Abstract
An approach to genetically engineered resistance to pseudorabies virus (PRV) infection was examined by using a transgene encoding a soluble form of nectin-1, also known as herpesvirus entry mediator C. Nectin-1 is an α-herpesvirus receptor that binds to virion glycoprotein D. Nectin-1 mediates entry of PRV, herpes simplex virus types 1 and 2, and bovine herpesvirus type 1. To assess the antiviral potential of an ectopic expression of the nectin-1 ectodomain in vivo, six transgenic mouse lines expressing a soluble form of nectin-1, consisting of an extracellular domain of porcine nectin-1 and the Fc portion of human IgG1, were generated. All of the transgenic mouse lines showed nearly complete resistance to PRV infection by means of both i.p. and intranasal routes. These results suggest that the introduction into farm animals of a transgene encoding a soluble form of nectin-1 would offer a potent biological approach to generating α-herpesvirus-resistant livestock.
Altering host susceptibility to viral infections through vaccination is a widespread strategy used to reduce associated pathologies and virus shedding. Viral disease control in farm animals also offers significant routes to improved host resistance through selection for existing resistant alleles or expression of transgenes, the products of which interfere with the viral replication cycle. Up to now, the development of disease-resistant farm animals by gene transfer has been almost nonexistent.
Pseudorabies virus (PRV), a representative member of the α-herpesvirius subfamily, causes lethal encephalitis in piglets, acute respiratory syndrome in growing pigs, abortion and infertility in breeding sows, and latent infection in surviving pigs. PRV infection is a major economic risk in the swine industry worldwide. In general, vaccination strategies alone usually suppress manifestation of the disease but do not stamp out viral infection from a population. Dominant-negative mutants of viral proteins have been demonstrated in cell culture systems as potent inhibitors of herpesvirus replication and proposed as a disease control strategy termed “intracellular immunization” (1). Intracellular immunization against herpesvirus infection in vivo was obtained by using mutated forms of the virus transcription factors (2, 3). Unfortunately, it appeared that these transgenes might exert adverse side effects, such as retarded growth, by hitting unknown host targets and thus illustrating a tight level of host-pathogen interactions during all steps of development. As an alternative to these previous strategies, we proposed to alter known host molecular components of the viral infection cycle.
We have set our sights on inhibition of viral entry to induce protection of the animals against PRV infection. Binding of α-herpesviruses to cells occurs primarily through an interaction of glycoprotein C and/or glycoprotein B with cell-surface heparan sulfate (4-7), whereas fusion between the virion envelope and cell membrane requires glycoproteins B, D, H, and L (8-11). Five α-herpesvirus receptors have been identified: herpesvirus entry mediator (Hve)A (HVEM), HveB (nectin-2), HveC (nectin-1), HveD (CD155), and 3-O-sulfated heparan sulfate (12-16). Recently, we reported that expression of a soluble form of HVEM in transgenic mice enhanced resistance to herpes simplex virus type 1 (HSV-1) infection (17). However, HVEM has no entry activity for PRV or bovine herpesvirus type-1 (12). In contrast, nectin-1 has the broadest specificity for mediating α-herpesvirus entry and is present in a broad range of tissues and cells (13, 14). The nectin family has the same Ig-like domain structures (18-22) (Fig. 1A). We demonstrated that transformed cell lines expressing a soluble form of porcine nectin-1 showed significant resistance to PRV infection (23). This in vitro model system provided a possible basis for the development of livestock with enhanced resistance to pseudorabies.
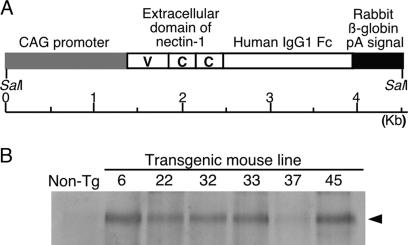
Fig. 1.
Generation of transgenic mice expressing PHveCIg. (A) Schematic representation of the transgene. The PHveCIg gene is under the control of the CAG promoter. The extracellular domain of nectin-1 consists of one V domain and two C domains. Cleavage sites for restriction enzyme SalI are indicated. (B) Western blot analysis of sera from the transgenic (Tg) mice. The arrow indicates the position of detected PHveCIg.
Here we demonstrate that transgenic mouse lines expressing a soluble form of porcine nectin-1 were highly resistant to PRV infection without any side effects. If this approach can be applied to farm animals, it could have a significant impact on animal production industries worldwide.
