Abstract
Mouse F9 cells differentiate to primitive endoderm (PrE) when treated with retinoic acid (RA). Differentiation is accompanied by increased reactive oxygen species (ROS) levels, and while treating F9 cells with antioxidants attenuates differentiation, H2O2 treatment alone is sufficient to induce PrE. We identified the NADPH oxidase (NOX) complexes as candidates for the source of this endogenous ROS, and within this gene family, and over the course of differentiation, Nox1 and Nox 4 show the greatest upregulation induced by RA. Gata6, encoding a master regulator of extraembryonic endoderm is also up-regulated by RA and we provide evidence that NOX1 and NOX4 protein levels increase in F9 cells overexpressing Gata6. Pan-NOX and NOX1-specific inhibitors significantly reduced the ability of RA to induce PrE, and this was recapitulated using a genetic approach to knockdown Nox1 and/or Nox4 transcripts. Interestingly, overexpressing either gene in untreated F9 cells did not induce differentiation, even though each elevated ROS levels. Thus, the data suggests that ROS produced during PrE differentiation is dependent in part on increased NOX1 and NOX4 levels, which is under the control of GATA6. Furthermore, these results suggest that the combined activity of multiple NOX proteins is necessary for the differentiation of F9 cells to primitive endoderm.
Introduction
Mouse extraembryonic endoderm formation can be studied in vitro using F9 teratocarcinoma cells, which are chemically induced to form primitive endoderm (PrE) when treated with retinoic acid (RA) [1] or to parietal endoderm (PE) when treated with RA and cAMP analog [2]. F9 cell differentiation, specifically to PrE is accompanied by the appearance of molecular markers, and morphological changes, many resulting from the activation of the canonical Wnt-β-catenin pathway [1]. In this pathway when Wnt is absent a destruction complex serves to phosphorylate β-catenin marking it for ubiquitination and degradation in the proteasome. When present, Wnt binds to a Frizzled receptor causing Dishevelled (DVL) to move towards the plasma membrane, where it recruits Axin out of the destruction complex making it non-functional and allowing β-catenin to accumulate and translocate to the nucleus where it binds to and activates the T-cell-factors-Lymphoid enhancer factors (TCF-LEF) family of transcription factors. We reported previously that differentiation is also accompanied by a burst of ROS, which is necessary as F9 cells treated with antioxidants or when treated with a non-specific NADPH oxidase inhibitor failed to form PrE [3]. That H2O2 treatment alone induces PrE indicates that ROS are sufficient to initiate differentiation [3]. To explore this further, we recently reported that DVL in undifferentiated F9 cells associates with nucleoredoxin (NRX) a redox sensitive protein that scavenges ROS, and is known to play a role in PrE differentiation [4]. This association and regulation of the Wnt-β-catenin pathway occurs in other systems [5–8], and we propose that this inhibition prevents aberrant canonical Wnt signaling when Wnt is absent as DVL in this state cannot recruit Axin away from a destruction complex. Thus, in the presence of ROS NRX dissociates from DVL and the Wnt pathway is primed awaiting the ligand.
The source of the ROS when F9 cells are treated with RA was investigated and the candidates identified are members of the NADPH oxidase (NOX) family, which are sources of superoxide anions and H2O2 [9]. In F9 cells Nox1-4 and Duox2 are upregulated following RA treatment [3]. Duox1 is not RA-responsive and may not be involved in PrE differentiation. Nox1 and Nox4 are up-regulated to the greatest extent following RA treatment, and given the previous reports suggesting a link to extraembryonic endoderm formation and stem cell differentiation, we specifically selected these members to interrogate as the candidates involved in the ROS production involved in RA-induced PrE formation. To address that the activity of NADPH oxidase 1 and/or 4 is/are responsible for producing the ROS that are necessary and sufficient to induce F9 cells to differentiate, we first tested and found Nox genes are under the control of GATA6, the master regulator of endoderm differentiation [10]. Inhibiting all NOX activity, or specifically inhibiting NOX1 was sufficient to block differentiation, and knocking down Nox1 or Nox4 expression using an siRNA approach complemented the chemical inhibitor data. Confident from these studies that both NOX proteins were necessary for differentiation, we expected that their overexpression would induce PrE. However, despite the overexpression of each having increased ROS levels, no significant difference in β-catenin-dependent TCF activity relative to controls was seen and neither would induce PrE. Together, these results indicate that RA-induced differentiation of F9 cells requires a coordinate increase in NOX activity that is due in part to the upregulation of the Nox genes by GATA6.
Materials and methods
Cell culture conditions and transfections
Mouse teratocarcinoma F9 cells (ATCC) were cultured in Dulbecco’s modified Eagles medium (Lonza) supplemented with 10% fetal bovine serum (Gibco) and 1% penicillin-streptomycin (Lonza), and incubated at 37°C and 5% CO2. Cells were treated with 10−7 M retinoic acid (RA all-trans; Sigma Aldrich) or dimethyl sulfoxide (DMSO; Caledon) as a negative control. Cells were co-treated with 1 μM VAS2870 (Sigma) and RA 24 hours after seeding and grown for 3 days, or co-treated with 250 nM ML171 (Tocris) and RA and grown for 4 days as described above. F9 cells were reverse transfected using Lipofectamine 2000 (Thermo Fisher Scientific). Freshly passaged cells were added to a 35 mm dish already containing a total of 4 μg of DNA plasmid. Culture media was replenished 6–8 h post-transfection and transfected cells were selected using antibiotics.
Plasmids
The following plasmids were used: pcDNA3.1-Gata6; pcDNA-mNox4, a gift from Dr. M. Jaconi, University of Geneva), pcDNA3.1-mNox1 (Addgene # 58340), pRL-TK, a gift from Dr. R. DeKoter (Western University), piLenti-siRNA-GFP-Nox1 “CTATTTAACTTCGAACGCTACAGAAGAAG”, piLenti-siRNA-GFP-Nox1 “TGCTTCCATCTTGAAATCTATCTGGTACA”, piLenti-siRNA-GFP-Nox4 “ACATTTGGTGTCCACTTTAAAGTAGTAGG”, piLenti-siRNA-GFP-Nox4 “TCCAGTGGTTTGCAGATTTACTCTGTGTG” (Applied Biological Materials, Richmond BC).
Immunoblot analysis
Protein lysates were collected in RIPA buffer containing 150 mM sodium chloride, 1.0% Triton X-100, 0.5% deoxycholate, 0.1% SDS and 50 mM Tris pH 8.0. Protein concentrations were determined using the DC™ Protein Assay (Bio-Rad), and 20–50 μg of protein lysate was mixed 2:1 with 3X SDS loading buffer containing 10% β-mercaptoethanol and separated on 10% polyacrylamide gels by electrophoresis for 2 h with 100 V at 4°C. Following electrophoresis, the proteins were wet-transferred electrophoretically to Immunoblot PVDF membrane (Bio-Rad) for 16 h with 20 V at 4°C using a Tris-glycine transfer buffer containing 20% methanol. Membranes were incubated in Tris buffered saline with 0.1% Tween-20 (TBS-T) containing 10% w/v skim milk powder for 1 h shaking at room temperature, then incubated with primary antibody overnight at 4°C. Following 3 washes 5 min each in TBS-T, membranes were incubated with secondary antibody for 2 h at room temperature, followed by 3 washes for 10 min each in TBS-T. SuperSignal West Pico Chemiluminescent Detection Kit (Thermo Fisher Scientific) was used to detect the presence of secondary antibodies conjugated to horseradish peroxidase (HRP). Signals were captured using a Molecular Imager Gel Doc XR system (Bio-Rad) with Quantity One Software. The primary antibodies used were directed against TROMA1 (1:10; 55 kDa, Developmental Studies Hybridoma Bank), OCT4 (1:1000; 42 kDa, Cell Signaling Technology), DAB2 (1:10000; 96 kDa, BD Transduction Laboratories), NOX1 (1:10000; 65 kDa, Thermo Fisher Scientific), NOX4 (1:10000; 67 kDa, Novus Biologicals) and ß-actin (1:10000; 47 kDa, Santa Cruz) dissolved in 3% Bovine Serum Albumin w/v in TBS-T. Secondary anti-rat (1:1000), anti-mouse (1:1000) and anti-rabbit (1:1000) antibodies were HRP-conjugated and dissolved in 3% skim milk w/v in TBS-T.
