these disorders.
In order to further expand our understanding of the biology of these disorders we carried out a GWAS of 12,160 IBD cases and 13,145 population controls of European ancestry that had not been included in any genome-wide meta-analysis of IBD to date (Supplementary Table 1, Online Methods). We imputed genotypes using a reference panel comprising whole genome sequences from 4,686 IBD cases9 and 6,285 publically available population controls10,11. Following quality control (Online Methods) we tested 9.7 million sites for association. At the 232 IBD associated SNPs in the latest meta-analysis by the International IBD Genetics Consortium1, 228 had effects in the same direction in our data, 188 showed at least nominal evidence of replication (P<0.05) and none showed significant evidence of heterogeneity of effect by Cochrane’s Q test. Among these replicated loci was a genome-wide significant association on chromosome 10q25 that was only previously significantly associated with Crohn’s disease in individuals of East Asian ancestry3,7, further supporting near complete sharing of genetic risk loci across populations1. We meta-analyzed our new GWAS data with previously published summary statistics from 12,882 IBD cases and 21,770 population controls imputed using the 1000 Genomes Project reference panel1 (Supplementary Figures 1-3, Supplementary Table 2). We observed inflation of the summary statistics (λGC = 1.23 and 1.29 for Crohn’s and ulcerative colitis, respectively), but LD score regression demonstrated that this was due to broad polygenic signal, rather than confounding population substructure (both intercepts = 1.09, Online Methods).
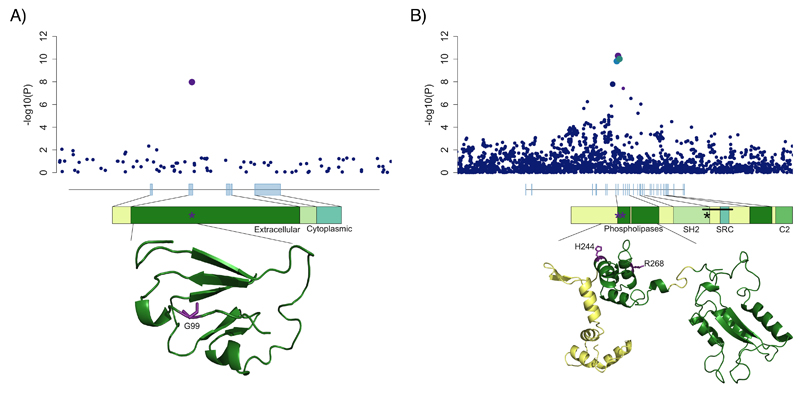
We identified 25 new loci at genome-wide significance (Table 1). In order to identify causal variants, genes and mechanisms, we performed a summary-statistic fine-mapping analysis on these loci, as well as 40 previously discovered loci that were genome-wide significant in our data but where fine-mapping had not yet been attempted12 (Online Methods, Supplementary Table 3). In order to be confident about fine-mapping inferences, we restricted subsequent analyses to 12 signals where we had high quality imputed data for all relevant variants (Online Methods). At 6 of these 12 loci we identified a single variant with >50% probability of being causal (Table 2, Supplementary Figures 4-6). Among these were two loci where a single variant had >99% probability of being causal: a missense variant predicted to affect protein function in SLAMF8, (rs34687326, p.Gly99Ser, Figure 1a), and an intronic variant in the key regulator of Th17 cell differentiation, RORC13. SLAMF8 is a cell surface receptor that is expressed on activated myeloid cells and has been reported to negatively regulate inflammatory responses by inhibiting their migration to sites of inflammation14 and repressing their production of reactive oxygen species (ROS)15. This, together with the observation that the risk-decreasing allele (MAF=0.1) is predicted to affect protein function (CADD=32.0, 92nd percentile of missense variants)16, suggests further experiments evaluating a possible gain-of-function mechanism may be worthwhile. RORC encodes RORγt, the master transcriptional regulator of Th17 cells13 and group 3 innate lymphoid cells17. Both of these cell types play important roles in defence at mucosal surfaces, especially in the intestine, and have been shown to contribute to the homeostasis between the intestinal immune system and gut microbiota18,19, an equilibrium that is known to be lost in inflammatory bowel disease20. Pharmacologic inhibition of RORγt has been shown to offer therapeutic benefit in mouse models of intestinal inflammation, and reduces the frequency of Th17 cells isolated from primary intestinal samples of IBD patients21.
Table 1. Novel IBD-associated loci.