Materials and Methods
Generation of Transgenic Mice. The cDNA of the extracellular domain of porcine nectin-1 was amplified as described by Milne et al. (24). The cDNA was inserted into the XhoI and BamHI sites of a plasmid carrying IgG1-Fc DNA (25). The chimeric gene was inserted into an XhoI site of pCXN2 vector (26) having a SalI site instead of HindIII site. The SalI transgene fragment (Fig. 1 A) containing the CAG promoter (cytomegalovirus IE enhancer and chicken β-actin promoter), the PHveCIg gene, and the rabbit β-globin polyA signal was isolated and purified. The DNA fragment was microinjected into C57BL/6 mouse eggs to generate transgenic mice. Transgenic founders were identified by PCR with genomic DNA isolated from mouse tail (27) and the specific primers (PHveC-F, 5′-GGACCCCTCGAGCGCCATGGCT-3′; 3IG1, 5′-TGC CCT GGA CTG GGG CTG CAT AG-3′). The transgenic founders were crossed with C57BL/6 mice. Heterozygous F1 transgenic mice were crossed with C57BL/6 mice, and their offspring (F2) was used for experimental infections. All mice were maintained in the animal facilities at our institute and treated according to the Laboratory Animal Control Guidelines of our institute, which are similar to those of the National Institutes of Health American Association of Laboratory Animal Control.
Statistical Analysis. Means test comparisons were done by using ANOVA (litter size) and general linear modeling (body weights) procedures from sas 8.12 software. (SAS Institute, Cary, NC). Means comparisons were done by using pairwise t tests.
Analysis of Transgene Expression. A reference PHveCIg protein sample was purified from a supernatant of the transformed Vero cell line (C-A6) expressing PHveCIg (23). To measure PHveCIg concentrations in sera of the transgenic mice, a competitive ELISA system using a rabbit anti-human nectin-1 antibody (28) was established as described in ref. 17. Western blotting with 1 μl of each serum of the transgenic mice and histopathological procedure was performed as described in ref. 17. The rehydrated sections were immunostained by the indirect immunoperoxidase technique with biotinylated anti-human IgG and avidinhorseradish peroxidase detection reagent.
Virus Infection in Mice. PRV strains YS-81, Kojnock, Chiba-03, a new field isolate from Japan (developed in 2003), and HSV-1 strain VR-3 were used for experimental infections. The LD50 of each virus strain were titrated on C57BL/6 mice. The mice at 6-8 weeks of age were infected i.p. with 200 μl of DMEM containing 20 LD50 of PRV strain YS-81 in Sapporo, Japan, or strain Kojnock in Paris. Experimental infection with HSV-1 was also performed as described above. Intranasal PRV infection was performed with 5 μl of DMEM containing 10 LD50 of PRV strain YS-81 or strain Chiba-03 under anesthesia. Survival of mice and signs of disease were recorded for 14 days. Anti-PRV antibodies in sera of surviving mice at least 1 month after the virus inoculation were measured by ELISA, with disrupted-purified PRV as the viral antigen (3).
Detection of the Virus DNA in Trigeminal Ganglia by PCR. Mice surviving intranasal infections were killed by decapitation at least 1 month after the virus inoculation, and trigeminal ganglia were immediately removed and frozen in liquid nitrogen. As a control experiment, transgenic mice and nontransgenic littermates were infected with PRV strain Begonia, an attenuated vaccine strain deleted for glycoprotein E and thymidine kinase genes (Intervet International, Boxmeer, The Netherlands). Genomic DNA was isolated from trigeminal ganglia and screened for PRV latency-associated transcript (LAT) sequences. PRV DNA was detected by PCR analysis with the specific primers for the PRV LAT gene (LAT-F, 5′-GAGGAGGAGGAGGACACGA-3′; LAT-R, 5′-TCCAGCTCCGGCACCAAGT-3′). PCR for the LAT gene was carried out as described in ref. 29. Digoxigenin-labeled DNA probes for detection of the virus DNA were derived from pG/KpnI-E (30) by using the specific primers described above and a PCR digoxigenin probe synthesis kit (Roche, Gipf-Oberfrick, Switzerland). Southern blotting of the PCR products was performed as described in ref. 3.
Virus Infection in Cultured Cells. Transgenic and nontransgenic embryonic fibroblasts were prepared as described in ref. 17. Immunofluorescence assay with FITC-labeled goat anti-human IgG (Fc), Western blotting, and the plaque assay were performed as described in ref. 23.
Results
Characterization of Transgenic Mice. To assess the antiviral potential of nectin-1 in vivo, we generated six transgenic mouse lines expressing a soluble form of porcine nectin-1 (PHveCIg) consisting of an extracellular domain of porcine nectin-1 and the Fc portion of human IgG1. In these transgenic mice, the PHveCIg gene was under the control of the CAG promoter (Fig. 1 A), which allows an expression in all cell types (26). Southern blot analysis performed on genomic DNA from these mice showed that the transgenic mouse lines harbored different transgene copy numbers (1 copy for line 6, 4 copies for line 22, 20 copies for line 32, 3 copies for line 33, 50 copies for line 37, and 2 copies for line 45) (Table 1). By using a rabbit anti-human nectin-1 antibody (28), Western blot analysis revealed that the transgene was expressed in all of the six transgenic mouse lines (Fig. 1B). As shown in Table 1, PHveCIg concentrations in serum samples from the different mouse lines were different in a large extent ranging from 5.0 ± 1.9 to 1,820.5 ± 188.3 μg/ml.