Quantitative reverse transcription polymerase chain reaction
Total RNA from treated and/or transfected cells was isolated and collected using the RNeasy Mini Kit (Qiagen). RNA was reverse transcribed into first strand cDNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) and the manufacturer's recommendations. The CFX Connect Real-Time PCR Detection System (Bio-Rad) was used for qRT-PCR analysis. Each reaction contained 500 nM of each primer, SensiFAST SYBR Mix (FroggaBio), and 1 μL of cDNA. Primers were designed to: L14 F (5’GGGAGAGGTGGCCTCGGACGC), L14 R (5’GGCTGGCTTTCACTCAAAGGCC), Gata6 F (5’ATGGCGTAGAAATGCTGAGG), Gata6 R (5’TGAGGTGGTCGCTTGTGTAG), Dab2 F (5’GGAGCATGTAGACCATGATG), Dab2 R (5’AAAGGATTTCCGAAAGGGCT), Nox1 F (5’AATGCCCAGGATCGAGGT), Nox1 R (5’GATGGAAGCAAAGGGAGTGA), Nox4 F (5’GATCACAGAAGGTCCCTAGCA), and Nox4 R (5’GTTGAGGGCATTCACCAAGT). Analysis of gene expression was determined using the comparative cycle threshold (Δ/ΔCt) method. Gene expression was normalized to the constitutively expressed L14 gene and relative values were normalized by comparing treatments to DMSO-treated and/or control plasmid transfected control cells to determine fold change.
TCF reporter assay
Cells were transfected with pGL3-BARL and pRL-TK (transfection control) and then treated with DMSO (vehicle control), RA, or co-transfected with Nox1 or Nox4 plasmids in equal amounts of DNA. Protein lysates were collected 3 days post-treatment or post-transfection using the Dual-Glo Luciferase Assay System and the manufacturer's recommendations (Promega). Luciferase expression was quantified using the GloMax Multi Detection System (Promega). Values were presented as relative luminescence derived from the quotient of firefly luminescence (pGL3-BARL) and Renilla luminescence (pRL-TK).
ROS detection
Intracellular ROS was detected using both an Amplex Red Assay and CM-H2DCFDA (Thermo Fisher Scientific). Briefly, Amplex Red was prepared in DMSO and cells were treated following manufacturer’s instructions with slight modifications. Following treatment or transfection, cells were detached and resuspended in 100 μM Amplex Red in measurement media buffer (120 mM KCl, 5 mM KH2PO4, 5 mM EDTA, 10 mM HEPES, 5 mM MgCl2 at pH 7.40) supplemented with 40 μg/mL Saponin (Sigma) and 0.2 units/mL HRP (Thermo Fisher Scientific). Immediately after incubation, cells were seeded onto an opaque 96-well plate and florescence was measured at Ex/Em of 530/590 nm using the GloMax Multi Detection System (Promega). Values were normalized to protein concentration. For CM-H2DCFDA staining cells following treatment with DMSO or RA were incubated in Hank’s Balanced Salt Solution for 15 min at 37°C and 5% CO2 with 2 μM CM-H2DCFDA dissolved in DMSO and 10 μg/ml Hoechst 33342 (Thermo Fisher Scientific). Immediately after incubation, cells were rinsed twice with PBS and images were captured using a Zeiss Axio Observer A1 inverted microscope with a QImaging Retiga CCD.
MTT assay
F9 cells were cultured and treated with DMSO, RA, VAS2870 or ML171 for 4 days followed by incubation in MTT reagent (Sigma) for 2–4 hours at 37°C and 5% CO2. Media was removed and DMSO was added to solubilize crystals, and then plates were incubated overnight in the dark. Absorbance values were measured at 570nm with a reference wavelength at 650nm using the GloMax®-Multi Detection System (Promega).
Statistical analysis
Data from qRT-PCR, densitometric analysis of immunoblots and Luciferase assays were gathered format least three independent biological replicates. Comparisons of data between control and experimental groups were performed using a one-way ANOVA with Tukey’s honest significant difference (HSD) post-hoc test or a Student’s t-Test (SPSS Statistics for Windows Version 19.0, IBM Corp. Released 2010, Armonk, NY). Student’s t-Test was used for statistical analysis of data when comparing control to only one experimental data set and (*) was used to denote significant difference. All other data were analyzed using a one-way ANOVA followed by Tukey’s HSD test and letters were used to indicate significant differences. P-values were considered statistically significant at the 0.05 level. Statistical data are presented as the mean ± SEM.
Results
RA increases NOX1 and NOX4 levels and ROS production
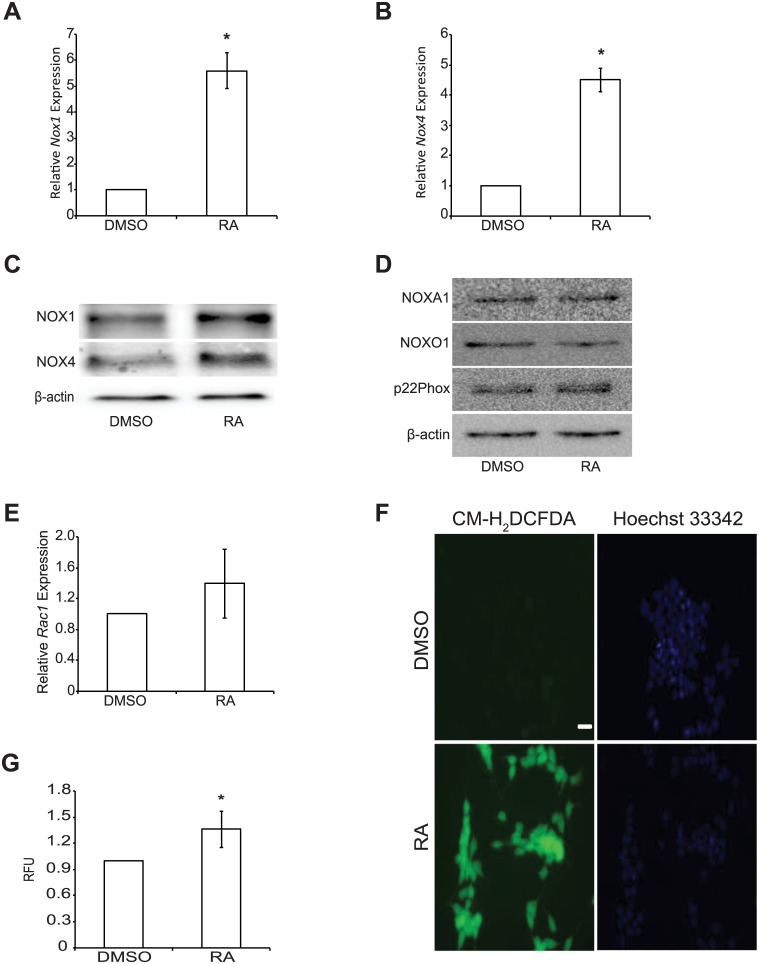
F9 cells treated with RA for 4 days showed a significant increase (P < 0.05) in the expression of Nox1 (Fig 1A) and Nox4 (Fig 1B), as we have shown previously [3]. Furthermore, NOX1 and NOX4 protein levels also increased after RA treatment (Fig 1C), however, the levels of accessory subunits required for NOX protein function, including NOXA1, NOXO1 and p22Phox (Fig 1D), or the transcripts encoding Rac1 (Fig 1E), did not appear to change during differentiation. Thus, the subunits to generate functional NOX1 and NOX4 were present in undifferentiated F9 cells. The transcripts encoding the NOX subunits, however, are in low abundance prior to differentiation to PrE (S1A–S1C Fig). More importantly, elevated ROS production accompanying differentiation was detected visually by CM-H2DCFDA (Fig 1F), and the significant increase in levels were validated quantitatively using Amplex Red (Fig 1G). When F9 cells were differentiated towards a PE lineage, Nox1 (S2A Fig) and Nox4 (S2B Fig) transcript levels decreased significantly (P < 0.05) when compared to DMSO- and RA-treated F9 cells. It should be noted, however, that a significant oxidative state (P < 0.05; S2C Fig) was maintained, thus suggesting another potential source of ROS when cells differentiate to PE.
