ABSTRACT
Triterpenoids produced by plants play important roles in the protection against biotic stress. Roots of Arabidopsis thaliana produce different triterpenoids, which include the tricyclic triterpene diol, arabidiol. In a degradation reaction induced by infection with the oomycete pathogen, Pythium irregulare, arabidiol is cleaved to the 11-carbon volatile homoterpene, (E)-4,8-dimethyl-1,3,7-nonatriene (DMNT), and the 19-carbon ketone, apo-arabidiol. The arabidiol pathway and its volatile breakdown product DMNT have been implicated in the defense against P. irregulare infection. Here we show that the non-volatile breakdown product apo-arabidiol is further converted to the acetylated derivative α-14-acetyl-apo-arabidiol via a presumed epimerization and subsequent acetylation reaction. α-14-acetyl-apo-arabidiol and the detected intermediates in the derivatization pathway are partially exuded from the root indicating possible defensive activities of these molecules in the rhizosphere. The conversion steps of apo-arabidiol vary among different Arabidopsis accessions and are present in only rudimentary form in the close relative Arabidopsis lyrata, which supports an intra- and inter-specific modularity in triterpenoid metabolism.
KEYWORDS: Accession variation, Arabidopsis, defense, homoterpene, root, triterpene, volatile
Plants produce a diversity of secondary or specialized metabolites that play various roles in long- and short-distance interactions with other organisms and the environment.1-4 Specialized metabolites released from plant roots are implicated in direct and indirect defensive activities. For instance benzoxazinoids, cyclic hydroxamic acids produced by grasses, act as effective belowground defenses against microbes, insects, or competing plants.5 In the class of terpenoids, twenty-carbon semivolatile or non-volatile diterpenes such as rhizathalenes and momilactones function as anti-feedants and allelopathic agents in the roots of Arabidopsis and rice, respectively.6,7 Moreover, 30-carbon triterpene glycosides exuded by plant roots are well known for their potent antifungal activities in belowground defense.8
Arabidopsis roots produce several different triterpenoids. For instance, thalianol and marneral and their designated or putative derivatives are synthesized by enzymes whose genes are located on metabolic gene clusters and co-expressed in epidermal and/or cortex cell layers of the root hair zone.9,10 Similarly, we showed recently that the genes responsible for the synthesis and oxidative breakdown of the triterpene diol, arabidiol, are positioned on an arabidiol/baroul biosynthetic gene cluster and share coexpression patterns in the pericycle of the root hair zone and the quiescent center.11 Arabidiol, and its downstream breakdown products contribute to the resistance of Arabidopsis roots against infection by the oomycete root rot pathogen Pythium irregulare.11 In a response induced by Pythium infection, arabidiol undergoes an oxidative C-C cleavage reaction catalyzed by the cytochrome P450 monooxygenase CYP705A1 to form the volatile 11-carbon homoterpene, (E)-4,8-dimethyl-1,3,7-nonatriene (DMNT), and the non-volatile 19-carbon ketone, apo-arabidiol (Fig. 1).11 DMNT represents one of the most common volatile organic compounds that are released by plants upon biotic stress.12 While homoterpenes are involved in indirect defense in aboveground interactions,13 our studies in Arabidopsis roots suggested a possible defensive role of DMNT at low concentrations in the early phase of Pythium infection by reducing oospore germination rates and retarding Pythium growth.11 In addition, arabidiol biosynthetic and breakdown mutants were more susceptible to inoculation with Pythium in potting substrate indicating a possible contribution of both volatile and non-volatile breakdown products of arabidiol to plant resistance throughout the infection process of the root. Since apo-arabidiol could not be detected in vivo, we reasoned that the compound could be further converted into derivatives that may remain in the tissue or be released into the rhizosphere. In this study, we examined whether any downstream products of apo-arabidiol could be identified in Arabidopsis root tissue or exudates and we investigated the possible intra- and interspecific variation of these steps.
Figure 1.
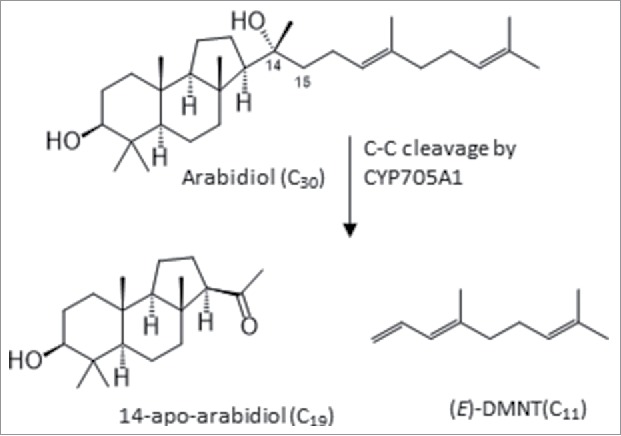
Degradation of the triterpenoid arabidiol catalyzed by CYP705A1.