Table 1. Characteristics of transgenic mouse lines expressing PHveClg.
| Line | Copy no. | PHveClg in serum, μg/ml | Body weight, g | Litter size |
|---|---|---|---|---|
| 6 | 1 | 1,820.5 ± 188.3 | 16.2 ± 1.8a (4) | 8.0 ± 1.9d |
| 22 | 4 | 258.0 ± 100.5 | 18.9 ± 2.3b (3) | 7.4 ± 1.9d |
| 32 | 20 | 742.9 ± 47.9 | 18.3 ± 1.6b,c (8) | 6.0 ± 1.0d |
| 33 | 3 | 1,283.0 ± 370.8 | 17.1 ± 1.2b,c (7) | 7.6 ± 1.7d |
| 37 | 50 | 5.0 ± 1.9 | 17.5 ± 1.6a,b,c (6) | 7.2 ± 1.3d |
| 45 | 2 | 1,180.1 ± 279.9 | 18.2 ± 1.6b,c (8) | 3.8 ± 2.2 |
| C57BL/6 | 0 | 1.2 ± 0.6 | 17.4 ± 1.5a,b,c (8) | 6.2 ± 1.3d |
Copy number was estimated by Southern blot analysis, and the amount of PHveClg in serum was measured by competitive ELISA with at least three transgenic offspring. Shown is the body weight of 8-week-old female mice, with the number of transgenic mice tested shown in parentheses. Litter size was measured for five litters. Means sharing the same superscript letter are not significantly different from each other at threshold P < 0.05.
One major concern with the transgenesis approach is possible adverse side effects generated by the transgene. However, no significant abnormalities in the transgenic mice were observed. They developed normally, and there were no significant differences in body weight between transgenic mice and control mice (Table 1). Moreover, the transgenic mouse lines had a normal capacity to reproduce and lactate, except for line 45 (Table 1). Histological studies with hematoxylin and eosin staining with tissue sections, including main organs, from the transgenic and the nontransgenic littermate mice of each transgenic mouse line at 7 weeks of age were performed to examine abnormalities of phenotypic appearance in the transgenic organs. No difference between the transgenic and the littermate mice was observed, and each tissue section observed was scored as normal for the tissue and the age considered. In a rotarod test (31), the motor coordination and balance of the transgenic mice was not significantly different from that of control mice (data not shown).
Resistance to i.p. Challenge with PRV. To find out whether the transgenic mice expressing PHveCIg were protected against PRV infection, 20 LD50 of PRV strain YS-81 was i.p. inoculated into transgenic mice and their nontransgenic littermates. The survival data (Table 2) demonstrate that the transgenic mice from all lines showed remarkable resistance to PRV infection. Only one transgenic mouse, from line 33, died after infection. In contrast, >90% of their littermates died within 14 days (Table 2). Specific antibodies to PRV were not detected in the surviving i.p. inoculated transgenic mice tested by ELISA (data not shown), indicating that the mice were not infected with PRV. To confirm this resistance, experimental infections were repeated in the French laboratories with 20 LD50 of a different strain of PRV, strain Kojnock. All transgenic mice of lines 22 and 32 that were tested survived, whereas almost all of their littermates died (Table 2). Furthermore, it is known that porcine nectin-1 is a glycoprotein D receptor for HSV-1 (24). To find out whether the transgenic mice expressing PHveCIg were protected against HSV-1 infection, 20 LD50 of HSV-1 strain VR-3 was i.p. inoculated into the transgenic mice of line 6 and their littermates. All transgenic mice survived, and six of seven nontransgenic littermates died within 14 days after HSV-1 infection (Table 2).