Fig 1. RA increases NOX1 and NOX4 levels and ROS production.
Total RNA and protein was harvested from F9 cells treated with either DMSO or RA for 4 days. (A) Nox1 and (B) Nox4 expression following DMSO or RA treatment. (C) NOX1 and NOX4 protein levels following DMSO and RA treatment. (D) Protein levels of NADPH oxidase complex accessory subunits NOXA1, NOXO1, and p22Phox following DMSO and RA treatment. (E) Rac1 expression following DMSO or RA treatment. (F) ROS detection using CM-H2DCFDA or (G) Amplex Red in F9 cells treated with either DMSO or RA. A total of 3–5 independent experiments were analyzed and results presented as mean ± SEM. * denotes significance (P < 0.05) tested by a Student’s t-Test.
Gata6 induction results in increased NOX1 and NOX4 levels
Gata6, a master regulator of endoderm and extraembryonic endoderm formation, is significantly upregulated (P < 0.05) in F9 cells treated with RA (Fig 2A). This evidence, together with the presence of GATA-binding sites in the Nox1 promoter [11], and that GATA6 is responsible for increased Nox1 transcription in Caco-2 cells [12] led us to propose that GATA6 might be responsible for the upregulation of Nox1 and Nox4 in F9 cells preceding their differentiation. To address this, we overexpressed Gata6 and then analyzed Nox1 and Nox4 expression and protein levels using qRT-PCR and immunoblot analysis, respectively. When Gata6 was overexpressed alone (Fig 2B), it induced PrE [10], as evident by the increase in the differentiation markers DAB2 and TROMA1, and the decrease in the pluripotency marker OCT4 (Fig 2C). Gata6 overexpression also induced a significant increase (P < 0.05) in Nox1 (Fig 2D) and Nox4 (Fig 2E) expression. Interestingly, these changes were associated with an apparent increase in the levels of NOX1 and NOX4 protein (Fig 2F), which were comparable to those seen in RA-treated F9 cells. Together, results indicate that changes in NOX1 and NOX4 levels accompany F9 cell differentiation to PrE, and would suggest that the increase in ROS is due in part to these changes. Thus, studies were conducted to determine if the activity of NOX1 and/or NOX4 is/are required for PrE differentiation.
Fig 2. Gata6 induction results in increased NOX1 and NOX4 levels.
Total RNA and protein was collected from F9 cells treated with DMSO or RA, or transfected with pcDNA3.1-EV (EV) and treated with either DMSO or RA, or transfected with pcDNA3.1-Gata6 (Gata6) and cultured 4 days. (A) Gata6 expression of F9 cells treated with DMSO or RA. (B) Gata6 expression of F9 cells transfected with EV or Gata6. (C) Immunoblot analysis for DAB2 (arrow), TROMA1 and OCT4 in F9 cells transfected with EV and treated with DMSO or RA or F9 cells ectopically expressing Gata6. (D) Nox1 and (E) Nox4 expression in F9 cells transfected with EV and treated with DMSO or RA, or F9 cells ectopically expressing Gata6. (F) Immunoblot analysis for NOX1 and NOX4 in F9 cells transfected with EV and treated with DMSO or RA, or F9 cells ectopically expressing Gata6. A total of 3 independent experiments were analyzed and results are presented as mean ± SEM. * denotes significance (P < 0.05) tested by a Student’s t-Test, whereas letters denote groups of significance (P < 0.05) tested by a One-Way ANOVA followed by a Tukey’s test.
Pan-NOX inhibition blocks RA-mediated differentiation
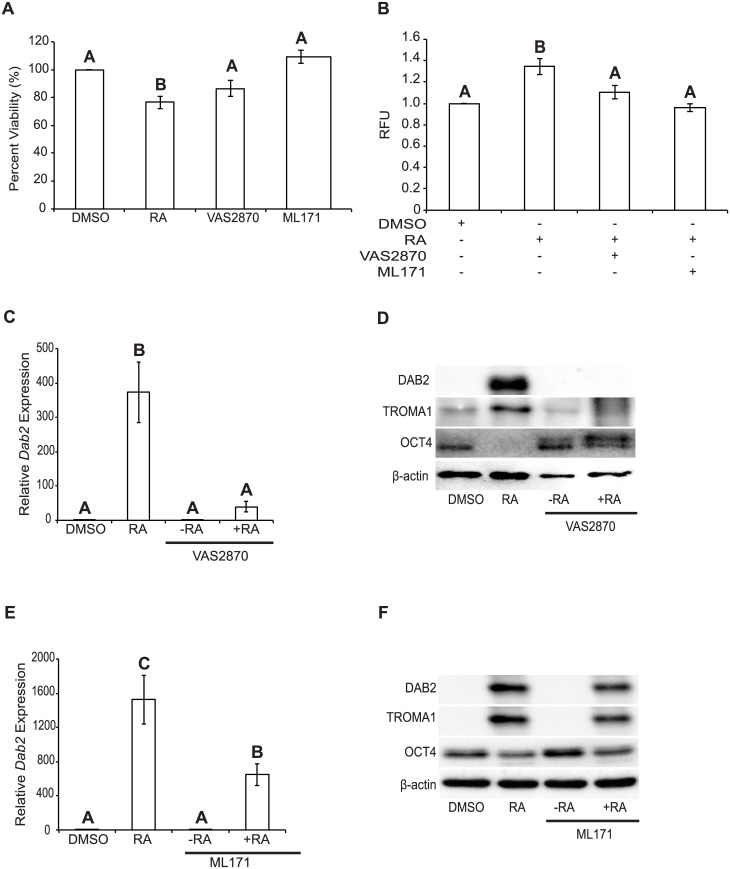
Our previous study revealed that Diphenyleneiodonium chloride (DPI), an inhibitor of flavoenzymes that produce ROS attenuates RA-induced PrE differentiation [3]. DPI is used frequently to inhibit ROS production, but since it acts on numerous oxidoreductases, including the NOX proteins [13] the source of the ROS may not be completely NOX-dependent. To test the hypothesis that the activity of the NOX proteins is necessary for PrE differentiation, F9 cells were treated with NOX inhibitors. First, the pan-NOX inhibitor, VAS2870 [14, 15] was used to determine specifically if members of the NOX family were necessary for F9 cells to differentiate to PrE. VAS2870 inhibits NOX1, NOX2 [15] and NOX4 [14], and although it can inhibit all NOX isoforms it does not serve as an antioxidant because it has no O2- scavenging effect [14]. VAS2870 treatment showed no significant effect on cell viability relative to the DMSO control (Fig 3A), but it did cause a significant attenuation (P < 0.05) in the amount of ROS produced in cells co-treated with RA (Fig 3B). Cells treated with DMSO-, RA-, VAS2870-, or co-treated with VAS2870 and RA were examined using qRT-PCR for Dab2 (Fig 3C), and immunoblot analysis for DAB2, TROMA1 and OCT4 (Fig 3D). Results show that Dab2 expression, relative to the loading control L14, was not significantly different between DMSO and VAS2870. The expression of Dab2 in F9 cells treated with RA alone was, as expected, significantly higher (P < 0.001) from the DMSO treatment, but surprisingly dramatically reduced in cells treated with RA and VAS2870 (Fig 3C). Immunoblot analysis showed comparable results with elevated DAB2 levels following RA treatment alone (Fig 3D). A similar increase was seen with the TROMA1 marker, while OCT4 levels decreased with RA treatment. The OCT4 signal, however, remained elevated in VAS2870 and RA-treated cells, and was comparable to that seen in DMSO-treated cells (Fig 3D). Thus, inhibiting NOX activity attenuates RA-mediated differentiation, which suggests strongly that NOX complexes are necessary for F9 cell differentiation.