| Rsid | Chr | Position bp | Left - right Mb | Risk Allele | Non - risk Allele | Risk Allele Frequency in 1000 Genomes CEU+GBR | PMeta | OR | 95% CI | Phenotype | Implicated gene |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs34687326 | 1 | 159799910 | 159.80 - 159.80 | G | A | 0.900 | 1.06 × 10-08 | 1.18 | 1.12 - 1.24 | CD | SLAMF8 |
| rs59043219 | 1 | 209970610 | 209.97 - 210.02 | A | G | 0.379 | 1.09 × 10-08 | 1.08 | 1.05 - 1.10 | IBD | - |
| rs6740847 | 2 | 182308352 | 182.31 - 182.33 | A | G | 0.508 | 1.22 × 10-13 | 1.10 | 1.07 - 1.12 | IBD | ITGA4 |
| rs144344067 | 2 | 187576378 | 187.50 - 187.68 | A | AT | 0.895 | 1.29 × 10-08 | 1.12 | 1.08 - 1.16 | IBD | - |
| rs1811711 | 2 | 228670476 | 228.67 - 228.67 | C | G | 0.826 | 6.09 × 10-09 | 1.14 | 1.10 - 1.18 | UC | - |
| rs76527535 | 2 | 242484701 | 242.47 - 242.49 | C | T | 0.745 | 2.87 × 10-08 | 1.09 | 1.06 - 1.12 | IBD | - |
| rs2581828 | 3 | 53133149 | 53.10 - 53.17 | C | G | 0.597 | 6.46 × 10-09 | 1.10 | 1.07 - 1.13 | CD | - |
| rs2593855 | 3 | 71175495 | 71.16 - 71.19 | C | T | 0.663 | 2.54 × 10-09 | 1.09 | 1.06 - 1.11 | IBD | - |
| rs503734 | 3 | 101023748 | 100.91 - 101.27 | A | G | 0.513 | 2.67 × 10-08 | 1.07 | 1.05 - 1.10 | IBD | - |
| rs56116661 | 3 | 188401160 | 188.40 - 188.40 | C | T | 0.795 | 5.67 × 10-10 | 1.14 | 1.10 - 1.18 | CD | - |
| rs11734570 | 4 | 38588453 | 38.58 - 38.59 | A | G | 0.368 | 4.80 × 10-08 | 1.07 | 1.05 - 1.10 | IBD | - |
| rs17656349 | 5 | 149605994 | 149.59 - 149.63 | T | C | 0.466 | 1.54 × 10-08 | 1.09 | 1.06 - 1.13 | UC | - |
| rs113986290 | 6 | 19781009 | 19.72 - 19.83 | C | T | 0.989 | 7.59 × 10-09 | 1.36 | 1.25 - 1.46 | UC | - |
| rs67289879 | 6 | 42007403 | 42.00 - 42.01 | T | C | 0.179 | 3.04 × 10-08 | 1.09 | 1.06 - 1.13 | IBD | - |
| rs11768365 | 7 | 6545188 | 6.50 - 6.55 | A | G | 0.816 | 3.88 × 10-08 | 1.09 | 1.06 - 1.12 | IBD | - |
| rs149169037 | 7 | 20577298 | 20.58 - 20.58 | G | A | 0.895 | 3.26 × 10-08 | 1.14 | 1.10 - 1.19 | IBD | ITGB8 |
| rs243505 | 7 | 148435339 | 148.40 - 148.58 | A | G | 0.624 | 3.04 × 10-10 | 1.08 | 1.06 - 1.11 | IBD | - |
| rs7911117 | 10 | 27179596 | 27.16 - 27.18 | T | G | 0.871 | 1.84 × 10-08 | 1.14 | 1.10 - 1.19 | UC | - |
| rs111456533 | 10 | 126439381 | 126.32 - 126.55 | G | A | 0.829 | 1.18 × 10-09 | 1.11 | 1.08 - 1.14 | IBD | - |
| rs80244186 | 13 | 42917861 | 42.84 - 42.94 | C | T | 0.111 | 3.66 × 10-08 | 1.13 | 1.09 - 1.18 | CD | - |
| rs11548656 | 16 | 81916912 | 81.91 - 81.92 | A | G | 0.961 | 5.18 × 10-11 | 1.27 | 1.20 - 1.34 | IBD | PLCG2 |
| rs10492862 | 16 | 82867456 | 82.87 - 82.92 | A | C | 0.308 | 1.26 × 10-09 | 1.11 | 1.08 - 1.15 | CD | - |
| rs4256018 | 20 | 6093889 | 6.08 - 6.10 | G | T | 0.250 | 1.23 × 10-08 | 1.08 | 1.05 - 1.11 | IBD | - |
| rs138788 | 22 | 35729721 | 35.72 - 35.74 | A | G | 0.418 | 2.95 × 10-08 | 1.09 | 1.06 - 1.13 | UC | - |
| rs4821544 | 22 | 37258503 | 37.26 - 37.26 | C | T | 0.321 | 1.76 × 10-08 | 1.10 | 1.07 - 1.13 | CD | - |
Table 2. Variants fine-mapped to >50% probability of being causal in their given signal.
| Rsid | Chr | Position (bp) | PCausal | Effect | Credible set size | Phenotype | PMeta | Locus type |
|---|---|---|---|---|---|---|---|---|
| rs34687326 | 1 | 159799910 | 1.000 | SLAMF8 p.Gly99Ser (missense) | 1 | CD | 1.06 × 10-08 | Novel |
| rs4845604 | 1 | 151801680 | 0.999 | RORC (intronic) | 1 | IBD | 7.09 × 10-14 | Known |
| rs1811711 | 2 | 228670476 | 0.914 | 2 | UC | 6.09 × 10-09 | Novel | |
| rs56116661 | 3 | 188401160 | 0.561 | LPP (intronic) | 11 | CD | 5.67 × 10-10 | Novel |
| rs11548656 | 16 | 81916912 | 0.502 | PLCG2 p.His244Arg (missense) | 3 | IBD | 5.18 × 10-11 | Novel |
| rs1143687 | 16 | 81922813 | 0.746 | PLCG2 p.Arg268Trp (missense) | 5 | IBD | 3.83 × 10-08 | Novel |
| rs4821544 | 22 | 37258503 | 0.804 | NCF4 (intronic) | 2 | CD | 1.76 × 10-08 | Novel |
Figure 1. Likely causal missense variants.