Table 2. Survival rates of mice after the challenge with α-herpesviruses.
| Line | Transgenic mice, % | Control littermates, % | Virus | Dose | Route | Location |
|---|---|---|---|---|---|---|
| 6 | 100 (5/5) | 14 (2/14) | PRV YS-81 | 20 LD50 | i.p. | Sapporo |
| 22 | 100 (12/12) | 0 (0/12) | PRV YS-81 | 20 LD50 | i.p. | Sapporo |
| 32 | 100 (10/10) | 14 (1/7) | PRV YS-81 | 20 LD50 | i.p. | Sapporo |
| 33 | 86 (6/7) | 0 (0/9) | PRV YS-81 | 20 LD50 | i.p. | Sapporo |
| 37 | 100 (10/10) | 25 (1/4) | PRV YS-81 | 20 LD50 | i.p. | Sapporo |
| 45 | 100 (6/6) | 8 (1/12) | PRV YS-81 | 20 LD50 | i.p. | Sapporo |
| 22 | 100 (7/7) | 0 (0/1) | PRV Kojnock | 20 LD50 | i.p. | Paris |
| 32 | 100 (7/7) | 14 (1/7) | PRV Kojnock | 20 LD50 | i.p. | Paris |
| 6 | 100 (6/6) | 14 (1/7) | HSV-1 VR-3 | 20 LD50 | i.p. | Sapporo |
| 6 | 50 (9/18) | 0 (0/9) | PRV YS-81 | 10 LD50 | i.n. | Sapporo |
| 22 | 78 (18/23) | 10 (2/21) | PRV YS-81 | 10 LD50 | i.n. | Sapporo |
| 32 | 61 (11/18) | 8 (1/12) | PRV YS-81 | 10 LD50 | i.n. | Sapporo |
| 33 | 65 (11/17) | 12 (1/8) | PRV YS-81 | 10 LD50 | i.n. | Sapporo |
| 37 | 90 (9/10) | 8 (1/12) | PRV YS-81 | 10 LD50 | i.n. | Sapporo |
| 45 | 100 (9/9) | 10 (1/10) | PRV YS-81 | 10 LD50 | i.n. | Sapporo |
| 37 | 100 (12/12) | 10 (1/10) | PRV Chiba-03 | 10 LD50 | i.n. | Sapporo |
Deaths were recorded daily until the termination of the experiments at day 14. Shown are the survival percentages for transgenic mice and their littermates, which were used as controls. The values in parentheses are the ratios of surviving mice to the number of mice tested. Locations where experimental infection were performed are also shown. i.n., intranasal.
Resistance to Intranasal Challenge with PRV. Because PRV usually enters the body in pigs by infection of mucosal epithelium, intranasal challenges with PRV were performed. Ten LD50 of YS-81 strain was intranasaly inoculated into the transgenic mice and their nontransgenic littermates. This challenge was lethal to >90% of all control mice (56 of 62 nontransgenic littermates died), as shown in Table 2. In contrast, the survival data (Table 2) demonstrate that three of the transgenic mouse lines showed remarkable resistance to PRV infection by means of mucosal inoculation (survival rates of 78% (18/23), 90% (9/10), and 100% (9/9) for lines 22, 37, and 45, respectively). In other transgenic mouse lines, a lower but still significant protection was observed: 50% (9/18), 61% (11/18), and 65% (11/17) of the animals from lines 6, 32, and 33 survived, respectively (Table 2). The same intranasal challenge with 10 LD50 of PRV strain Chiba-03, a new field isolate, was performed by using transgenic mice of line 37 and their nontransgenic littermates, with identical results: 100% (12/12) of transgenic mice survived, and 9 of 10 nontransgenic littermates died, as observed for the Japanese reference, strain YS-81 (Table 2).
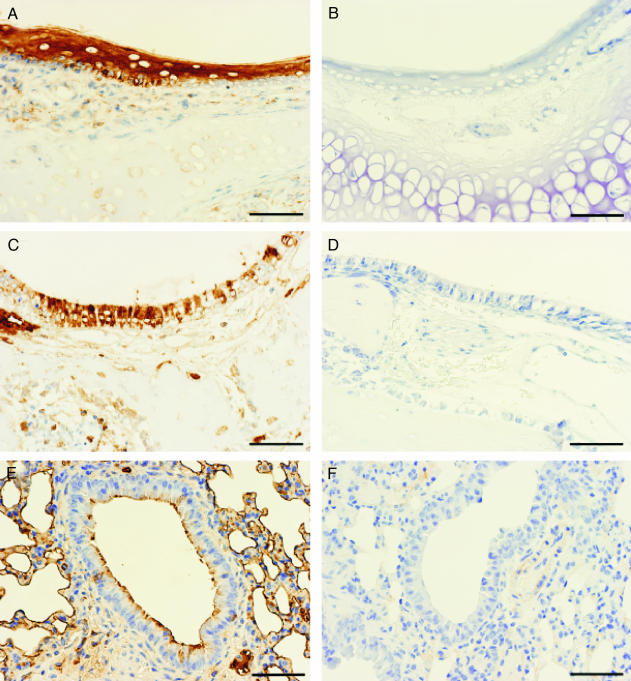
Expression of PHveCIg in nasal stratified squamous epithelium (Fig. 2A) and pseudostratified columnar respiratory epithelium (Fig. 2C) of transgenic mice (line 22) and control littermates was examined by immunohistochemical staining by using anti-human IgG antibodies. Specific staining was noted on the surface of alveolar and bronchiolar epithelium (Fig. 2E). PHveCIg staining tended to be located in the periphery of neurons in brainstem and trigeminal ganglia (data not shown). No staining was detected in any organs from nontransgenic littermates (Fig. 2).
