ABSTRACT
The architecture determining grain production in rice is mainly affected by factors such as lamina angle, tillering, plant height and panicle morphology. In particular, leaf angle, the degree of bending between the leaf blade and leaf sheath directly affects crop architecture and grain yields. Balancing activities between the 2 antagonistic groups of proteins, atypical helix-loop-helix (HLH) and basic HLH (bHLH) proteins have been regarded as one of the major molecular machineries regulating lamina angles through the control of cell elongation in the lamina joint of rice. Recently, formation of a complex consisting of atypical HLH protein, OsBUL1, a KxDL motif-containing protein, LO9–177 and a bHLH protein, OsBC1 has been reported to play a positive role in lamina inclination of rice unraveling a novel layer of cell elongation control in rice lamina joint. Here, we demonstrate a trimeric complex formation in rice cells by a combination of BiFC and FRET-FLIM assays.
KEYWORDS: Bimolecular florescence complementation (BiFC), fluorescence resonance energy transfer (FRET)-fluorescence lifetime imaging (FLIM), leaf angle, rice architecture, trimeric protein complex
Rice is one of the most important food crops and feeds more than half of the world population.1 Plant architecture, a complex of the key agronomic traits that determines grain yield, is a primary target of artificial selection for rice domestication. Yield-related plant architecture includes tillering pattern, plant height, leaf angle and so on.1,2 ‘The Green Revolution’ is a successful example of developing high-yield rice and wheat with shorter and sturdier stems. Researchers have tried to increase yield by dense planting plants with erect leaves.2,3 In rice, erect leaves have a higher leaf area index that increases photosynthetic carbon assimilation rates through increased light capture and nitrogen use efficiency.2,3 Especially, the angle of rice lamina inclination is an important trait for the determination of rice architecture. A rice leaf is composed of a leaf sheath, leaf blade, and a lamina joint (area between leaf blade and leaf sheath, also called the collar) with auricles and ligules.4
Previously, it was shown that brassinosteroid (BR) is an important phytohormone for controlling lamina inclination implicated in rice architecture and grain yield.5,6,7,8 In particular, controlling the balance of activities between the 2 groups of antagonistic proteins, atypical helix-loop-helix (HLH) and basic HLH (bHLH) proteins has been regarded as a key step in determination of rice lamina inclination at the molecular level. BRASSINOSTEROID UPREGULATED1 (BU1),9 INCREASED LAMINA INCLINATION1 (ILI1)10 and Oryza sativa BU1-like1 (OsBUL1)11 genes upregulated by brassinolide (BL) treatment encode rice atypical HLH proteins and overexpression of those genes caused increased lamina inclination with enlarged grain size whereas ectopic expression of ILI1 BINDING bHLH1 (OsIBH1)10 encoding a bHLH protein produced erect leaves. Of note, reduction of OsIBH1 transcript levels by BL treatment was also reported. Thus, BR regulation of lamina inclination is likely due to the antagonizing HLH-bHLH transcription factors acting in the BR signaling pathways.10,12 Moreover, in Arabidopsis, a similar observation was also reported: PACLOBUTRAZOL RESISTANCE1 (PRE1),13 KIDARI14 and ACTIVATION-TAGGED Bri1-SUPPRESSOR1 (ATBS1)15 encode atypical HLH proteins and Arabidopsis IBH110 and ATBS1-INTERACTING FACTORs (AIFs)15,16 are members of the bHLH family. However, it was recently reported that the bHLH domain of IBH1 was diverged from that of typical bHLH proteins since it lacks amino acids essential for binding to the E-box and G-box in the basic motif.17 Thus, a triantagonistic model through the balance of PRE1, IBH1 and newly identified IBH1-interacting bHLH proteins, ACTIVATORS FOR CELL ELONGATION (ACEs) has been suggested for cell elongation regulation in Arabidopsis.17
In this system, ACE bHLH transcription factors activate the expression of enzyme genes for cell elongation by interacting with their promoter regions. IBH1 negatively regulates cell elongation by interacting with the ACEs thus interfering with their DNA binding. PRE1 interacts with IBH1 and counteracts the ability of IBH1 to affect ACEs. Thus, PRE1 restores the transcriptional activity of ACEs, resulting in cell elongation.