For A) SLAMF8 and B) PLCG2, local association results are plotted with point size corresponding to LD to our lead variant and color to fine-mapping probability (purple > 50%, intermediate blue 10-50%, navy blue <10%). Gene body diagrams and protein domain annotations are taken from ENSEMBL, and partial predicted crystal structures for both proteins are obtained from the SWISS-MODEL repository.
In loci where fine-mapping was less clearly resolved, we searched for likely functional variants, observing a missense variant (CADD=16.5, 50.2% probability of causality) in PLCG2. Furthermore, after conditioning on this variant, we discovered a second, independent, likely functional (CADD=34.0, 74.6% probability of causality) missense variant in the same gene (P=2x10-8). PLCG2 encodes a phospholipase enzyme that plays a critical role in regulating immune pathway signalling22, and has previously been implicated in two autosomal dominant immune disorders. Intragenic deletions in its autoinhibitory domain cause antibody deficiency and immune dysregulation (familial cold autoinflammatory syndrome 3, MIM 614468)23 and heterozygous missense variants (e.g. p.Ser707Tyr) lead to a phenotype that includes intestinal inflammation24 (Figure 1b).
A more general overlap between candidate IBD GWAS genes and Mendelian disorders of inflammation and immunity has been previously observed in 163 loci discovered at that time25. We replicated this finding in our list of 241 loci (p < 10-6, Supplementary Table 4), and observed that this enrichment is even stronger when considering just the 26 loci where a gene can be confidently implicated by fine-mapping to a coding variant or colocalisation with an eQTL (27% vs 3%, p=2x10-5). In addition to PLCG2 we identified an association between Crohn’s disease and an intronic variant in NCF4 (P=1.76x10-8). This gene encodes p40phox, a component of the NADPH-oxidase system that is responsible for the oxidative burst in innate immune cells and which is a key mechanism of killing phagocytosed bacteria. Rare pathogenic variants in NCF4 cause autosomal recessive chronic granulomatous disease, characterized by Crohn’s disease-like intestinal inflammation and defective ROS production in neutrophils26. Our associated variant, rs4821544, had previously been suggestively associated with small bowel Crohn’s disease27,28, and when we stratified patients by disease location we found that the effect was consistently stronger for small bowel compared to large bowel disease (Supplementary Figure 7).
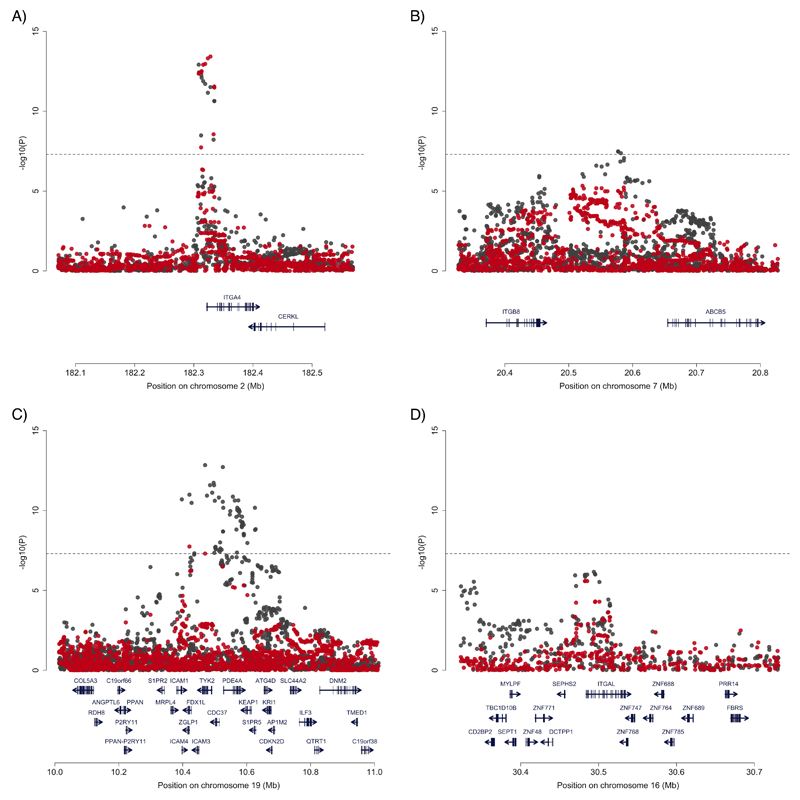
Among the remaining 21 novel loci we noted three that were within 150kb of integrin genes (ITGA4, ITGAV and ITGB8), while a previously associated locus overlaps with a fourth integrin, ITGAL. Furthermore, a recent study demonstrated that there is an IBD specific association that affects expression of ICAM1, which encodes the binding partner of ITGAL29. Integrins are cell adhesion mediators with bi-directional signalling capabilities that play a crucial role in leukocyte homing and cell differentiation in inflammation and cancer30. Given the strong candidacy of these genes, we sought potentially causal molecular mechanisms that would connect the IBD associated SNPs to integrin regulation. Our fine-mapping analysis excluded the possibility that these associations are caused by protein-coding changes, so we next tested for effects of IBD risk SNPs on integrin gene expression in immune cells using twelve publicly available eQTL datasets. While many eQTLs and GWAS signals show some degree of correlation, inferences about causality require more robust statistical co-localization of the two signals. Remarkably, we observed three of our five associations had >90% probability of being driven by the same variants as monocyte-specific stimulus response eQTLs (ITGA4, PLPS_24hr=0.984; ITGAL, PLPS_24hr=0.980; ICAM1, PLPS_2hr=0.961; Supplementary Table 5). A fourth association, ITGB8, is difficult to map due to extended linkage disequilibrium in the locus, but shows intermediate evidence of co-localization (PLPS_24hr=0.712) in response to the same stimulus (Figure 2). All four of the IBD risk increasing alleles upregulate expression of their respective genes, suggesting that increased levels of pro-inflammatory cell surface markers in response to stimulus may be a consistent mechanism of action. Proving this hypothesis would require showing that IBD risk alleles causally change stimulus-response expression (e.g. by targeted editing of each allele in cell lines homozygous for the low risk haplotype), and moreover that such changes have physiological relevance to disease processes.