Fig. 2.
Immunohistochemical staining of nasal mucosa and lung with anti-human IgG antibody of transgenic (A, C, and E) and nontransgenic (B, D, and F) littermate mice of line 22. (Bar, 50 μm.) (A and B) Nasal squamous epithelium. (C and D) Nasal respiratory epithelium. (E and F) Lung alveolar and bronchiolar epitheliums.
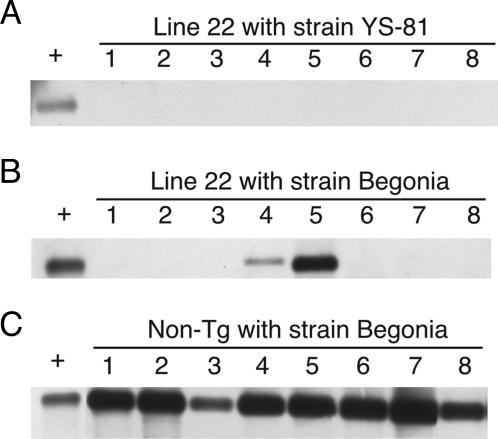
To test whether mice were latently infected with PRV, PCR analysis was performed to detect PRV DNA in the trigeminal ganglia of the surviving mice. Because lethal challenge with the virulent strain YS-81 does not lead to latency in control mice but to death, positive control for this test was provided by detecting PRV DNA in the trigeminal ganglia of control mice infected with the attenuated strain Begonia [250 plaque-forming units (the same no. of plaque-forming units as 10 LD50 of YS-81 strain)]. As shown in Fig. 3A, PRV DNA was not detected in the surviving transgenic mice (line 22) after intranasal inoculation with 10 LD50 of YS-81 strain. However, PRV DNA was detected in all of eight nontransgenic controls after intranasal inoculation with Begonia strain (Fig. 3C). Furthermore, no specific antibodies to PRV were found in the surviving transgenic mice by using an ELISA test (data not shown).
Fig. 3.
Detection of PRV DNA in the trigeminal ganglia of mice after intranasal inoculation of PRV. Shown are transgenic mice (line 22) inoculated with PRV strain YS-81 (A) or Begonia (B) and nontransgenic (non-Tg) littermate inoculated with strain Begonia (C). The positive control (+) is a DNA extracted from PRV-infected Vero cells. The number of each lane corresponds to the mouse number tested in each experiment.
We also checked how PHveCIg expression would protect against latent infections observed with the attenuated PRV strain. Neither transgenic or nontransgenic littermate mice showed any symptoms or died, because Begonia strain is attenuated. PRV DNA was detected in two of eight transgenic mice inoculated with Begonia strain (Fig. 3B) and in all nontransgenic littermates (Fig. 3C). The 25% of latently infected transgenic animals is about the same as the 22% of transgenic animals that succumbed to the virulent strain (Table 2; intranasal challenge with 10 LD50 PRV strain YS-81 for transgenic mouse line 22). Specific antibodies to PRV were detected in the latently infected transgenic and nontransgenic littermate mice by using an ELISA test (data not shown). Taken together, these findings demonstrate that transgenic mice that survived after i.p. and intranasal challenge with YS-81 virulent strain were not infected, suggesting that expression of PHveCIg protects transgenic mice against PRV primary infection and not only against disease symptoms.
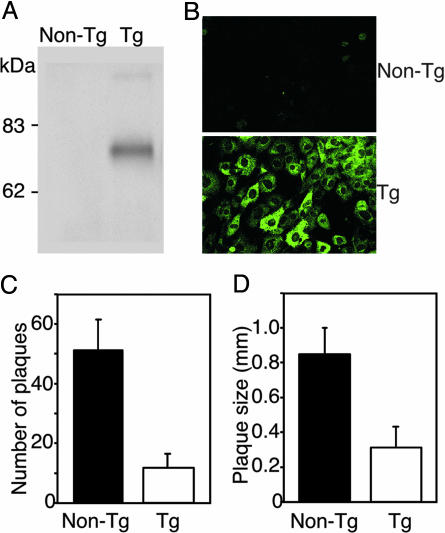
Inhibition of PRV Infection in Cultured Cells. To assess whether cells isolated from the transgenic mice would reflect the in vivo resistance to PRV infection, primary cultured embryonic fibroblasts from transgenic and control mice were infected with PRV. Embryonic fibroblasts were prepared from embryos of heterozygous transgenic (line 22) and nontransgenic parents. PHveCIg expression in the fibroblasts from the transgenic mouse was confirmed by Western blot analysis and indirect immunofluorescence assay, but was not found in the control fibroblasts (Fig. 4 A and B). Immunofluorescent staining was mainly observed in the cytoplasm (Fig. 4B) but was slightly detectable on the cell surface (data not shown). The transgenic and nontransgenic fibroblasts were washed twice with DMEM just before the viral inoculation to remove secreted PHveCIg in the medium and infected with PRV. In the dishes containing the transgenic fibroblasts, the number of plaques was markedly lower than in those containing the control fibroblasts 2 days after infection (Fig. 4C). The plaque sizes were also definitely smaller than those observed in the control fibroblasts (Fig. 4D). These results confirm that isolated cells from transgenic animals expressing PHveCIg are less sensitive to PRV infection than corresponding wild-type cells, even when the secreted PHveCIg molecules, which have neutralizing activity, were washed away.