Fig 3. Chemical inhibition of NOX proteins attenuates RA-mediated differentiation of F9 cells.
(A) MTT cell viability assay of F9 cells treated with DMSO, RA, VAS2870 or ML171. (B) F9 cells were treated with DMSO, RA, VAS2870 and RA, or ML171 and RA and assayed for ROS production using Amplex Red. (C) Dab2 expression of F9 cells treated with DMSO, RA, VAS2870, or VAS2870 and RA. (D) Immunoblot analysis for DAB2, TROMA1, and OCT4 in F9 cells treated with DMSO, RA, VAS2870, or VAS2870 and RA. (E) Dab2 expression of F9 cells treated with DMSO, RA, ML171, or ML171 and RA. (F) Immunoblot analysis for DAB2, TROMA,1 and OCT4 in F9 cells treated with DMSO, RA, ML171, or ML171 and RA. β-actin was used as a loading control. A total of 4 independent experiments were analyzed and results are presented as mean ± SEM. Letters denote groups of significance (P < 0.05) tested by a One-Way ANOVA followed by a Tukey’s test.
NOX1-specific inhibition attenuates RA-induced differentiation
F9 cells were treated with 250 nM of ML171, the IC50 of this NOX1-specific inhibitor [16]. ML171 has no ROS scavenging effects [16], and at this concentration it did not have an effect on F9 cell viability (Fig 3A). Co-treatment with RA, however, showed ROS levels that were significantly lower than those seen in RA-treated F9 cells (Fig 3B). qRT-PCR analysis revealed that Dab2 expression in DMSO- and ML171-treated F9 cells were not significantly different from each other, but both were significantly different (P < 0.001) from that in RA-treated F9 cells, and F9 cells co-treated with RA and ML171 (P < 0.05) (Fig 3E). Since Dab2 expression in cells co-treated with RA and ML171 was significantly lower (P < 0.05) than that in RA-treated F9 cells (Fig 3E), but not as low as that in cells co-treated with RA and VAS2870 (Fig 3C) this suggest that inhibiting NOX1 alone was not sufficient in completely attenuating Dab2 expression. Immunoblot analysis for DAB2 and TROMA1 showed no signals in DMSO- or ML171-treated F9 cells (Fig 3F). F9 cells co-treated with ML171 and RA, however, showed DAB2 and TROMA1 signals (Fig 3F), but they appeared to be of less intensity than that seen in the RA alone lane. OCT4 signals were seen in DMSO-, ML171 and RA- and ML171-treated F9 cells, but again the intensity appeared reduced in RA-treated F9 cells with or without ML171 (Fig 3F). Since NOX4 was not targeted chemically, as there are no known specific inhibitor(s) that do not have ROS scavenging effects or additionally target other NOX proteins [16], we can only conclude that NOX proteins are required for PrE differentiation, and that NOX1 specifically plays a role in this process.
Nox1 and Nox4 knockdown decreases ROS production
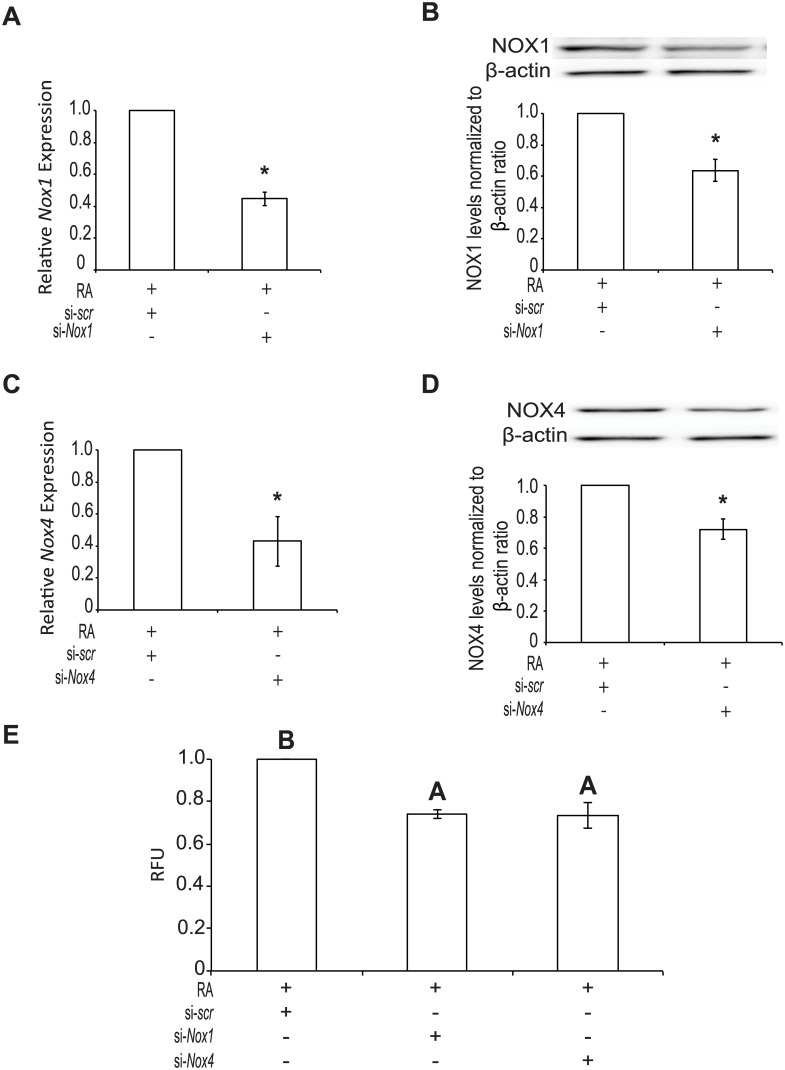
To corroborate the chemical inhibitor data, F9 cells were transfected with a siRNA vector to knockdown Nox1, Nox4 expression or a scrambled siRNA and treated with DMSO or RA as negative and positive controls, respectively. Since there is a significant increase (P < 0.05) in Nox1 and 4 expressions after RA treatment (Fig 1A and 1B and S1A–S1C Fig), F9 cells were treated with RA and the siRNA knockdown efficiency was compared to that seen with the RA-treated scrambled siRNA control. F9 cells containing Nox1 (Fig 4A) and Nox4 (Fig 4C) siRNAs showed significantly less expression of the respective Nox genes (P < 0.05) when compared to the RA-treated scrambled control. Both siRNA targeting vectors transfected into cells did not affect the individual expression of Nox1 or Nox4 (data not shown). The levels of NOX1 (Fig 4B) and NOX4 (Fig 4D) were examined to test if the knockdown effects were seen at the protein level. Results show that levels were significantly lower (P < 0.05) in cells containing the targeting siRNAs and treated with RA, compared to RA-treated F9 cells transfected with the scrambled siRNA. Furthermore, knocking down NOX1 and NOX4 in F9 cells treated with RA showed a significant reduction (P < 0.05) in ROS levels compared to those seen in cells transfected with the scrambled siRNA and treated with RA (Fig 4E).
Fig 4. Nox1 and Nox4 knockdown reduces ROS production.
Total RNA was collected from F9 cells transfected with scrambled (scr) si-RNA, si-Nox1 and/or si-Nox4 siRNA and cultured 4 days with RA treatment. (A) Expression of Nox1 and (B) NOX1 protein levels following transfection with scr or si-Nox1 and RA treatment. (C) Expression of Nox4 and (D) NOX4 protein levels following transfection with scr or si-Nox1 and RA treatment. (E) ROS production detected using Amplex Red of F9 cells transfected with scr, si-Nox1, or si-Nox4 and treated with RA. A total of 3 independent experiments were analyzed and results are presented as mean ± SEM. * denotes significance (P < 0.05) tested by a Student’s t-Test, whereas letters denote groups of significance (P < 0.05) tested by a One-Way ANOVA followed by a Tukey’s test.