Figure 2. Co-localization of disease association and stimulus response eQTLs in monocytes.
The local pattern of disease association (IBD: (A) ITGA4, (B) ITGB8, (C) ICAM1; (D) UC: ITGAL) in grey, and the association of that variant with response to LPS (lipopolysaccharide) stimulation in red. Evidence of co-localization (probability > 70%) is observed for all for signals.
One line of evidence that supports such disease relevance for integrins and their counter-receptors is their recent emergence as important therapeutic targets in IBD. Most promisingly, the monoclonal antibodies vedolizumab and etrolizumab, which target the components of the α4β7 dimer (encoded by ITGA4 and ITGB7, and responsible for the gut-homing specificity of certain leukocytes), have demonstrated efficacy in IBD31–33. Additionally, an antisense oligonucleotide targeting ICAM1 has shown promise in the treatment of ulcerative colitis and pouchitis34. The importance of gut-selectivity for therapeutic approaches is highlighted by the antibodies that bind the αL and α4 integrin subunits (encoded by ITGAL and ITGA4, respectively). Therapies targeting αL (efalizumab) and α4 (natalizumab) demonstrated potential in Crohn’s disease35,36, but both medications have been associated with progressive multifocal leukoencephalopathy (PML)37. This potentially fatal condition is likely mediated by binding to integrin dimers that are not gut-specific, leading to impaired leukocyte migration to the central nervous system and JC virus infection of the brain. Owing to the risk of PML, efalizumab has been withdrawn from the market and natalizumab is not licensed for Crohn’s disease in Europe.
Integrins are not only important in cell trafficking, but can also participate in cellular signalling. For example, the αVβ8 heterodimer – both subunits of which are encoded by genes which are now within confirmed IBD loci (ITGAV and ITGB8, respectively) – is a potent activator of TGFβ38, with a range of cell-type specific effects. Furthermore, mice with dendritic-cell specific deletion of this complex had impaired regulatory T cell function and severe colitis39, whereas deleting the complex in regulatory T cells themselves prevented them from suppressing pathogenic T cell responses during active inflammation40. While no current IBD therapeutics target αVβ8 directly, promising early results of an oral antisense oligonucleotide to the inhibitory TGFβ-signalling protein SMAD741, itself encoded by a locus identified by genetic association studies25, demonstrate the therapeutic potential of modifying TGFβ signaling in Crohn’s disease.
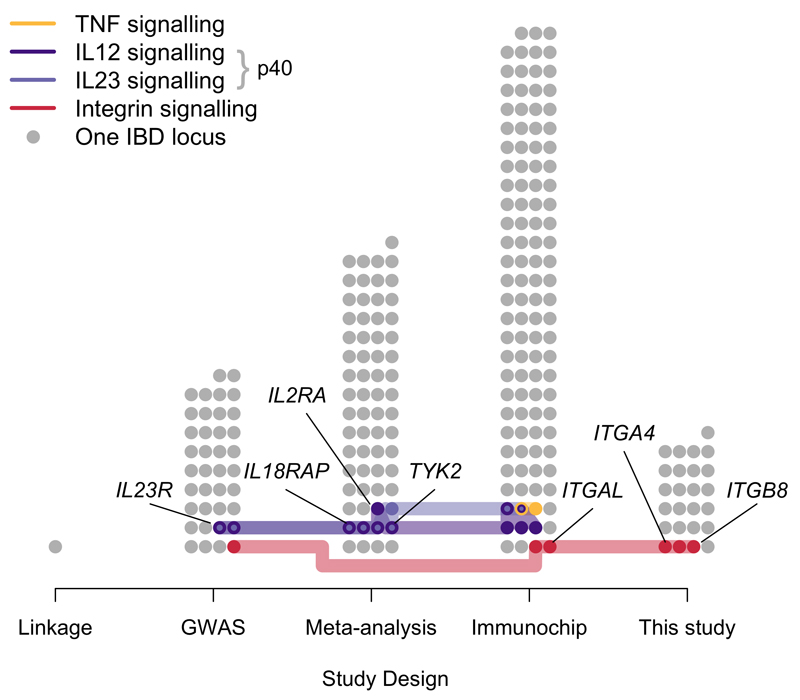
In addition to the connections to anti-integrin and anti-TGFβ therapies described above, IBD GWAS have previously implicated loci containing other therapeutically relevant genes, such as those in signalling pathways relevant to the targets of anti-TNF and anti-p40 IBD therapies (Figure 3, Supplementary Table 6). These discoveries have demonstrated that the importance of the biological pathways underlying associations, and their potential therapeutic relevance, are not necessarily reflected in their GWAS effect sizes. For example, the modest odds ratios of the signals near integrin genes (1.10-1.12) required tens of thousands of samples to detect at genome-wide significance. Furthermore, analyses aimed at understanding the specific cellular contexts in which these genes are active in IBD, as well as the risk-increasing direction of effect (e.g. consistent up-regulation of integrins in response to LPS stimulus), are only beginning to bear fruit.