Even though it is not yet obvious whether the triantagonistic model can be applied to other plant species, recently a novel layer of cell elongation regulation through the HLH-bHLH system has been reported in rice.11 A trimeric complex composed of OsBUL1, LO9–177 and OsBUL1 COMPLEX1 (OsBC1) was shown to positively affect lamina inclination through cell elongation. For the complex formation, an atypical HLH protein, OsBUL1 interacted with a typical bHLH transcription factor, OsBC1 only in the presence of LO9–177, a KxDL motif-containing protein that is able to interact directly with OsBUL1 and OsBC1.11,18
Since the trimeric complex formation was shown previously by in vitro pull-down assays and the yeast system, the assembly of the trimeric complex in rice cells was examined here by a combination of bimolecular florescence complementation (BiFC) and fluorescence resonance energy transfer (FRET)-fluorescence lifetime imaging (FLIM) assays.19 Each N-terminal and C-terminal half of the yellow florescent protein (YFP)20 was fused to OsBUL1 and LO9–177, respectively for BiFC, and OsBC1 was fused to the cyan florescent protein (CFP).21 Those 3 constructs were introduced into rice protoplasts simultaneously.22 A plasmid for the CFP:OsMADS34 fusion protein was also introduced into rice protoplasts together with YFPn:OsBUL1 and YFPc:LO9–177 plasmids as a negative control for FRET-FLIM. OsMADS34 is a rice MADS-domain transcription factor that interacts with neither OsBUL1 nor LO9–177 in yeast systems (data not shown).23
Of note, we could observe the YFP signals preferentially from the nucleus when we introduced YFPn:OsBUL1 and YFPc:LO9–177 together with CFP:OsBC1, which is distinct from the results either of the combination YFPn:OsBUL1 and YFPc:LO9–17711or YFPn:OsBUL1 and YFPc:LO9–177 together with CFP:OsMADS34 suggesting the possibility of trimeric complex formation as was shown in yeast systems and in vitro pull-down assays (Fig. 1A–F). Next, we performed FRET-FLIM experiments19 to test whether physical interactions between the reconstituted YFP by OsBUL1 and LO9–177 interaction and the CFP from the CFP:OsBC1 fusion are available and detected significantly shorter lifetime of the CFP fluorophore compared with controls that co-express YFPn:OsBUL1, YFPc:LO9–177 and CFP:OsMADS34 in the nuclei where YFP and CFP fusion proteins are co-localized (Fig. 1G–I). Therefore, we demonstrated here that formation of a trimeric complex comprised of OsBUL1, LO9–177 and OsBC1 occurs in rice cells, as well.
Figure 1.
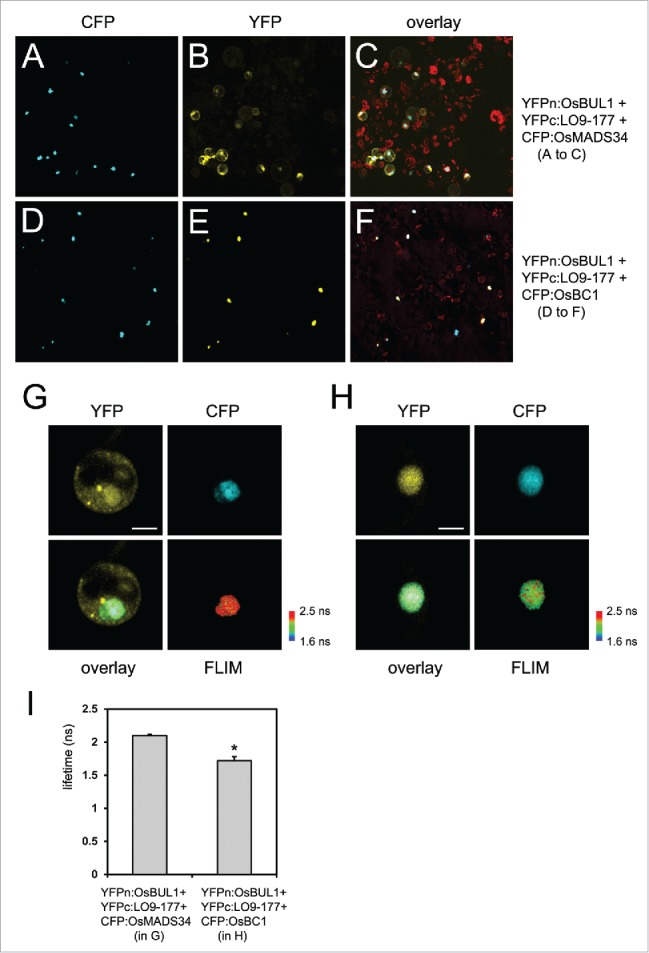
Protein localization and interaction. (A-C) CFP:OsMADS34, YFPn:OsBUL1 and YFPc:LO9–177 were introduced into rice protoplasts. CFP signals were exclusively detected in the nucleus (A) while florescent signals from reconstituted YFP are in the cytoplasm as well as the nucleus (B). (C) Merged image of CFP and YFP signals with auto-florescence from chloroplasts. (D-F) CFP:OsBC1, YFPn:OsBUL1 and YFPc:LO9–177 were introduced into rice protoplasts. YFP signals from YFPn:OsBUL1 and YFPc:LO9–177 