Nox1 and Nox4 knockdown attenuates RA-induced differentiation
qRT-PCR and immunoblotting were used to determine if the knockdowns would affect Dab2 expression (Fig 5A) or levels of DAB2, TROMA1, and/or OCT4 protein (Fig 5B–5D). Analysis showed that compared to RA treatment, a significant reduction (P < 0.05) in Dab2 expression (Fig 5A) was seen when either Nox1 or Nox4 expression was knocked down. Also, there was no significant difference between the individual knockdowns compared to that due to the knockdown of both genes together. As expected, DAB2 and TROMA1 levels were low or not detected in cells transfected with the scrambled siRNA and treated with DMSO, however they did increase significantly in cells treated with RA (Fig 5B and 5C). Also, it is interesting to note that DAB2 levels in cells transfected with the targeted siRNAs and treated with RA were not significantly different between those transfected with the scr siRNA and treated with either DMSO or RA. However, TROMA1 levels were significantly lower in cells transfected with targeted siRNAs and treated with RA than RA treated cells transfected with scr siRNA (P < 0.05) (Fig 5C). OCT4 levels were high in the DMSO-treated scr siRNA control cells, but these were significantly reduced (P < 0.05) in F9 cells transfected with either the scr siRNA and treated with RA or those transfected with the siRNA targeting vectors and treated with RA (Fig 5B and 5D). Together with the inhibitor data, these results would indicate that NOX1 and NOX4 play a role in PrE differentiation.
Fig 5. Nox1 and Nox4 knockdown attenuates RA-induced differentiation.
RNA was collected from F9 cells transfected with scrambled (scr) si-RNA, si-Nox1 and/or si-Nox4 siRNA and cultured 4 days with RA treatment. (A) Dab2 expression of F9 cells transfected with scr, si-Nox1, or si-Nox4 and treated with RA. (B) Immunoblot analysis for DAB2, TROMA1, and OCT4 in F9 cells transfected with scr, si-Nox1, or si-Nox4 and treated with RA. (C) Densitometric analysis for DAB2, TROMA1, and (D) OCT4 in F9 cells transfected with scr, si-Nox1, or si-Nox4 and treated with RA. β-actin was used as a loading control. A total of 3 independent experiments were analyzed and results are presented as mean ± SEM. Letters and symbols denote groups of significance (P < 0.05) tested by a One-Way ANOVA followed by a Tukey’s test.
Overexpressing Nox1 and Nox4 increases ROS production
Since the inhibitor and genetic knockdown studies attenuated the formation of PrE, and our earlier study showing F9 cells treated with H2O2 develop into PrE [3], it seemed logical to propose that Nox overexpression should induce F9 cells. To test if Nox1 or Nox4 overexpression induced differentiation, F9 cells were transfected with pcDNA3.1-EV and treated with either DMSO or RA or transfected with pcDNA3.1-Nox1 or -Nox4. The relative degree of overexpression was determined by qRT-PCR, and results showed an approximate 700 and 1800-fold increase for Nox1 (Fig 6A) and Nox4 (Fig 6B), respectively. Similarly, NOX1 and NOX4 protein levels were higher in F9 cells transfected with the respective Nox construct (Fig 6C and 6D). Having established that transfection resulted in elevated NOX1 and NOX4 levels, we examined if this would affect ROS levels. For the positive control, F9 cells transfected with pcDNA3.1-EV and treated with RA showed a significant increase (P < 0.05) in ROS levels over those seen in transfected cells treated with DMSO (Fig 6E). Note that the levels seen following RA treatment were comparable to those in cells transfected with either Nox1 or Nox4.
Fig 6. Overexpressing Nox1 and Nox4 increases ROS production.
Total RNA was collected from F9 cells transfected with an empty vector (EV) control, pcDNA3.1-Nox1 or pcDNA3.1-Nox4 and cultured 4 days. (A) Nox1 and (B) Nox4 expression following transfection of pcDNA3.1-Nox1 or pcDNA3.1-Nox4, respectively, and relative to EV transfected cells. (C) NOX1 and (D) NOX4 protein levels following transfection of pcDNA3.1-Nox1 or pcDNA3.1-Nox4, respectively, relative to EV transfected cells. (E) ROS production using Amplex Red in F9 cells transfected with EV and treated with DMSO or RA, or transfected with pcDNA3.1-Nox1 or pcDNA3.1-Nox4. A total of 3 independent experiments were analyzed and results are presented as mean ± SEM. * denotes significance (P < 0.05) tested by a Student’s t-Test, whereas letters denote groups of significance (P < 0.05) tested by a One-Way ANOVA followed by a Tukey’s test.
Nox overexpression activates canonical Wnt signaling, but does not promote differentiation
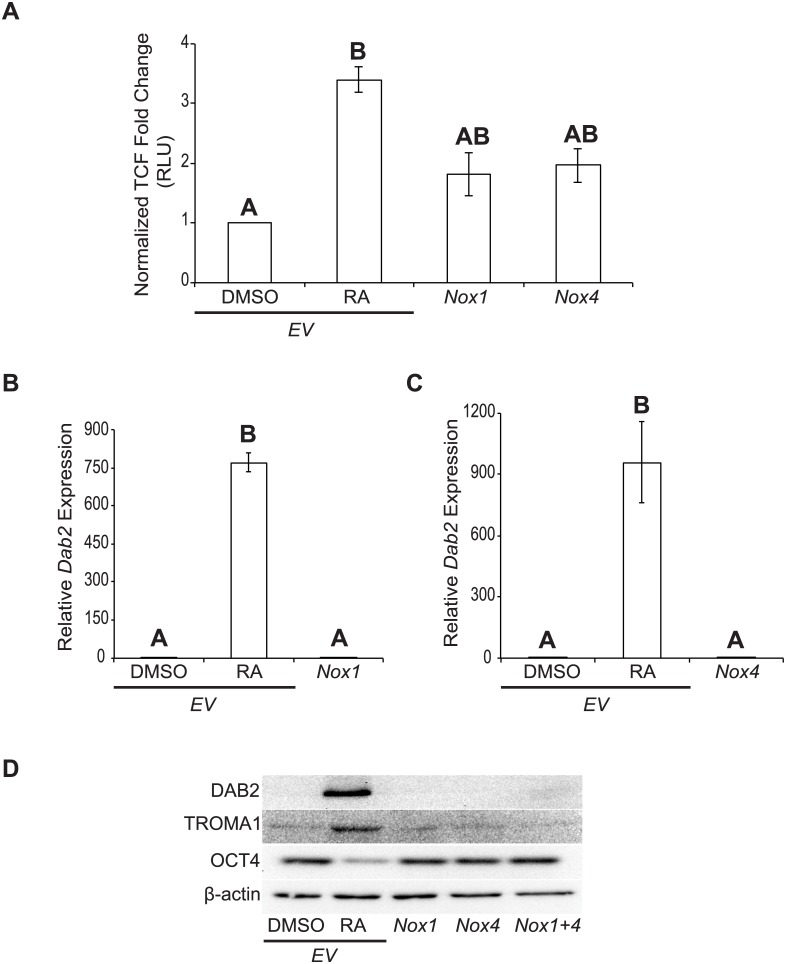
The overexpression results and those from earlier studies [3, 4] suggested that the increased levels in ROS due to ectopic Nox1 or Nox4 expression would activate canonical Wnt-β-catenin signaling. To test this F9 cells were co-transfected with Nox1 or Nox4, and the firefly luciferase pGL3-BARL reporter that is used as readout for active Wnt-β-catenin signaling [17]. All cells were transfected with the Renilla luciferase construct, TK-RL to normalize for luciferase expression. RA-treated F9 cells transfected with pcDNA3.1-EV showed a significant increase (P < 0.05) in TCF activity compared to the DMSO control (Fig 7A). F9 cells overexpressing Nox1 or Nox4 also showed increased TCF activity, however, this increase was less than that seen in RA-treated F9 cells, and was not significant from either RA or DMSO treatments (Fig 7A). Nevertheless, F9 cells overexpressing Nox1 or Nox4 were analyzed for PrE differentiation, and as expected Dab2 expression relative to the L14 loading control was significantly higher (P < 0.001) in RA-treated F9 cells than that in DMSO treated cells (Fig 7B and 7C). Dab2 expression resulting from either the overexpression of Nox1 (Fig 7B) or Nox4 (Fig 7C) was not significantly different from that in DMSO-treated F9 cells, suggesting that differentiation had not occurred. Similarly, DAB2 protein was only present in RA-treated cells, and this evidence of differentiation was seen in the corresponding increase in TROMA1 and concomitant decrease in OCT4 levels (Fig 7D). TROMA1 signals were not seen in DMSO-treated, Nox1-, Nox4-, or Nox1 and Nox4-overexpressing cells, and the appearance of OCT4 in the same samples was indicative that PrE had not differentiated as a result of Nox1 and/or Nox4 overexpression (Fig 7D). Thus, even though overexpressing Nox1 or Nox4 in F9 cells increased ROS levels, there was no significant activation of Wnt-β-catenin signaling or hallmark molecular indicators of PrE to suggest that the overexpression of Nox1, Nox4 or both was sufficient to induce differentiation.