Figure 3. IBD-associated loci containing genes in immune pathways related to classes of approved therapeutics.
All IBD loci are divided into the studies where they were first identified1. Loci that contain a gene in one of four signalling pathways related to targets of three classes of approved IBD therapeutics (Online Methods) are highlighted, with those where the pathway gene has been confidently identified as the causal IBD gene labelled. Despite the general pattern that effect size decreases from left to right, therapeutically relevant associations continue to be found.
Our study has demonstrated that continuing to pursue GWAS, even in a well studied complex disease like IBD, has the potential to complement other powerful approaches, such as targeted genotyping (via the Immunochip) and large-scale genome and exome sequencing. In two cases we have implicated genes in which different variants have previously been shown to cause immune-related Mendelian disorders, echoing a connection made to the very first Crohn’s disease risk gene, NOD2, in which rare missense mutations cause the autosomal dominant granulomatous disorder Blau syndrome42. Finally, while the individual effect sizes of our newly discovered associations are modest, our results show that GWAS continues to deliver new loci, which help understand many aspects of disease biology, including possible mechanisms of known therapies. For example, four IBD associations that plausibly co-localize with changes in integrin expression underscore the value of comprehensive catalogs of the regulatory consequences of GWAS variants in specific cells and contexts. Even when specific genes are implicated, cellular assays with relevance to disease physiology (for example, protein response to bacterial stimulus in colonic organoids) will be needed to achieve the ultimate payoff from prospectively mining these signals for promising targets for new therapeutics.
Data availability
Genotype data that supports this study has been deposited in the European Genome-phenome Archive (EGA) under the accession code EGAS00001000924. Association summary statistics are available from ftp://ftp.sanger.ac.uk/pub/project/humgen/summary_statistics/human/2016-11-07/.
Online Methods
New genome-wide genetic data
GWAS samples and genotyping
Following ethical approval by Cambridge MREC (reference: 03/5/012), 11,768 British IBD cases, diagnosed using accepted endoscopic, histopathological and radiological criteria, were consented into the study and genotyped on the Human Core Exome v12.1. 10,484 population control samples genotyped on the Human Core Exome v12.0 were obtained from the Understanding Society Project. Genotypes were called using optiCall43.
GWAS quality control
We removed variants that did not overlap between the two versions of the chip, had missingness > 5%, a significant difference in call rate between cases and controls (P < 1x10-5), deviated from Hardy-Weinberg equilibrium (HWE) in controls (P < 1x10-5), or that were affected by a genotyping batch effect (significant association [P < 1x10-5] between an outlier group of cases discovered using principal component analysis [PC1 < -0.005], and the remainder of the samples). We then removed samples with missingness > 1%, heterozygosity ±3 standard deviations from the mean, mismatch between reported and genotypic sex, first-degree relatives or closer (kinship coefficient > 0.177), and non-European samples identified through principal component analysis with HapMap3 populations. After quality control, data were available for 4,474 Crohn’s disease, 4,173 ulcerative colitis, 592 IBD-unclassified cases and 9,500 controls for 296,203 variants.
Whole-genome sequenced samples
We generated low-coverage whole genome sequences for 4,686 IBD cases and 3,781 population controls from the UK IBD Genetics Consortium (UKIBDGC) and UK10K Consortium, respectively. Detailed information on sequencing, genotype refinement and quality control are described elsewhere9.
Imputation
These sequences were combined with 2,504 samples from the Phase 3 v5 release of the 1000 Genomes project (2013-05-02 sequence freeze) to create a phased imputation reference panel enriched in IBD-associated variants. We used PBWT44 to impute from this reference panel (114.2 million total variants) into our new GWAS described above.
Association testing, meta-analysis, and quality control
Association testing
Prior to association testing, we removed all samples that were included in previous IBD GWAS meta-analyses (Supplementary Table 1). We then tested for association to ulcerative colitis, Crohn’s disease and IBD separately within the sequenced samples and new GWAS using SNPTEST v2.5, performing an additive frequentist association test conditioned on the first ten principal components for each cohort. We filtered out variants with minor allele frequency (MAF) < 0.1%, INFO < 0.4, or strong evidence for deviations from HWE in controls (pHWE<1x10-7).
Meta-analysis
We used METAL (release 2011-03-05) to perform a standard error weighted meta-analysis of our sequencing and GWAS cohorts with the publicly available International Inflammatory Bowel Disease Genetics Consortium (IIBDGC) meta-analysis summary statistics1, after applying the additional MAF ≥ 0.1%, and INFO ≥ 0.4 filters to the IIBDGC data.
Quality control
The output of the fixed-effects meta-analysis was further filtered, and sites with high evidence for heterogeneity (I2>0.90) were discarded. Only sites for which all cohorts passed our quality control filters were included in our analysis. In addition, we discarded genome-wide significant variants for which the meta-analysis p-value was not lower than all of the cohort-specific p-values.
LD score regression
We performed LD score regression using LDSC v1.0.0 and European linkage disequilibrium (LD) scores from the 1000 Genomes Project (downloaded from https://data.broadinstitute.org/alkesgroup/LDSCORE/eur_w_ld_chr.tar.bz2) on our filtered meta-analysis summary statistics for all sites with INFO > 0.95. This INFO threshold is to avoid confounding due to poor imputation, as recommended by the authors45.