as well as CFP signals were only found in the nucleus (D, E) suggesting the formation of a trimeric complex in rice cells. (F) Merged image of CFP and YFP signals with auto-florescence from chloroplasts. FRET-FLIM images in the nucleus containing YFPn:OsBUL1, YFPc:LO9–177 and CFP:OsMADS34 (G) and YFPn:OsBUL1, YFPc:LO9–177 and CFP:OsBC1 (H). (G-I) FRET-FLIM analyses show significant changes in the lifetime of CFP from the CFP:OsBC1 fusion when OsBUL1 and LO9–177 were co-transformed compared with the control, CFP:OsMADS34 was transformed together with OsBUL1 and LO9–177 (*, Student's t test: P < 0.01). ns; nanoseconds. Scale Bars, 10 µm in (G, H). Image and data acquisition was obtained with a Leica TCS SP5, combined with a PicoHarp 300 TCSPC Module and a Ti:Sapphire multiphoton laser (Coherent). The samples were excited with a 840 nm pulsed laser (80 MHz). The emission was recorded from 470 nm to 510 nm in 512 × 512 pixel images with at least 500 counts/pixel. The fluorescence lifetime measurements were analyzed using the PicoQuant SymphoTime Software (ver. 5.3.2.2). For each nucleus average fluorescence decay profiles were plotted and lifetimes were estimated by fitting the data with a double-exponential decay function.
Proper cellular or developmental processes in plants demand precise, robust and efficient regulation of gene expression. Recently, several reports have been published showing that such a sophisticated control for gene expression is exerted by trimeric transcriptional complexes: Microproteins such as B-box 30 (BBX30) and BBX31 interact with flowering activator, CONSTANS (BBX1) and additionally engage in a large protein complex involving the co-repressor protein TOPLESS resulting in delayed flowering by reduced FLOWERING LOCUS T (FT) expression.24 In addition, trichome‐promoting genes encode the R2R3 MYB transcription factors such as GLABRA1 (GL1) and MYB23, the bHLH factors including GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) and the WD40‐repeat protein TRANSPARENT TESTA GLABRA1 (TTG1) while a class of 6 single‐repeat MYB‐related transcription factors such as TRIPTYCHON (TRY) and CAPRICE (CPC) are trichome inhibitors.25 Trichome activators form a R2R3 MYB/bHLH/WD40 trimeric complex, which is counteracted by the inhibitors through competition with R2R3MYB for binding to bHLH proteins.25
Components of the OsBUL1 transcriptional activator complex such as OsBUL1 and OsBC1 are able to interact with OsIBH1, a negative regulator of cell elongation.11 Thus, competition may occur between LO9–177 and OsIBH1 for binding to OsBUL1 and/or OsBC1 suggesting that various cellular and developmental processes are controlled by a complex transcriptional network involving different groups of proteins.
Disclosure of potential conflicts of interest
No conflict of interest declared.
Acknowledgments
The author would like to thank the core facility of the Institute of Cellular and Organismic Biology, Academia Sinica for the technical support for FRET-FLIM, and Ms. Miranda Loney for help with English editing.