Fig. 4.
Resistance to PRV infection of embryonic fibroblasts prepared from transgenic (Tg) and nontransgenic mice. (A) Western blot analysis of cell extracts. (B) Immunofluorescent staining of fibroblasts. (C) Number of plaques of infected fibroblasts. (D) Plaque size of infected fibroblasts.
Discussion
The animals described expressing PHveCIg showed a high resistance to PRV challenge by means of both i.p. and intranasal routes. It is especially noteworthy that protection against PRV entry in the sites of primary infection was observed in the transgenic mice after intranasal inoculation of PRV. The transgenic embryonic fibroblasts expressing PHveCIg showed marked resistance to PRV infection. PRV DNA in the trigeminal ganglia was not detected in the surviving transgenic mice inoculated with PRV strain YS-81 by means of the intranasal route. In the transgenic nasal mucosa and respiratory tract, expression of PHveCIg was confirmed by immunohistochemical staining. These findings suggest that the epithelial cells of the nasal mucosa and respiratory tract expressing PHveCIg are also resistant to PRV infection. Transgenic mouse line 37, expressing only 5 μg/ml PHveCIg in the serum, showed significant resistance to PRV infection by means of intranasal route, indicating that such a low level of expression of the transgene is sufficient to inhibit the virus entry into cells and that circulating PHveCIg may not play an important role in the resistance to PRV infection. There are several possible effects of PHveCIg on the suppression of virus replication, as seen in transgenic primary fibroblasts. Firstly, cell-bound PHveCIg may inhibit the virus entry into fibroblasts as described previously (23). Secondly, intracellular PHveCIg may bind to newly synthesized glycoprotein D and inhibit secondary infections as intrabodies (32, 33). Thirdly, secreted PHveCIg may inhibit secondary infections of the fibroblasts, neutralizing free virus released in the first round of infection. Fourthly, cell-bound and intracellular PHveCIg may inhibit secondary infection in the fibroblasts mediated by cell-to-cell spread. These possible effects of PHveCIg, expressed in the nasal mucosa and respiratory tract, may account for the protection against intranasal PRV infection.
Nectin-1 is a known component of intercellular junctions and plays a major role in several cell functions, such as morphogenesis, differentiation, proliferation, and migration, and in the molecular mechanisms that underlie junctional disorders associated diseases. Thus, the issue of possible adverse effects needs to be addressed carefully. However, detailed histological examination of main tissues from 7-week-old transgenic mice did not reveal any differences in cell morphology induced by PHveCIg expression. Functional neuronal disorders were not suggested because no differences were observed between transgenic and control mice in the rotarod test. Transgenic mice developed normally with normal body weights and litter sizes. The smaller litter size in line 45 and the slightly lower body weights in line 6 may be due to insertional mutation of the transgene, because similar findings were not observed in other lines. Nonetheless, these effects were not correlated to higher expression levels. Primary structure of nectin-1 is well conserved across mammalian species, including mouse and pig, in which they share 95% of amino acid residues in common regarding their extracellular domain (24). In a different context, a soluble form of murine HVEM expressed in transgenic mice enhanced resistance to HSV-1 infection (17). In those transgenic mice, no gross abnormality was observed, and overexpression of a homologous form of other α-herpesvirus receptors did not produce side effects expected on the immune system. HVEM is a member of the tumor necrosis factor receptor family (12) and plays an important role on the immune systems. Taken together, these data suggest that overexpressing a soluble form of homologous nectin-1, such as PHveCIg described here in pigs, may not raise host safety issues.