Locus definition
Computing LD windows
An LD window was calculated for every genome-wide significant variant in any of the three traits (Crohn’s disease, ulcerative colitis, IBD), defined by the left-most and right-most variants that are correlated with the main variant with an r2 of 0.6 or more. The LD was calculated in the GBR and CEU samples from the 1000 Genomes Phase 3, release v5 (based on 20130502 sequence freeze and alignments). Loci with overlapping LD windows, as well as loci whose lead variants were separated by 500kb or less, were subsequently merged, and the variant with the strongest evidence of being associated was kept as the lead variant for each merged locus.
Identifying novel loci
A locus was annotated as known if it contained at least one variant previously reported at genome-wide significance (irrespective of the LD between that variant and the most associated variants in the locus). To ensure that putatively novel signals were not due to long-range LD with variants in previously reported loci, we conducted conditional analysis in our new GWAS for all variants in loci which were less than 3Mb away from a known locus. Putatively novel loci already known in a lower order IBD trait (e.g. a previously known Crohn’s disease locus coming up as an IBD locus) were also removed from this list. This did not apply where, for example, a known Crohn’s disease locus was now associated with ulcerative colitis, or vice versa.
Fine-mapping
Approximate Bayes factors were calculated from the meta-analysis effect sizes and standard errors described above by applying equation (2) of Wakefield46, assuming a prior variance on the log odds ratios of 0.04 (the default prior used by the software SNPTest, and used by Maller et al47). We then performed fine-mapping using these Bayes factors as described in Maller et al to calculate the posterior that each variant is causal, and the 95% credible set for each association (the smallest set of variants with posteriors that sum to at least 95%). For each association we use the meta-analysis results for the phenotype (Crohn’s disease, ulcerative colitis or IBD) specified in Supplementary Table 2. We only consider a locus to be confidently fine-mapped if there are no variants in the Phase 3 v5 release of the 1000 Genomes project (2013-05-02 sequence freeze) in high LD (r2 ≥ 0.6) with our hit SNP, but missing from our dataset, and no variants in our data within high LD (r2 > 0.8) that fail during our QC procedure.
eQTL overlap
Identifying eQTL overlaps
Twelve eQTL datasets were searched to identify variants within the 25 newly identified IBD risk loci that are associated with variation in gene expression (Supplementary Table 7). Splice-QTLs based on exon-ratio48 and transcript-ratio49–51 were also included in the search where available (Supplementary Table 7). The most significant variant-gene associations were extracted from each eQTL/splice-QTL dataset and were reported as candidates if that variant had r2 > 0.8 with any of the lead SNPs in the 25 IBD risk loci.
Testing for co-localization
We tested for co-localization between IBD association signals and eQTLs using the coloc2 method52, implemented in the R package coloc. We used a window size of 250kb on either side of the IBD association, and implemented the default settings as recommended. Each test was repeated using two different values for the prior probability of co-localization, p12: 1x10-5 and 1x10-6.
Signalling pathway definitions
We identify the following immune pathways as relevant to classes of approved IBD therapeutics: the IL12 and IL23 signalling pathways (ustekinumab53), the TNFa signalling pathway (infliximab54, adalimumab55), and the integrin signalling pathway (vedolizumab31,32). Genes involved in these pathways were identified from the Molecular Signatures Database canonical pathways gene sets (C2; http://software.broadinstitute.org/gsea/msigdb/genesets.jsp?collection=CP). These gene lists had been previously curated by the Pathway Interaction Database56. The integrin signalling gene list was comprised of all unique genes from the following gene sets: integrin beta1 pathway (PID_INTEGRIN1_PATHWAY), integrin beta7 pathway (PID_INTEGRIN5_PATHWAY) and integrin cell surface interactions (PID_INTEGRIN_CS_PATHWAY). The list of TNFa signalling genes was obtained from PID_TNF_PATHWAY and the list of IL-23/IL-12 p40 signalling genes was comprised of all unique genes from the PID_IL12_PATHWAY and PID_IL23_PATHWAY.
Supplementary Material
Acknowledgements
We would like to thank all individuals who contributed samples to the study. This work was co-funded by the Wellcome Trust [098051] and the Medical Research Council, UK [MR/J00314X/1]. Case collections were supported by Crohn’s and Colitis UK. KMdL, LM, CAL, YL, DR, JG-A, NJP, CAA and JCB are supported by the Wellcome Trust [098051; 093885/Z/10/Z; 094491/Z/10/Z]. KMdL is supported by a Woolf Fisher Trust scholarship. CAL is a clinical lecturer funded by the NIHR. We thank Anna Stanton for co-ordinating the Guy’s and St Thomas’ patient recruitment. We acknowledge support from the Department of Health via the NIHR comprehensive Biomedical Research Centre awards to Guy’s and St Thomas’ NHS Foundation Trust in partnership with King’s College London and to Addenbrooke’s Hospital, Cambridge in partnership with the University of Cambridge. This research was also supported by the NIHR Newcastle Biomedical Research Centre. The UK Household Longitudinal Study is led by the Institute for Social and Economic Research at the University of Essex and funded by the Economic and Social Research Council. The survey was conducted by NatCen and the genome-wide scan data were analysed and deposited by the Wellcome Trust Sanger Institute. Information on how to access the data can be found on the Understanding Society website https://www.understandingsociety.ac.uk/.