Funding
This work was supported by a core grant from the Biotechnology Center in Southern Taiwan (BCST) of the Agricultural Biotechnology Research Center (ABRC), Academia Sinica, Taiwan.
References
- 1.Van Camp W. Yield enhancement genes: seeds for growth. Curr Opin Biotechnol 2005; 16:147-53; PMID:15831379; http://dx.doi.org/ 10.1016/j.copbio.2005.03.002 [DOI] [PubMed] [Google Scholar]
- 2.Sinclair TR, Sheehy JE. Erect leaves and photosynthesis in rice. Science 1999; 283:1456-7; PMID:10206873; http://dx.doi.org/ 10.1126/science.283.5407.1455c10206873 [DOI] [Google Scholar]
- 3.Sakamoto T, Morinaka Y, Ohnishi T, Sunohara H, Fujioka S, Ueguchi-Tanaka M, Mizutani M, Sakata K, Takatsuto S, Yoshida S, et al.. Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat Biotech 2006; 24:105-9; PMID:16369540; http://dx.doi.org/12445121 10.1038/nbt1173 [DOI] [PubMed] [Google Scholar]
- 4.Hoshikawa K. The growing rice plant: an anatomical monograph. Nobunkyo 1989 [Google Scholar]
- 5.Cao H, Chen S. Brassinosteroid-induced rice lamina joint inclination and its relation to indole-3-acetic acid and ethylene. Plant Growth Regulation 1995; 16:189-96; http://dx.doi.org/ 10.1007/BF00029540 [DOI] [Google Scholar]
- 6.Hong Z, Ueguchi-Tanaka M, Shimizu-Sato S, Inukai Y, Fujioka S, Shimada Y, Takatsuto S, Agetsuma M, Yoshida S, Watanabe Y, et al.. Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J 2002; 32:495-508; PMID:12445121; http://dx.doi.org/ 10.1046/j.1365-313X.2002.01438.x [DOI] [PubMed] [Google Scholar]
- 7.Wu CY, Trieu A, Radhakrishnan P, Kwok SF, Harris S, Zhang K, Wang J, Wan J, Zhai H, Takatsuto S, et al.. Brassinosteroids regulate grain filling in rice. Plant Cell 2008; 20:2130-45; PMID:18708477; http://dx.doi.org/ 10.1105/tpc.107.055087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng Z, Wu C, Wang C, Roh J, Zhang L, Chen J, Zhang S, Zhang H, Yang C, Hu J, et al.. SLG controls grain size and leaf angle by modulating brassinosteroid homeostasis in rice. J Exp Botany 2016; 67:4241-53; PMID:27252468; http://dx.doi.org/ 10.1093/jxb/erw204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanaka A, Nakagawa H, Tomita C, Shimatani Z, Ohtake M, Nomura T, Jiang CJ, Dubouzet JG, Kikuchi S, Sekimoto H, et al.. Brassinosteroid upregulated1, encoding a Helix-Loop-Helix Protein, is a novel gene involved in Brassinosteroid signaling and controls bending of the lamina joint in rice. Plant Physiology 2009; 151:669-80; PMID:19648232; http://dx.doi.org/ 10.1104/pp.109.140806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang LY, Bai MY, Wu J, Zhu JY, Wang H, Zhang Z, Wang W, Sun Y, Zhao J, Sun X, et al.. Antagonistic HLH/bHLH transcription factors mediate Brassinosteroid regulation of cell elongation and plant development in rice and arabidopsis. Plant Cell 2009; 21:3767-80; PMID:20009022; http://dx.doi.org/ 10.1105/tpc.109.070441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jang S, An G, Li HY. Rice leaf angle and grain size are affected by the OsBUL1 transcriptional activator complex. Plant Physiol. 2017; 173:688-702; PMID:27879391; http://dx.doi.org/22363621 10.1104/pp.16.01653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heang D, Sassa H. Antagonistic actions of HLH/bHLH proteins are involved in grain length and weight in rice. PLoS One 2012; 7:e31325; PMID:22363621; http://dx.doi.org/ 10.1371/journal.pone.0031325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee S, Lee S, Yang KY, Kim YM, Park SY, Kim SY, Soh MS. Overexpression of PRE1 and its homologous genes activates Gibberellin-dependent responses in Arabidopsis thaliana. Plant Cell Physiol 2006; 47:591-600; PMID:16527868; http://dx.doi.org/ 10.1093/pcp/pcj026 [DOI] [PubMed] [Google Scholar]
