ABSTRACT
Oxalic acid is the simplest of the dicarboxylic acids. In addition to its role in biological and metabolic processes, oxalate has been implicated in biotic and abiotic stresses. Being a strong chelator of Al, oxalate also has pivotal role in Al resistance mechanisms. However, we demonstrated that cytoplasmic oxalate accumulation is a critical event leading to root growth inhibition under Al stress. Transcriptome analysis from three crop plants identified Acyl Activating Enzyme3 (AAE3) genes to be upregulated by Al stress. These AAE3 proteins display high sequence identity to known AAE3 proteins, suggesting they are oxalyl-CoA synthetases specifically involved in oxalate degradation. However, phylogenetic analysis revealed divergence of AAE3 between monocots and dicots, pointing to the necessity for functional characterization of AAE3 proteins from other plant species with respect to Al stress.
KEYWORDS: Acyl activating enzyme, aluminum toxicity, acid soil, organic acids, oxalate, transcriptome
Aluminum toxicity constraints severely crop production on acid soils which comprise approximately 50% of world potentially arable lands.1 However, many native plants and some crops thrive on these acidic soils, implicating that they have evolved sophisticated mechanisms to deal with Al toxicity. In general, plant Al resistance mechanisms can be classified into external exclusion mechanism or internal tolerance mechanism.2,3 The external exclusion mechanisms are those that prevent Al from entering the root apex (both apoplasm and symplasm), and internal tolerance mechanisms refer to those that detoxify and sequester Al once it enters the plant.2,3 Having strong binding capacity to Al and being ubiquitous in plant cells, organic acid anions, mainly citrate, malate and oxalate, are key component of both mechanisms.3,4,5
Before the cloning and characterization of genes encoding transporters that facilitate malate and citrate exudation, many studies have focused on improvement of plant Al resistance through genetic manipulation of malate and citrate biosynthesis.6 Although some reports remain uncertain,7 others believe that increasing OA content will improve Al resistance either through increased exudation or internal detoxification.8,9,10,11,12 However, the information on the relationship between oxalate content and Al resistance is still lacking.
Intuitively, increasing oxalate content in root cells will be beneficial for Al resistance. However, we previously demonstrated that oxalate accumulation is harmful for root growth in response to Al stress in rice bean (Vigna umbellata).13 We further identified a gene VuAAE3 (Vigna umbellata Acyl Activating Enzyme3) that converts oxalate to oxalyl-CoA, thereby preventing oxalate toxicity induced by Al stress.13 Although in silico analysis revealed that AAE3 proteins seem to be conserved among plant species, more evidence is required to support the viewpoint that AAE3 proteins-dependent Al resistance mechanism is conserved among plant species. In the present study, we provided more compelling evidence that regulating of cytoplasmic oxalate homeostasis by AAE3 proteins provides additional layer of Al resistance mechanisms in plants.
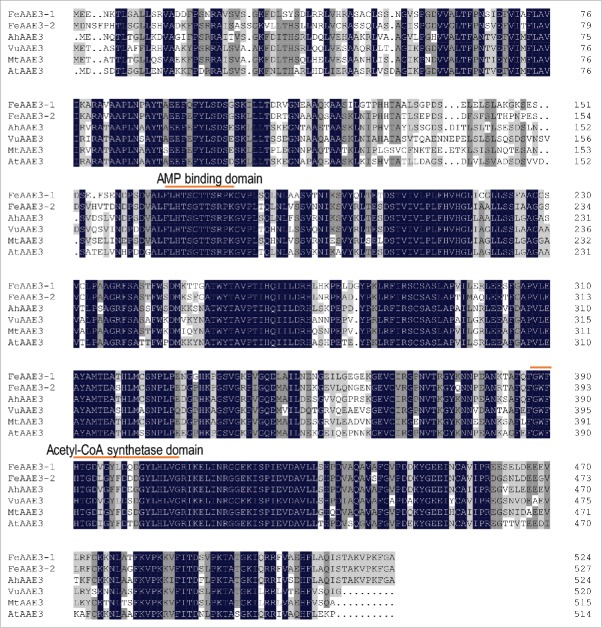
We have carried out transcriptome analysis of Al-responsive genes from crops including rice bean, common buckwheat (Fagopyrum esculentum), and amaranth (Amaranthus hypochondriacus) (unpublished data). From upregulated genes, one of consistently identified were AAE3 genes (Fig. 1). In a previous study, we found that VuAAE3 transcripts were frequently identified in the upregulated suppression subtractive hybridization libraries both under low and high Al stress conditions.14 To date, AAE3 proteins from rice bean, Medicago truncatula, and Arabidopsis (Arabidopsis thaliana) have been characterized as oxalyl-CoA synthetases.13,15,16 Here, amino acid sequence alignment revealed that AAE3 proteins from common buckwheat and amaranth display high identity with known AAE3 proteins, all having a conserved AMP binding domain and acetyl-CoA synthetase domain (Fig. 2). Although the biochemical and molecular biological characterization of FeAAE3 and AhAAE3 proteins has to be investigated, it is reasonable to suggest that they also function as oxalyl-CoA synthetases. Given that AAE3 protein is characterized by having activities specific to oxalate,13,15,16 the upregulation of AAE3 genes suggests that oxalate accumulates under Al stress.