Footnotes
Author contributions
KMdL, LM, YL, LJ, DLR, CAA, and SGJ performed statistical analysis. KMdL, LM, YL, LJ, JCL, JGA, SGJ, CAL, NAK, and CAA analysed the data. GH, ERN, CE, CM, AS, DCW, MT, AH, CGM, MP, WGM, CWL, HU, CH, NJP, TA, JCM, JackS, JerS, and PH contributed samples/materials. CAA, JCB, KMdL, LM, JCL, CGM, MP, CAL, NAK, YL, and PH wrote the paper. JCB, CAA, JCM, MP, CWL, TA, and NJP conceived & designed experiments.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Liu JZ, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–989. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parkes M, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamazaki K, et al. A Genome-Wide Association Study Identifies 2 Susceptibility Loci for Crohn’s Disease in a Japanese Population. Gastroenterology. 2013;144:781–788. doi: 10.1053/j.gastro.2012.12.021. [DOI] [PubMed] [Google Scholar]
- 4.Anderson CA, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kenny EE, et al. A genome-wide scan of Ashkenazi Jewish Crohn’s disease suggests novel susceptibility loci. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Julià A, et al. A genome-wide association study identifies a novel locus at 6q22.1 associated with ulcerative colitis. Hum Mol Genet. 2014;23:6927–6934. doi: 10.1093/hmg/ddu398. [DOI] [PubMed] [Google Scholar]
- 7.Yang S-K, et al. Genome-wide association study of Crohn’s disease in Koreans revealed three new susceptibility loci and common attributes of genetic susceptibility across ethnic populations. Gut. 2014;63:80–87. doi: 10.1136/gutjnl-2013-305193. [DOI] [PubMed] [Google Scholar]
- 8.Ellinghaus D, et al. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat Genet. 2016;48:510–518. doi: 10.1038/ng.3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo Y, et al. Exploring the genetic architecture of inflammatory bowel disease by whole genome sequencing identifies association at ADCY7. Nat Genet. doi: 10.1038/ng.3761. (In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walter K, et al. The UK10K project identifies rare variants in health and disease. Nature. 2015;526:82–90. doi: 10.1038/nature14962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang H, et al. Association mapping of inflammatory bowel disease loci to single variant resolution. bioRxiv. 2015:028688. doi: 10.1101/028688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ivanov II, et al. The Orphan Nuclear Receptor RORγt Directs the Differentiation Program of Proinflammatory IL-17+ T Helper Cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 14.Wang G, et al. Migration of myeloid cells during inflammation is differentially regulated by the cell surface receptors Slamf1 and Slamf8. PLoS One. 2015;10:e0121968. doi: 10.1371/journal.pone.0121968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang G, et al. Cutting edge: Slamf8 is a negative regulator of Nox2 activity in macrophages. J Immunol. 2012;188:5829–5832. doi: 10.4049/jimmunol.1102620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luci C, et al. Influence of the transcription factor RORgammat on the development of NKp46+ cell populations in gut and skin. Nat Immunol. 2009;10:75–82. doi: 10.1038/ni.1681. [DOI] [PubMed] [Google Scholar]
- 18.Yang Y, et al. Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature. 2014;510:152–156. doi: 10.1038/nature13279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sawa S, et al. RORγt+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nat Immunol. 2011;12:320–326. doi: 10.1038/ni.2002. [DOI] [PubMed] [Google Scholar]
- 20.Gevers D, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Withers DR, et al. Transient inhibition of ROR-[gamma]t therapeutically limits intestinal inflammation by reducing TH17 cells and preserving group 3 innate lymphoid cells. Nat Med. 2016;22:319–323. doi: 10.1038/nm.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu G, Chen Y, Schuman J, Wang D, Wen R. Phospholipase Cγ2 plays a role in TCR signal transduction and T cell selection. J Immunol. 2012;189:2326–2332. doi: 10.4049/jimmunol.1103458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ombrello MJ, et al. Cold urticaria, immunodeficiency, and autoimmunity related to PLCG2 deletions. N Engl J Med. 2012;366:330–338. doi: 10.1056/NEJMoa1102140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Q, et al. A hypermorphic missense mutation in PLCG2, encoding phospholipase Cγ2, causes a dominantly inherited autoinflammatory disease with immunodeficiency. Am J Hum Genet. 2012;91:713–720. doi: 10.1016/j.ajhg.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matute JD, et al. A new genetic subgroup of chronic granulomatous disease with autosomal recessive mutations in p40 phox and selective defects in neutrophil NADPH oxidase activity. Blood. 2009;114:3309–3315. doi: 10.1182/blood-2009-07-231498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rioux JD, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts RL, et al. Confirmation of association of IRGM and NCF4 with ileal Crohn’s disease in a population-based cohort. Genes Immun. 2008;9:561–565. doi: 10.1038/gene.2008.49. [DOI] [PubMed] [Google Scholar]
- 29.Dendrou CA, et al. Resolving TYK2 locus genotype-to-phenotype differences in autoimmunity. Sci Transl Med. 2016;8:363ra149–363ra149. doi: 10.1126/scitranslmed.aag1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 31.Sandborn WJ, et al. Vedolizumab as induction and maintenance therapy for Crohn’s disease. N Engl J Med. 2013;369:711–721. doi: 10.1056/NEJMoa1215739. [DOI] [PubMed] [Google Scholar]