Fig 7. Nox overexpression activates canonical Wnt signaling, but does not promote differentiation.
(A) Dual-luciferase assay of TCF activity in F9 cells transfected with EV and treated with DMSO or RA, or transfected with pcDNA3.1-Nox1 or pcDNA3.1-Nox4. (B) Dab2 expression in F9 cells transfected with EV and treated with DMSO or RA, or transfected with pcDNA3.1-Nox1. (C) Dab2 expression in F9 cells transfected with EV and treated with DMSO or RA, or transfected with pcDNA3.1-Nox4. (D) Immunoblot analysis for DAB2, TROMA1, and OCT4 in F9 cells transfected with EV and treated with DMSO or RA, or transfected with pcDNA3.1-Nox1, pcDNA3.1-Nox4 or both vectors. β-actin was used as a loading control. A total of 3 independent experiments were analyzed and results are presented as mean ± SEM. Letters denote groups of significance (P < 0.05) tested by a One-Way ANOVA followed by a Tukey’s test.
Discussion
ROS, which are by-products of metabolic processes produced through the incomplete reduction of oxygen, create oxidative stress on cells, and damage nucleic acids, proteins and lipids. Although this damage is linked to age-related and other human disease pathogenesis, recent studies now show cells benefitting from ROS as a second messenger to modulate biological processes such as differentiation, apoptosis and proliferation [18, 19]. F9 cells are induced to form primitive endoderm when treated with RA, an in vitro process that mimics a very early event in mouse embryogenesis. Differentiation is accompanied by numerous changes in gene expression and morphology and requiring several signaling pathways. One of these pathways involves canonical Wnt-β-catenin signaling [1], where Wnt6 is induced by GATA6 [10] a master regulator of extraembryonic and embryonic endoderm. Regulation of this Wnt pathway occurs in a redox-regulated manner [3] involving NRX and DVL, the latter serving as a lynchpin in the three major Wnt signaling pathways [4]. We have shown previously that cytosolic ROS plays a role in F9 cell differentiation to form primitive endoderm [3]. ROS impact positively on the canonical Wnt-β-catenin signaling pathway, and the Nox genes that are upregulated in response to RA were identified as candidates responsible for this cytosolic ROS [3]. Increases in mitochondrial ROS have been also observed in F9 cells differentiated to primitive and parietal endoderm, a consequence of what appears to be a metabolic transition from aerobic glycolysis towards oxidative phosphorylation (manuscript in preparation). This increase in mitochondrial ROS might explain the maintained oxidative state of F9 cells at PE, despite the reduction in Nox1 and Nox4 expression (S2A and S2B Fig).
The expression profiles of the six mouse Nox genes led us to interrogate Nox1 and Nox4, which were the most up-regulated in response to RA in F9 cells [3]. In addition to RA (Fig 1A–1D), both genes are up-regulated, and the level of the corresponding proteins increase in response to GATA6 (Fig 2D–2F). Previous studies provide evidence for a link between GATA6 and Nox genes, specifically Nox1, which is activated directly by GATA6 [11, 12]. NOX1 impacting on Wnt signaling occurs in colonic and intestinal epithelial cells [8, 20], and it is known to post-transcriptionally increase the levels of Keratin 18 [21], a cytokeratin encoded by transcripts that increase during RA-induced F9 cell differentiation [22]. Furthermore, NOX1 and NOX4 activate the p38 MAPK pathway in mouse and human cells [23, 24], which is also required to inhibit GSK-3β thereby maintaining Wnt in the on-state during PrE differentiation [25]. Evidence for NOX4 interacting with Wnt signaling or regulated by GATA transcription factors is not as compelling as NOX1. However, it is interesting to note a study showing the opposite scenario exists in cardiomyocytes, where NOX4 increases Gata4 expression [26]. Other reports have linked NOX4 to cell proliferation, cytoskeletal reorganization, migration and differentiation [27–30], which incidentally are processes that F9 cells participate in during differentiation [31].
Having identified that NOX1 and NOX4 were under the indirect or direct control of GATA6, we next used chemical inhibitors to test whether or not the activity of both NOX proteins were required for PrE differentiation. As noted above, a previous study in our lab using the non-specific flavoenzyme inhibitor DPI, demonstrated the necessity for ROS in the differentiation of F9 cells [3]. Since DPI acts on other non-NOX flavoenzymes [13], we refined the approach and used a pan-NOX and NOX1-specific inhibitor to test their effects on differentiation (Fig 3). Both inhibitors affected differentiation at the gene and protein levels, but VAS2870, the pan-NOX inhibitor was more potent than ML171, the selective inhibitor of NOX1 activity (Fig 3). Although there could be many reasons to explain this, the fact that all the NOX proteins were being inhibited by VAS2870 would suggest that multiple NOX proteins contribute to the ROS required for differentiation. This argument would be tested later as these pharmacological inhibitor studies proved informative, but required a genetic approach to corroborate the data. Mouse knockout models exist for all 6 Nox genes [14, 32, 33], but disrupting any one of them individually or in combination does not appear to have obvious effects in embryos. Therefore, an alternative strategy was to use siRNAs, which have had some success in knocking down the expression of specific Nox genes [14]. This approach was used to knockdown Nox1 and/or Nox4 gene expression, and it resulted in reduced expression and protein levels of NOX1 and NOX4 (Fig 4A–4D). Knockdown led to the attenuation of differentiation, as evident by the decrease in the markers of PrE, and the maintenance of pluripotency as noted by the persistence of the OCT4 signal (Fig 5A–5D). Together with the inhibitor results, this data adds support that NOX activity, specifically from NOX1 and NOX4, is necessary for F9 cells to form PrE. While encouraged by these results, the contribution to the differentiation process by the two other Nox genes and Duox2 that are upregulated by RA in F9 cells needs to be considered and is currently under investigation.