To detect any derivatives of apo-arabidiol, we treated roots of 21 d old axenically grown Arabidopsis wild type plants (accession Col-0, ABRC stock center) and the mutant of the arabidiol degrading enzyme CYP705A1 (cyp705a1–1; SALK_043195, ABRC stock center) with 100 µM jasmonic acid (JA) to mimic Pythium infection as described before.11 Treatments were performed for 24 h since the emission rate of DMNT was highest at this time point (with approximately 50% emission at 12 h of treatment)11 and, consequently, highest production of apo-arabidiol and its possible derivatives was expected at this time. Apo-arabidiol derivatives were extracted with 10 mL ethyl acetate from 1 g of ground root material. One volume of water was added for phase separation and collection of the organic layer and the extraction procedure was repeated twice in the same way. The combined organic phases were concentrated and purified over a small silica gel column (2 cm). Samples were dried, re-suspended in ethyl acetate and analyzed by GC-MS.14 Extractions from root exudates were performed by lyophilizing 15 ml of growth medium prior to resuspension in 2 mL of distilled water followed by three times extraction with 10 mL of ethyl acetate. The combined organic extracts were prepared and analyzed as described above. Analysis of root tissue extracts showed a compound with a mass spectrum similar to that of apo-arabidiol but with a different retention time (TR = 27.8 min). This compound, named D-R (for derivative in root tissue), was not found in the cyp705a1-1 mutant (Fig. 2). When we analyzed organic solvent extracts of the medium from axenic cultures, we detected D-R and a second compound (retention time of TR = 26.3 min) with a mass spectrum similar to that of apo-arabidiol. This second compound, named D-M (for derivative in the medium), was absent in exudates of the cyp705a1–1 mutant and the arabidiol biosynthetic mutant abds-1 (SALK_018285, ABRC stock center) (Fig. 3). Based on these results, we concluded that apo-arabidiol is converted into at least two other products, which are in part exuded by the root tissue.
Figure 2.

Detection of arabidiol-derived compounds in Arabidopsis root tissue. (A) GC chromatograms of ethyl acetate extracts from roots of wild type Col-0 and cyp705a1–1 plants and a 14-apo-arabidiol standard. 14-apo-arabidiol was not detected in Col-0 roots upon 24 h of JA treatment (traces of the 274 ion at RT = 26.8 min correspond to other metabolites). A putative derivative named D-R was detected at RT = 27.8 min in roots of Col-0 but not cyp705a1–1 plants [left panel TIC, right panel single ion monitoring (SIM) for m/z 274]. (B) Mass spectra 14-apo-arabidiol and D-R.
Figure 3.
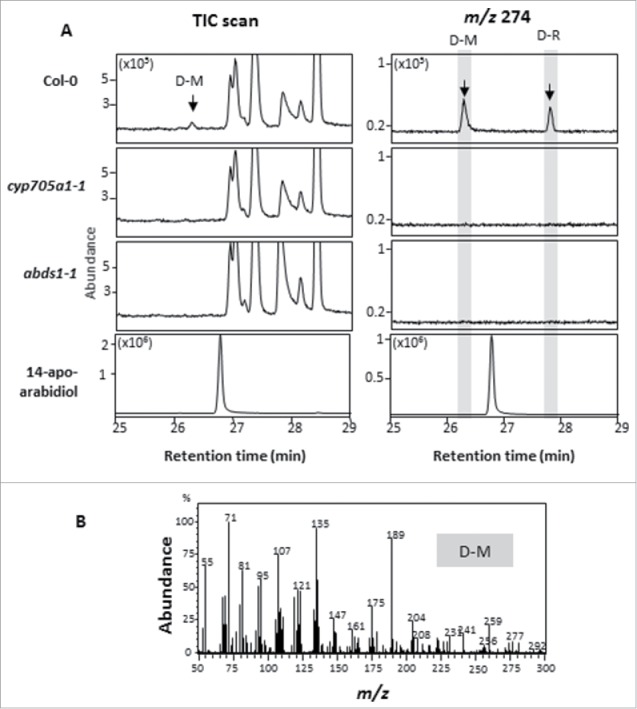
Detection of arabidiol-derived compounds in Arabidopsis root exudates. (A) GC chromatograms of ethyl acetate extracts from the culture medium of axenically grown roots of Col-0 wild type, cyp705a1–1, and abds1–1 plants and a 14-apo-arabidiol standard [left panel TIC, right panel single ion monitoring (SIM) for m/z 274]. After 24 h of JA treatment a compound named D-M was detected in addition to D-R with a retention time different from that of 14-apo-arabidiol. Both compounds were absent in exudates of the cyp705a1–1 and abds1–1 mutants. (B) Mass spectrum of D-M.