Figure 1.
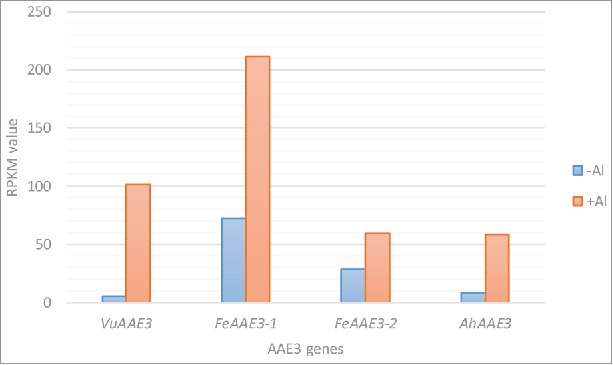
AAE3 gene transcription in response to Al stress in rice bean (VuAAE3), buckwheat (FeAAE3-1 and FeAAE3-2), and amaranth (AhAAE3). The data was based on the transcriptome analysis of roots under Al stress (25 µM, 6 h for rice bean, 20 µM, 6 h for buckwheat, and 10 µM, 6 h for amaranth). The expression level expressed as RPKM (the Reads Per kb Million reads) value.
Figure 2.
Amino acid sequence alignment of AAE3 proteins from rice bean (VuAAE3), Arabidopsis (AtAAE3), Medicago truncatula (MtAAE3), Amaranthus hypochondriacus (AhAAE3), and Fagopyrum esculentum (FeAAE3-1 and FeAAE3-2). The conserved AMP binding domain and acetyl-CoA synthetase domain are indicated.
It is worth to note that common buckwheat and amaranth plants are oxalate accumulators.17 However, both plant species have evolved AAE3-dependent regulation of cytoplasmic oxalate content under Al stress, suggesting that cytoplasmic oxalate content must be tightly controlled. Clearly, free oxalate in cytosol is toxic, because it represents a strong acid, reductant, and chelator.18 In line with our expectations, transgenic Arabidopsis plants overexpressing a bacterial oxalic acid biosynthetic enzyme gene displayed not only significant increase in oxalate content, but also a reduction in plant stature as well as a pronounced delay in bolting and seed set.19
In addition to AAE3-dependent degradation of oxalate, oxalate can be oxidized into CO2 and H2O2 by oxalate oxidase that belongs to germin protein family. However, it seems that oxalate oxidase is only present in monocots. Thus, question arises as to whether AAE3 proteins from monocots also play important role in regulating cytoplasmic oxalate homeostasis, because phylogenetic analysis clearly indicated that AAE3 proteins are evolutionally separated between dicots and monocots (Fig. 3). In the future, it would be necessary to characterize the role of AAE3 proteins from buckwheat and amaranth as well as those from monocots in Al resistance.
Figure 3.