- 32.Feagan BG, et al. Vedolizumab as induction and maintenance therapy for ulcerative colitis. N Engl J Med. 2013;369:699–710. doi: 10.1056/NEJMoa1215734. [DOI] [PubMed] [Google Scholar]
- 33.Vermeire S, et al. Etrolizumab as induction therapy for ulcerative colitis: a randomised, controlled, phase 2 trial. Lancet. 2014;384:309–318. doi: 10.1016/S0140-6736(14)60661-9. [DOI] [PubMed] [Google Scholar]
- 34.Hosten TA, Zhao K, Han HQ, Liu G, He XH. Alicaforsen: An Emerging Therapeutic Agent for Ulcerative Colitis and Refractory Pouchitis. Gastroenterol Res Pract. 2014;7:51–55. doi: 10.14740/gr599w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.James DG, Seo DH, Chen J, Vemulapalli C, Stone CD. Efalizumab, a human monoclonal anti-CD11a antibody, in the treatment of moderate to severe Crohn’s disease: An open-label pilot study. Dig Dis Sci. 2011;56:1806–1810. doi: 10.1007/s10620-010-1525-6. [DOI] [PubMed] [Google Scholar]
- 36.Sandborn WJ, et al. Natalizumab induction and maintenance therapy for Crohn’s disease. N Engl J Med. 2005;353:1912–1925. doi: 10.1056/NEJMoa043335. [DOI] [PubMed] [Google Scholar]
- 37.Carson KR, et al. Monoclonal antibody-associated progressive multifocal leucoencephalopathy in patients treated with rituximab, natalizumab, and efalizumab: a Review from the Research on Adverse Drug Events and Reports (RADAR) Project. Lancet Oncol. 2009;10:816–824. doi: 10.1016/S1470-2045(09)70161-5. [DOI] [PubMed] [Google Scholar]
- 38.Travis MA, Sheppard D. TGF-β activation and function in immunity. Annu Rev Immunol. 2014;32:51–82. doi: 10.1146/annurev-immunol-032713-120257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Travis MA, et al. Loss of integrin alpha(v)beta8 on dendritic cells causes autoimmunity and colitis in mice. Nature. 2007;449:361–365. doi: 10.1038/nature06110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Worthington JJ, et al. Integrin αvβ8-Mediated TGF-β Activation by Effector Regulatory T Cells Is Essential for Suppression of T-Cell-Mediated Inflammation. Immunity. 2015;42:903–915. doi: 10.1016/j.immuni.2015.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Monteleone G, et al. Mongersen, an oral SMAD7 antisense oligonucleotide, and Crohn’s disease. N Engl J Med. 2015;372:1104–1113. doi: 10.1056/NEJMoa1407250. [DOI] [PubMed] [Google Scholar]
- 42.Miceli-Richard C, et al. CARD15 mutations in Blau syndrome. Nat Genet. 2001;29:19–20. doi: 10.1038/ng720. [DOI] [PubMed] [Google Scholar]
- 43.Shah TS, et al. optiCall: a robust genotype-calling algorithm for rare, low-frequency and common variants. Bioinformatics. 2012;28:1598–1603. doi: 10.1093/bioinformatics/bts180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Durbin R. Efficient haplotype matching and storage using the positional Burrows-Wheeler transform (PBWT) Bioinformatics. 2014;30:1266–1272. doi: 10.1093/bioinformatics/btu014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bulik-Sullivan BK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wakefield J. Bayes factors for genome-wide association studies: comparison with P-values. Genet Epidemiol. 2009;33:79–86. doi: 10.1002/gepi.20359. [DOI] [PubMed] [Google Scholar]
- 47.Wellcome Trust Case Control Consortium et al. Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat Genet. 2012;44:1294–1301. doi: 10.1038/ng.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhernakova D, et al. Hypothesis-free identification of modulators of genetic risk factors. bioRxiv. 2015:033217. doi: 10.1101/033217. [DOI] [Google Scholar]
- 49.Battle A, et al. Characterizing the genetic basis of transcriptome diversity through RNA-sequencing of 922 individuals. Genome Res. 2014;24:14–24. doi: 10.1101/gr.155192.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Monlong J, Calvo M, Ferreira PG, Guigó R. Identification of genetic variants associated with alternative splicing using sQTLseekeR. Nat Commun. 2014;5:4698. doi: 10.1038/ncomms5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ongen H, Buil A, Brown AA, Dermitzakis ET, Delaneau O. Fast and efficient QTL mapper for thousands of molecular phenotypes. Bioinformatics. 2016;32:1479–1485. doi: 10.1093/bioinformatics/btv722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Giambartolomei C, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sandborn WJ, et al. Ustekinumab induction and maintenance therapy in refractory Crohn’s disease. N Engl J Med. 2012;367:1519–1528. doi: 10.1056/NEJMoa1203572. [DOI] [PubMed] [Google Scholar]
- 54.Hanauer SB, et al. Maintenance infliximab for Crohn’s disease: the ACCENT I randomised trial. Lancet. 2002;359:1541–1549. doi: 10.1016/S0140-6736(02)08512-4. [DOI] [PubMed] [Google Scholar]
- 55.Colombel J-F, et al. Adalimumab for maintenance of clinical response and remission in patients with Crohn’s disease: the CHARM trial. Gastroenterology. 2007;132:52–65. doi: 10.1053/j.gastro.2006.11.041. [DOI] [PubMed] [Google Scholar]
- 56.Schaefer CF, et al. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009;37:D674–9. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genotype data that supports this study has been deposited in the European Genome-phenome Archive (EGA) under the accession code EGAS00001000924. Association summary statistics are available from ftp://ftp.sanger.ac.uk/pub/project/humgen/summary_statistics/human/2016-11-07/.