Farm animals that are effectively protected against infectious disease through the use of transgenes are still concepts only, although several cellular and molecular mechanisms can theoretically be used to block the infection of cells or living organisms. Although the possibility of disease resistance in transgenic animals has been envisioned for many years, few, if any, convincing models have been validated by using in vivo challenges. Transgenic mice secreting coronavirus neutralizing antibodies in their milk were reported (34-36). An example of transgene-mediated lactogenic immunity in vivo was provided by this approach (36). A recent review (37) suggests a coronavirus receptor knockout strategy but also mentions potential side effects. The same report underlines the wide range of viral pathogens that could be targeted by using RNA interference-mediated knockdown of viral genes. However, in vivo demonstration of efficiency for any of these strategies is still to come. Here, we showed that a transgene could efficiently protect animals against a major viral infectious disease by expressing a soluble form of a viral receptor.
Although animal breeding is continuously demonstrating its ability to improve production traits, it does little to improve animal health and disease resistance. The present work proves the efficiency of a previously envisaged alternative strategy by building on acquired knowledge of infection pathogeny and hitting on critical components of known host pathogen interactions to achieve a practical genetic immunization.
Acknowledgments
We thank G. H. Cohen and R. J. Eisenberg (University of Pennsylvania, Philadelphia) for anti-human HveC rabbit serum, R. S. B. Milne (University of Pennsylvania, Philadelphia) for pRM361, J. Miyazaki (Osaka University, Suita, Japan) for pCXN2, T. Suzutani (Fukushima Medical University, Fukushima, Japan) for HSV-1 strain VR-3, Y. Shimizu and S. Katayama (Kyoto Biken Laboratories, Uji, Japan) for PRV strain Chiba-03, and F. Van Otegem and M. Chopping for manuscript preparation. This work was supported by Grant-in-Aid for Scientific Research (B)(2) from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Author contributions: E.O. designed research; E.O., K.A., S.T., Y.T., S.Y., Y.W., and M.A. performed research; M.I. and T.U. contributed new reagents/analytical tools; E.O., P.C., L.-M.H., and M.E. analyzed data; and E.O., P.C., L.-M.H., and M.E. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Hve, herpesvirus entry mediator; HVEM, HveA; PRV, pseudorabies virus; HSV-1, herpes simplex virus 1; LAT, latency-associated transcript.
References
- 1.Baltimore, D. (1988) Nature 335, 395-396. [DOI] [PubMed] [Google Scholar]
- 2.Smith, C. A. & DeLuca, N. A. (1992) Virology 191, 581-588. [DOI] [PubMed] [Google Scholar]
- 3.Ono, E., Tasaki, T., Kobayashi, T., Taharaguchi, S., Nikami, H., Miyoshi, I., Kasai, N., Arikawa, J., Kida, H. & Shimizu, Y. (1999) Virology 262, 72-78. [DOI] [PubMed] [Google Scholar]
- 4.Herold, B. C., Visalli, R. J., Susmarski, N., Brandt, C. R. & Spear, P. G. (1994) J. Gen. Virol. 75, 1211-1222. [DOI] [PubMed] [Google Scholar]
- 5.Mettenleiter, T. C., Zsak, L., Zuckermann, F., Sugg, N., Kern, H. & Ben-Porat, T. (1990) J. Virol. 64, 278-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shieh, M.-T., WuDunn, D., Montgomery, R. I., Esko, J. D. & Spear, P. G. (1992) J. Cell Biol. 116, 1273-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.WuDunn, D. & Spear, P. G. (1989) J. Virol. 63, 52-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai, W., Gu, B. & Person, S. (1988) J. Virol. 62, 2596-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forrester, A., Farrell, H., Wilkinson, G., Kaye, J., Davis-Poynter, N. & Minson, T. (1992) J. Virol. 66, 341-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ligas, M. W. & Johnson, D. C. (1988) J. Virol. 62, 1486-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roop, C., Hutchinson, L. & Johnson, D. C. (1993) J. Virol. 67, 2285-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Montgomery, R. I., Warner, M. S., Lum, B. J. & Spear, P. G. (1996) Cell 87, 427-436. [DOI] [PubMed] [Google Scholar]