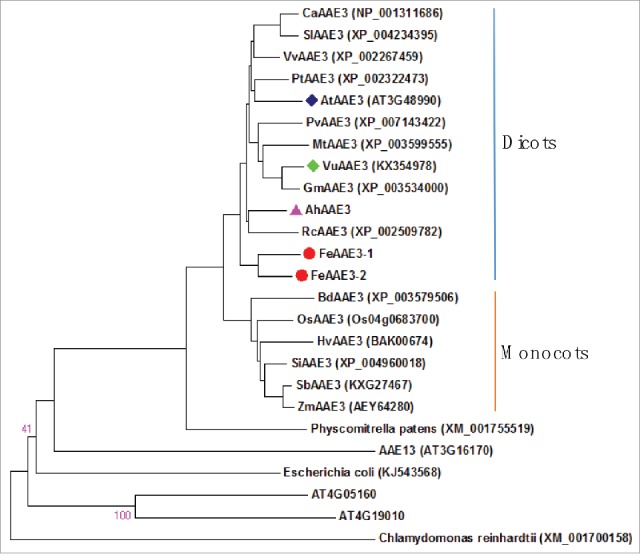
Evolutional relationship of AAE3 proteins. AAE3 proteins are derived from dicots: Capsicum annuum (CaAAE3), Solanum lycopersicum (SlAAE3), Vitis vinifera (VvAAE3), Amaranthus hypochondriacus (AhAAE3), Populus trichocarpa (PtAAE3), Arabidopsis thaliana (AtAAE3, AtAAE13, At4g05160, and At4g19010), Phaseolus vulgaris (PvAAE3), Ricinus communis (RcAAE3), Medicago truncatula (MtAAE3), Vigna umbellate (VuAAE3), Glycine max (GmAAE3), Fagopyrum esculentum (FeAAE3-1 and FeAAE3-2); monocots: Brachypodium distachyon (BdAAE3), Oryza sativa (OsAAE3), Setaria italic (0SiAAE3), Hordeum vulgare (HvAAE3), Sorghum bicolor (SbAAE3), Zea mays (ZmAAE3); Embrophyte (Physcomitrella patens); Bacteria (Escherichia coli); Chlorophyte (Chlamydomonas reinhardtii).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by grant from the 973 Project (2014CB441002), the Natural Science Foundation of China (31572193; 31222049; 31601765), and China Scholarship Council.
References
- 1.Kochian LV. Cellular mechanisms of aluminium toxicity and resistance in plants. Annu Rev Plant Physiol Plant Mol Biol 1995; 46:237-60; http://dx.doi.org/ 10.1146/annurev.pp.46.060195.001321 [DOI] [Google Scholar]
- 2.Taylor GJ. Current views of the aluminium stress response: the physiological basis of tolerance. Curr Top Plant Biochem Physiol 1991; 10:57-93 [Google Scholar]
- 3.Kochian LV, Hoekenga OA, Piñeros MA. How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu Rev Plant Biol 2004; 55:459-93; PMID:15377228; http://dx.doi.org/ 10.1146/annurev.arplant.55.031903.141655 [DOI] [PubMed] [Google Scholar]
- 4.Ryan PR, Delhaize E, Jones DL. Function and mechanism of organic anion exudation from plant roots. Annu Rev Plant Physiol Plant Mol Biol 2001; 52:527-60; PMID:11337408; http://dx.doi.org/ 10.1146/annurev.arplant.52.1.527 [DOI] [PubMed] [Google Scholar]
- 5.Kochian LV, Piñeros MA, Liu J, Magalhaes JV. Plant adaptation to acid soils: the molecular basis for crop aluminum resistance. Annu Rev Plant Biol 2015; 66:571-98; PMID:25621514; http://dx.doi.org/ 10.1146/annurev-arplant-043014-114822 [DOI] [PubMed] [Google Scholar]
- 6.Ryan PR, Tyerman SD, Sasaki T, Furuichi T, Yamamoto Y, Zhang WH, Delhaize E. The identification of aluminium-resistance genes provides opportunities for enhancing crop production on acid soils. J Exp Bot 2011; 62:9-20; PMID:20847099; http://dx.doi.org/ 10.1093/jxb/erq272 [DOI] [PubMed] [Google Scholar]
- 7.Delhaize E, Hebb DM, Ryan PR. Expression of a Pseudomonas aeruginosa citrate synthase gene in tobacco is not associated with either enhanced citrate accumulation or efflux. Plant Physiol 2001; 125:2059-67; PMID:11299385; http://dx.doi.org/ 10.1104/pp.125.4.2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de la Fuente JM, Ramirez-Rodriguez V, Cabrera-Ponce JL, Herrera-Estrella L. Aluminum tolerance in transgenic plants by alteration of citrate synthesis. Science 1997; 276:1566-8; PMID:9171061; http://dx.doi.org/ 10.1126/science.276.5318.1566 [DOI] [PubMed] [Google Scholar]