- 14.Hyun Y, Lee I. KIDARI, encoding a Non-DNA binding bHLH protein, represses light signal transduction in Arabidopsis thaliana. Plant Mol Biol 2006; 61:283-96; PMID:16786307; http://dx.doi.org/ 10.1007/s11103-006-0010-2 [DOI] [PubMed] [Google Scholar]
- 15.Wang H, Zhu Y, Fujioka S, Asami T, Li J, Li J. Regulation of Arabidopsis Brassinosteroid signaling by Atypical basic helix-loop-helix proteins. Plant Cell 2009; 21:3781-91; PMID:20023194; http://dx.doi.org/ 10.1105/tpc.109.072504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ikeda M, Mitsuda N, Ohme-Takagi M. ATBS1 interacting factors negatively regulate Arabidopsis cell elongation in the triantagonistic bHLH system. Plant Signal Behavior 2013; 8:e23448; PMID:23333962; http://dx.doi.org/ 10.4161/psb.23448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ikeda M, Fujiwara S, Mitsuda N, Ohme-Takagi M. A triantagonistic basic Helix-Loop-Helix system regulates cell elongation in Arabidopsis. Plant Cell 2012; 24:4483-97; PMID:23161888; http://dx.doi.org/ 10.1105/tpc.112.105023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayes MJ, Bryon K, Satkurunathan J, Levine TP. Yeast homologues of 3 BLOC-1 subunits highlight KxDL proteins as conserved interactors of BLOC-1. Traffic (Copenhagen, Denmark) 2011; 12:260-8; PMID:21159114; http://dx.doi.org/ 10.1111/j.1600-0854.2010.01151.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bücherl CA, Bader A, Westphal AH, Laptenok SP, Borst JW. FRET-FLIM applications in plant systems. Protoplasma 2014; 251:383-94; PMID:24390247; http://dx.doi.org/ 10.1007/s00709-013-0595-7 [DOI] [PubMed] [Google Scholar]
- 20.Jang S. Functional characterization of PhapLEAFY, a Floricaula/Leafy Ortholog in Phalaenopsis aphrodite. Plant Cell Physiol 2015; 56:2234-47; PMID:26493518; http://dx.doi.org/ 10.1093/pcp/pcv130 [DOI] [PubMed] [Google Scholar]
- 21.Jang S, Marchal V, Panigrahi KCS, Wenkel S, Soppe W, Deng XW, Valverde F, Coupland G. Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J 2008; 27:1277-88; PMID:18388858; http://dx.doi.org/ 10.1038/emboj.2008.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Su J, Duan S, Ao Y, Dai J, Liu J, Wang P, Li Y, Liu B, Feng D. A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 2011; 7:30; PMID:21961694; http://dx.doi.org/ 10.1186/1746-4811-7-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin X, Wu F, Du X, Shi X, Liu Y, Liu S, Hu Y, Theißen G, Meng Z. The pleiotropic SEPALLATA-like gene OsMADS34 reveals that the ‘empty glumes’ of rice (Oryza sativa) spikelets are in fact rudimentary lemmas. New Phytol 2014; 202:689-702; PMID:24372518; http://dx.doi.org/ 10.1111/nph.12657 [DOI] [PubMed] [Google Scholar]
- 24.Graeff M, Straub D, Eguen T, Dolde U, Rodrigues V, Brandt R, Wenkel S. MicroProtein-mediated recruitment of constans into a TOPLESS Trimeric complex represses flowering in Arabidopsis. PLoS Genetics 2016; 12:e1005959; PMID:27015278; http://dx.doi.org/ 10.1371/journal.pgen.1005959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pattanaik S, Patra B, Singh SK, Yuan L. An overview of the gene regulatory network controlling trichome development in the model plant, Arabidopsis. Front Plant Sci 2014; 5:259; PMID:25018756; http://dx.doi.org/ 10.3389/fpls.2014.00259 [DOI] [PMC free article] [PubMed] [Google Scholar]