- 13.Cocchi, F., Menotti, L., Mirandola, P., Lopez, M. & Campadelli-Fiume, G. (1998) J. Virol. 72, 9992-10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geraghty, R. J., Krummenacher, C., Eisenberg, R. J., Cohen, G. H. & Spear, P. G. (1998) Science 280, 1618-1620. [DOI] [PubMed] [Google Scholar]
- 15.Shukla, D., Liu, J., Blaiklock, P., Shworak, N. W., Bai, X., Esko, J. D., Cohen, G. H., Eisenberg, R. J., Rosenberg, R. D. & Spear, P. G. (1999) Cell 99, 13-22. [DOI] [PubMed] [Google Scholar]
- 16.Warner, M. S., Martinez, W., Geraghty, R. J., Montgomery, R. I., Whitbeck, J. C., Xu, R., Eisenberg, R. J., Cohen, G. H. & Spear, P. G. (1998) Virology 246, 179-189. [DOI] [PubMed] [Google Scholar]
- 17.Ono, E., Yoshino, S., Amagai, K., Taharaguchi, S., Kimura, C., Morimoto, J., Inobe, M., Uenishi, T. & Uede, T. (2004) Virology 320, 267-275. [DOI] [PubMed] [Google Scholar]
- 18.Eberle, F., Dubreuil, P., Mattei, M.-G., Devilard, E. & Lopez, M. (1995) Gene 159, 267-272. [DOI] [PubMed] [Google Scholar]
- 19.Mendelsohn, C. L., Wimmer, E. & Racaniello, V. R. (1989) Cell 56, 855-865. [DOI] [PubMed] [Google Scholar]
- 20.Lopez, M., Eberle, F., Mattei, M.-G., Gabert, J., Birg, F., Bardin, F., Maroc, C. & Dubreuil, P. (1995) Gene 155, 261-265. [DOI] [PubMed] [Google Scholar]
- 21.Reymond, N., Borg, J., Lecoco, E., Adelaide, J., Campadelli-Fiume, G., Dubreuil, P. & Lopez, M. (2000) Gene 255, 347-355. [DOI] [PubMed] [Google Scholar]
- 22.Satoh-Horikawa, K., Nakanishi, H., Takahashi, K., Miyahara, M., Ikeda, W., Yokoyama, S., Peng, Y. F., Yamanishi, K. & Takai, Y. (2000) J. Biol. Chem. 275, 10291-10299. [DOI] [PubMed] [Google Scholar]
- 23.Ono, E., Amagai, K., Yoshino, S., Taharaguchi, S., Inobe, M. & Uede, T. (2004) J. Gen. Virol. 85, 173-178. [DOI] [PubMed] [Google Scholar]
- 24.Milne, R. S. B., Connolly, S. A., Krummenacher, C., Eisenberg, R. J. & Cohen, G. H. (2001) Virology 281, 315-328. [DOI] [PubMed] [Google Scholar]
- 25.Nakagawa, I., Murakami, M., Ijima, K., Chikuma, S., Saitoh, I., Kanegae, Y., Ishikura, H., Yoshiki, T., Okamoto, H., Kitabatake, A. & Uede, T. (1998) Hum. Gene Ther. 9, 1739-1745. [DOI] [PubMed] [Google Scholar]
- 26.Niwa, H., Yamamura, K. & Miyazaki, J. (1991) Gene 108, 193-200. [DOI] [PubMed] [Google Scholar]
- 27.Hogan, B., Constantini, F. & Kacy, E. (1986) Manipulating the Mouse Embryo: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 28.Krummenacher, C., Nicola, A. V., Whitbeck, J. C., Lou, H., Hou, W., Lambris, J. D., Geraghty, R. J., Spear, P. G., Cohen, G. H. & Eisenberg, R. J. (1998) J. Virol. 72, 7064-7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taharaguchi, S., Kon, Y., Yoshino, S. & Ono, E. (2003) Virology 307, 243-254. [DOI] [PubMed] [Google Scholar]
- 30.Ono, E., Sakoda, Y., Taharaguchi, S., Watanabe, S., Tonomura, N., Kida, H. & Shimizu, Y. (1995) Virology 210, 128-140. [DOI] [PubMed] [Google Scholar]
- 31.Jones, B. J. & Roberts, D. J. (1968) Naunyn-Schmiedebergs Arch. Exp. Pathol. Pharmakol. 259, 211. [DOI] [PubMed] [Google Scholar]
- 32.Marasco, W. A., Haseltine, W. A. & Chen, S.-Y. (1993) Proc. Natl. Acad. Sci. USA 90, 7889-7893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen, S.-Y., Khouri, Y., Bagley, J. & Marasco, W. A. (1994) Proc. Natl. Acad. Sci. USA 91, 5932-5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Castilla, J., Pintado, B., Sola, I., Sanchez-Morgado, J. M. & Enjuanes, L. (1998) Nat. Biotechnol. 16, 349-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sola, I., Castilla, J., Pintado, B., Sánchez-Morgado, J. M., Whitelaw, C. B. A., Clark, A. J. & Enjuanes, L. (1998) J. Virol. 72, 3762-3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kolb, A. F., Pewe, L., Webster, J., Perlman, S., Whitelaw, C. B. A. & Siddell, S. G. (2001) J. Virol. 75, 2803-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clark, J. & Whitelaw, B. (2003) Nat. Rev. Genet. 4, 825-834. [DOI] [PMC free article] [PubMed] [Google Scholar]