Although NOX1 and NOX4 are members of the NOX subfamily that require p22phox, the former generates primarily superoxide, while the major product of the latter is H2O2 [34]. Nox4 is widely expressed in human, while Nox1 is much more restricted [32]. Nevertheless, there is overlap in places where functional redundancy might explain why the in vivo knockout models do not show obvious phenotypes. This does not appear to be the case for the F9 in vitro cell model as there was a common effect seen when knocking down either Nox1 or Nox4. Thus, it is possible that attenuating the activity of one NOX isoform in vitro reduces the total amount of ROS required for differentiation, and this cannot be compensated by the other members. The overexpression of either Nox1 and/or Nox4 (Fig 6A–6D) in F9 cells nevertheless resulted in increased ROS levels (Fig 6E), and what appeared to be an increase in canonical Wnt signaling (Fig 7A). However, these increases were not sufficient to induce differentiation as seen following the application of exogenous H2O2 [3]. Although there is much debate centered on the recently reported Nox1, Nox2 and Nox4 triple knockout mouse model [33], our information would indicate that ROS produced individually by at least two of the NOX isoforms is necessary in the F9 cell model of primitive extraembryonic endoderm formation. Building from previous studies our model now adds to the signaling hierarchy and crosstalk within these networks where RA induces Gata6 expression, which in turn regulates several genes including Wnt6 [10], Nox1 and Nox4 (this study). The subsequent increase in ROS permits NRX to dissociate from DVL, thereby priming the Wnt pathway [4] prior to the appearance of the WNT6 ligand that is sufficient to promote PrE differentiation [1]. WNT6 signaling is attenuated in a negative feedback loop involving Dickkopf-1 [35], and this is required for PrE cells to complete their differentiation to PE [36]. Given the report showing canonical Wnt signaling activates RAC1 and this in turn increases RAC1-dependent NOX1 activity that produces the ROS needed to oxidize NRX and dissociate it from DVL in order to keep the pathway in the on-state [8], it is tempting to speculate that a similar mechanism occurs in F9 cells in response to RA. However, since NOX4 is not RAC1-dependent [9], and is required for PrE differentiation (this study), canonical Wnt signaling must be stimulating NOX4 activity in another manner. Given that the production of ROS is increased as a direct result of increased NOX4 expression [37], the possibility exists that increased NOX4 activity is the result of an upregulation in the Nox4 gene by active Wnt signaling. Towards that end, experiments are underway to test if the Nox genes are WNT target genes or simply downstream in the pathway. Whatever the case, that aberrant Wnt signaling is prevalent in cancer biology [38, 39] underpins the importance of better understanding the crosstalk imparted by NOX proteins and ROS on the Wnt-β-catenin signaling pathway in both development and cancer research.
Supporting information
Total RNA was collected from F9 cells treated with DMSO or RA for 4 days. (A) Ct values for Nox1 and Nox4 under DMSO or RA treatment. (B and C) Graphical illustration of Ct values of Nox1 and Nox4 under DMSO or RA treatment over 40 cycles. L14 served as a control.
(EPS)
Total RNA was collected from F9 cells induced to differentiate into PrE and PE. Expression of Nox1 (A) and Nox4 (B) following DMSO, RA, and RA with db-cAMP treatment. (C) Oxidative state of F9 cells following treatment with DMSO, RA, RA with db-cAMP. A total of 4 independent experiments were analyzed and results are presented as mean ± SEM. Letters denote groups of significance (P < 0.05) tested by a One-Way ANOVA followed by a Tukey’s test.
(EPS)
Acknowledgments
The authors would like to thank Drs. R. J. Rylett and J. F. Staples for reagents.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Support was provided by the Natural Sciences and Engineering Research Council of Canada, Discovery Grant R2615A02004 [http://www.nserc-crsng.gc.ca/Professors-Professeurs/Grants-Subs/DGIGP-PSIGP_eng.asp].
References
- 1.Krawetz R, Kelly GM. Wnt6 induces the specification and epithelialization of F9 embryonal carcinoma cells to primitive endoderm. Cell Signal. 2008;20(3):506–17. 10.1016/j.cellsig.2007.11.001 [DOI] [PubMed] [Google Scholar]
- 2.Strickland S, Smith KK, Marotti KR. Hormonal induction of differentiation in teratocarcinoma stem cells: generation of parietal endoderm by retinoic acid and dibutyryl cAMP. Cell. 1980;21(2):347–55. [DOI] [PubMed] [Google Scholar]
- 3.Wen JW, Hwang JT, Kelly GM. Reactive oxygen species and Wnt signalling crosstalk patterns mouse extraembryonic endoderm. Cell Signal. 2012;24(12):2337–48. 10.1016/j.cellsig.2012.07.024 [DOI] [PubMed] [Google Scholar]
- 4.Sandieson L, Hwang JT, Kelly GM. Redox regulation of canonical Wnt signaling affects extraembryonic endoderm formation. Stem Cells Dev. 2014;23(10):1037–49. 10.1089/scd.2014.0010 [DOI] [PubMed] [Google Scholar]
- 5.Funato Y, Miki H. Redox regulation of Wnt signalling via nucleoredoxin. Free Radic Res. 2010;44(4):379–88. 10.3109/10715761003610745 [DOI] [PubMed] [Google Scholar]
- 6.Funato Y, Terabayashi T, Sakamoto R, Okuzaki D, Ichise H, Nojima H, et al. Nucleoredoxin sustains Wnt/beta-catenin signaling by retaining a pool of inactive dishevelled protein. Curr Biol. 2010;20(21):1945–52. 10.1016/j.cub.2010.09.065 [DOI] [PubMed] [Google Scholar]
- 7.Bahn YJ, Lee KP, Lee SM, Choi JY, Seo YS, Kwon KS. Nucleoredoxin promotes adipogenic differentiation through regulation of Wnt/beta-catenin signaling. J Lipid Res. 2015;56(2):294–303. 10.1194/jlr.M054056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kajla S, Mondol AS, Nagasawa A, Zhang Y, Kato M, Matsuno K, et al. A crucial role for Nox 1 in redox-dependent regulation of Wnt-beta-catenin signaling. FASEB J. 2012;26(5):2049–59. 10.1096/fj.11-196360 [DOI] [PubMed] [Google Scholar]
- 9.Brandes RP, Weissmann N, Schroder K. Nox family NADPH oxidases: Molecular mechanisms of activation. Free Radic Biol Med. 2014;76:208–26. 10.1016/j.freeradbiomed.2014.07.046 [DOI] [PubMed] [Google Scholar]
- 10.Hwang JT, Kelly GM. GATA6 and FOXA2 regulate Wnt6 expression during extraembryonic endoderm formation. Stem Cells Dev. 2012;21(17):3220–32. 10.1089/scd.2011.0492 [DOI] [PubMed] [Google Scholar]
- 11.Brewer AC, Sparks EC, Shah AM. Transcriptional regulation of the NADPH oxidase isoform, Nox1, in colon epithelial cells: role of GATA-binding factor(s). Free Radic Biol Med. 2006;40(2):260–74. 10.1016/j.freeradbiomed.2005.08.022 [DOI] [PubMed] [Google Scholar]
- 12.Adachi Y, Shibai Y, Mitsushita J, Shang WH, Hirose K, Kamata T. Oncogenic Ras upregulates NADPH oxidase 1 gene expression through MEK-ERK-dependent phosphorylation of GATA-6. Oncogene. 2008;27(36):4921–32. 10.1038/onc.2008.133 [DOI] [PubMed] [Google Scholar]
- 13.Riganti C, Gazzano E, Polimeni M, Costamagna C, Bosia A, Ghigo D. Diphenyleneiodonium inhibits the cell redox metabolism and induces oxidative stress. J Biol Chem. 2004;279(46):47726–31. 10.1074/jbc.M406314200 [DOI] [PubMed] [Google Scholar]