- 9.López-Bucio J, Martínez de la Vega O, Guevara-García A, Herrera-Estrella L. Enhanced phosphorus uptake in transgenic tobacco plants that overproduce citrate. Nat Biotech 2000; 18:450-3; PMID:10748530; http://dx.doi.org/ 10.1038/74531 [DOI] [PubMed] [Google Scholar]
- 10.Tesfaye M, Temple SJ, Allan DL, Vance CP, Samac DA. Overexpression of malate dehydrogenase in transgenic alfalfa enhances organic acid synthesis and confers tolerance to aluminum. Plant Physiol 2001; 127:1836-44; PMID:11743127; http://dx.doi.org/ 10.1104/pp.010376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anoop VM, Basu U, McCammon MT, McAlister-Henn L, Taylor GJ. Modulation of citrate metabolism alters aluminum tolerance in yeast and transgenic canola overexpressing a mitochondrial citrate synthase. Plant Physiol 2003; 132:2205-17; PMID:12913175; http://dx.doi.org/ 10.1104/pp.103.023903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng W, Luo K, Li Z, Yang Y, Hu N, Wu Y. Overexpression of Citrus junos mitochondrial citrate synthase gene in Nicotiana benthamiana confers aluminum tolerance. Planta 2009; 230:355-65; PMID:19466450; http://dx.doi.org/ 10.1007/s00425-009-0945-z [DOI] [PubMed] [Google Scholar]
- 13.Lou HQ, Fan W, Xu JM, Gong YL, Jin JF, Chen WW, Liu LY, Hai MR, Yang JL, Zheng SJ. An oxalyl-CoA synthetase is involved in oxalate degradation and aluminum tolerance. Plant Physiol 2016; 172:1679-90; PMID:27650448; http://dx.doi.org/ 10.1104/pp.16.01106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan W, Lou HQ, Gong YL, Liu MY, Wang ZQ, Yang JL, Zheng SJ. Identification of early Al-responsive genes in rice bean (Vigna umbellata) roots provides new clues to molecular mechanisms of Al toxicity and tolerance. Plant Cell Environ 2014; 37:1586-97; PMID:24372448; http://dx.doi.org/ 10.1111/pce.12258 [DOI] [PubMed] [Google Scholar]
- 15.Foster J, Kim HU, Nakata PA, Browse J. A previously unknown oxalyl-CoA synthetase is important for oxalate catabolism in Arabidopsis. Plant Cell 2012; 24:1217-29; PMID:22447686; http://dx.doi.org/ 10.1105/tpc.112.096032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foster J, Luo B, Nakata PA. An oxalyl-CoA dependent pathway of oxalate catabolism plays a role in regulating calcium oxalate crystal accumulation and defending against oxalate-secreting phytopathogens in Medicago trungatula. PLoS One 2016; 11:e0149850; PMID:26900946; http://dx.doi.org/ 10.1371/journal.pone.0149850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang JL, Zhang L, Zheng SJ. Aluminum-activated oxalate secretion does not associate with internal content among some oxalate accumulators. J Integr Plant Biol 2008; 50:1103-11; PMID:18924279; http://dx.doi.org/ 10.1111/j.1744-7909.2008.00687.x [DOI] [PubMed] [Google Scholar]
- 18.Libert B, Franceschi VR. Oxalate in crop plants. J Agric Food Chem 1987; 35:926-38; http://dx.doi.org/ 10.1021/jf00078a019 [DOI] [Google Scholar]
- 19.Nakata PA. An assessment of engineered calcium oxalate crystal formation on plant growth and development as a step toward evaluating its use to enhance plant defense. PLoS One 2015; 10:e0141982; PMID:26517544; http://dx.doi.org/ 10.1371/journal.pone.0141982 [DOI] [PMC free article] [PubMed] [Google Scholar]