- 14.Altenhofer S, Kleikers PW, Radermacher KA, Scheurer P, Rob Hermans JJ, Schiffers P, et al. The NOX toolbox: validating the role of NADPH oxidases in physiology and disease. Cell Mol Life Sci. 2012;69(14):2327–43. 10.1007/s00018-012-1010-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wingler K, Altenhoefer SA, Kleikers PW, Radermacher KA, Kleinschnitz C, Schmidt HH. VAS2870 is a pan-NADPH oxidase inhibitor. Cell Mol Life Sci. 2012;69(18):3159–60. 10.1007/s00018-012-1107-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drummond GR, Selemidis S, Griendling KK, Sobey CG. Combating oxidative stress in vascular disease: NADPH oxidases as therapeutic targets. Nat Rev Drug Discov. 2011;10(6):453–71. 10.1038/nrd3403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biechele TL, Moon RT. Assaying beta-catenin/TCF transcription with beta-catenin/TCF transcription-based reporter constructs. Methods Mol Biol. 2008;468:99–110. 10.1007/978-1-59745-249-6_8 [DOI] [PubMed] [Google Scholar]
- 18.Andersen JK. Oxidative stress in neurodegeneration: cause or consequence? Nat Med. 2004;10 Suppl:S18–25. [DOI] [PubMed] [Google Scholar]
- 19.Landry WD, Cotter TG. ROS signalling, NADPH oxidases and cancer. Biochem Soc Trans. 2014;42(4):934–8. 10.1042/BST20140060 [DOI] [PubMed] [Google Scholar]
- 20.Coant N, Ben Mkaddem S, Pedruzzi E, Guichard C, Treton X, Ducroc R, et al. NADPH oxidase 1 modulates WNT and NOTCH1 signaling to control the fate of proliferative progenitor cells in the colon. Mol Cell Biol. 2010;30(11):2636–50. 10.1128/MCB.01194-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sattayakhom A, Ittiwat W, Stremmel W, Chamulitrat W. Redox regulation of cytokeratin 18 protein by NADPH oxidase 1 in preneoplastic human epithelial cells. J Cancer Res Clin Oncol. 2011;137(11):1669–78. 10.1007/s00432-011-1041-x [DOI] [PubMed] [Google Scholar]
- 22.Inoue A, Nagafuchi A, Kikuchi A. Retinoic acid induces discrete Wnt-signaling-dependent differentiation in F9 cells. Biochem Biophys Res Commun. 2009;390(3):564–9. 10.1016/j.bbrc.2009.10.005 [DOI] [PubMed] [Google Scholar]
- 23.Kodama R, Kato M, Furuta S, Ueno S, Zhang Y, Matsuno K, et al. ROS-generating oxidases Nox1 and Nox4 contribute to oncogenic Ras-induced premature senescence. Genes Cells. 2013;18(1):32–41. 10.1111/gtc.12015 [DOI] [PubMed] [Google Scholar]
- 24.Pekarcikova L, Knopfova L, Benes P, Smarda J. c-Myb regulates NOX1/p38 to control survival of colorectal carcinoma cells. Cell Signal. 2016;28(8):924–36. 10.1016/j.cellsig.2016.04.007 [DOI] [PubMed] [Google Scholar]
- 25.Bikkavilli RK, Feigin ME, Malbon CC. p38 mitogen-activated protein kinase regulates canonical Wnt-beta-catenin signaling by inactivation of GSK3beta. J Cell Sci. 2008;121(Pt 21):3598–607. 10.1242/jcs.032854 [DOI] [PubMed] [Google Scholar]
- 26.Murray TV, Smyrnias I, Shah AM, Brewer AC. NADPH oxidase 4 regulates cardiomyocyte differentiation via redox activation of c-Jun protein and the cis-regulation of GATA-4 gene transcription. J Biol Chem. 2013;288(22):15745–59. 10.1074/jbc.M112.439844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Stouffs M, Serrander L, Banfi B, Bettiol E, Charnay Y, et al. The NADPH oxidase NOX4 drives cardiac differentiation: Role in regulating cardiac transcription factors and MAP kinase activation. Mol Biol Cell. 2006;17(9):3978–88. 10.1091/mbc.E05-06-0532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hecker L, Vittal R, Jones T, Jagirdar R, Luckhardt TR, Horowitz JC, et al. NADPH oxidase-4 mediates myofibroblast activation and fibrogenic responses to lung injury. Nat Med. 2009;15(9):1077–81. 10.1038/nm.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lyle AN, Deshpande NN, Taniyama Y, Seidel-Rogol B, Pounkova L, Du P, et al. Poldip2, a novel regulator of Nox4 and cytoskeletal integrity in vascular smooth muscle cells. Circ Res. 2009;105(3):249–59. 10.1161/CIRCRESAHA.109.193722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao Q, Luo Z, Pepe AE, Margariti A, Zeng L, Xu Q. Embryonic stem cell differentiation into smooth muscle cells is mediated by Nox4-produced H2O2. Am J Physiol Cell Physiol. 2009;296(4):C711–23. 10.1152/ajpcell.00442.2008 [DOI] [PubMed] [Google Scholar]
- 31.Hagiwara N, Kadono N, Miyazaki T, Maekubo K, Hirai Y. Extracellular syntaxin4 triggers the differentiation program in teratocarcinoma F9 cells that impacts cell adhesion properties. Cell Tissue Res. 2013;354(2):581–91. 10.1007/s00441-013-1680-0 [DOI] [PubMed] [Google Scholar]
- 32.Sirokmany G, Donko A, Geiszt M. Nox/Duox Family of NADPH Oxidases: Lessons from Knockout Mouse Models. Trends Pharmacol Sci. 2016;37(4):318–27. 10.1016/j.tips.2016.01.006 [DOI] [PubMed] [Google Scholar]
- 33.Rezende F, Lowe O, Helfinger V, Prior KK, Walter M, Zukunft S, et al. Unchanged NADPH Oxidase Activity in Nox1-Nox2-Nox4 Triple Knockout Mice: What Do NADPH-Stimulated Chemiluminescence Assays Really Detect? Antioxid Redox Signal. 2016;24(7):392–9. 10.1089/ars.2015.6314 [DOI] [PubMed] [Google Scholar]
- 34.Lambeth JD, Neish AS. Nox enzymes and new thinking on reactive oxygen: a double-edged sword revisited. Annu Rev Pathol. 2014;9:119–45. 10.1146/annurev-pathol-012513-104651 [DOI] [PubMed] [Google Scholar]
- 35.Golenia G G M.I. Kelly G.M. Frizzled gene expression and negative regulation of canonical WNT-β-catenin signaling in mouse F9 teratocarcinoma cells. Biochem Cell Biol. 2016;Bcb-2016-0150. In Press. [DOI] [PubMed] [Google Scholar]
- 36.Schlupf J, Steinbeisser H. IGF antagonizes the Wnt/beta-Catenin pathway and promotes differentiation of extra-embryonic endoderm. Differentiation. 2014;87(5):209–19. 10.1016/j.diff.2014.07.003 [DOI] [PubMed] [Google Scholar]
- 37.Ellmark SH, Dusting GJ, Fui MN, Guzzo-Pernell N, Drummond GR. The contribution of Nox4 to NADPH oxidase activity in mouse vascular smooth muscle. Cardiovasc Res. 2005;65(2):495–504. 10.1016/j.cardiores.2004.10.026 [DOI] [PubMed] [Google Scholar]
- 38.Polakis P. Wnt signaling in cancer. Cold Spring Harb Perspect Biol. 2012;4(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heuberger J, Birchmeier W. Interplay of cadherin-mediated cell adhesion and canonical Wnt signaling. Cold Spring Harb Perspect Biol. 2010;2(2):a002915 10.1101/cshperspect.a002915 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Total RNA was collected from F9 cells treated with DMSO or RA for 4 days. (A) Ct values for Nox1 and Nox4 under DMSO or RA treatment. (B and C) Graphical illustration of Ct values of Nox1 and Nox4 under DMSO or RA treatment over 40 cycles. L14 served as a control.
(EPS)
Total RNA was collected from F9 cells induced to differentiate into PrE and PE. Expression of Nox1 (A) and Nox4 (B) following DMSO, RA, and RA with db-cAMP treatment. (C) Oxidative state of F9 cells following treatment with DMSO, RA, RA with db-cAMP. A total of 4 independent experiments were analyzed and results are presented as mean ± SEM. Letters denote groups of significance (P < 0.05) tested by a One-Way ANOVA followed by a Tukey’s test.
(EPS)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.