To provide further evidence for the conversion of apo-arabidiol into the observed putative derivatives, we performed feeding experiments with purified apo-arabidiol (3 μM)11 using axenically grown arabidiol synthase (abds-1) plants (Fig. 4). Preliminary experiments showed that the conversion of apo-arabidiol to downstream metabolites was independent of the presence of JA. Therefore, we performed these feeding studies without JA to obtain less complex extracts by reducing the production of other JA-induced metabolites. Analysis of organic extracts obtained from root medium at different time points showed the presence of the two already known compounds (D-R and D-M) and one new putative derivative of apo-arabidiol (Fig. 4). The compounds were found at different ratios depending on the duration of the precursor application with the amount of derivative D-R increasing over time leading to the same ratio of both derivatives (D-R and D-M) as that observed in jasmonate-treated wild type plants (Fig. 3). The third compound was detected after 1.5 h at a retention time of 26.1 min with a mass spectrum similar to that of apo-arabidiol (Fig. 4). The amount of this compound, named D-I (for intermediate), declined over 3 h to trace amounts at 30 h after application suggesting that it is a possible intermediate in the apo-arabidiol modification pathway. To evaluate whether apo-arabidiol could be directly modified by root exudates, filtered exudates (0.22 µM filter to remove possible root cap cells) were obtained 24 h after growth medium replacement from axenically grown Arabidopsis roots and inoculated for 30 h with apo-arabidiol. No modification of apo-arabidiol was observed under these experimental conditions indicating that its conversion occurs in the root tissue most likely by specific enzymatic activities (Fig. 4).
Figure 4.
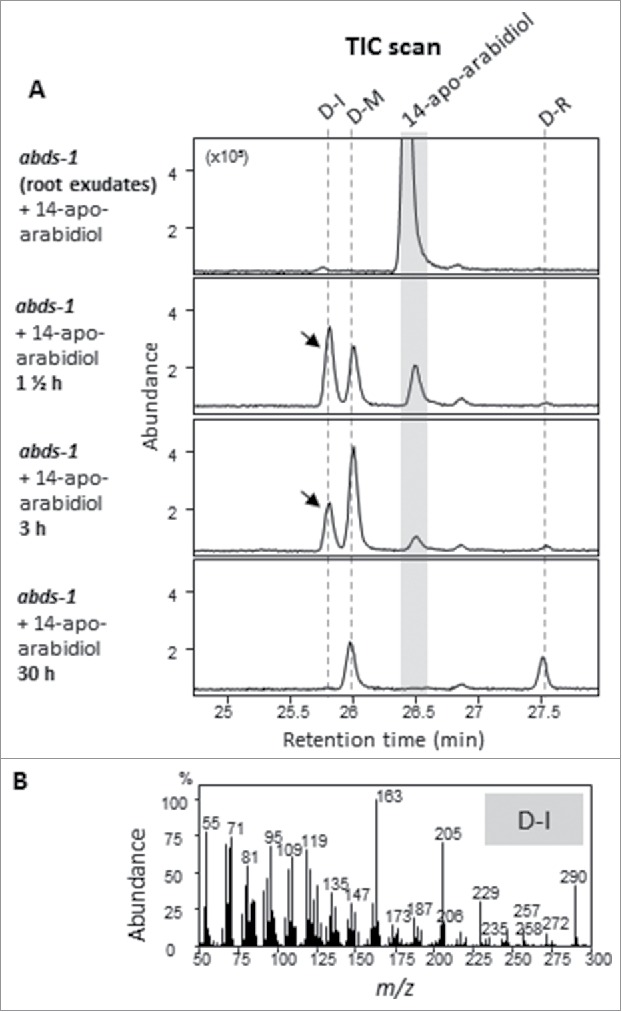
14-apo-arabidiol is converted into several modification products in planta. (A) Feeding experiments with 3 μM 14-apo-arabidiol were performed in the presence of axenically grown roots of the abds-1 mutant in comparison to root exudates only. GC chromatograms of ethyl acetate extracts from the culture medium are shown. 14-apo-arabidiol was not converted by root exudates from plants grown for 24 h in MS medium. In the presence of root tissue, 14-apo-arabidiol was converted to D-M and a putative intermediate D-I at 1–3 h of application. At 30 h of incubation, D-I was absent and levels of D-M were reduced while D-R accumulated. (B) Mass spectrum of D-I.
Since we found apo-arabidiol to be efficiently converted into downstream products in axenic plant cultures, we used this bioconversion approach to produce the derivatives for NMR analysis (Supplemental Methods and Results). Samples of D-R and D-I were analyzed with more than 90% purity by NMR spectroscopy (1H-NMR, 13C-NMR, HSQC, HMBC, and TOCSY) (Figs. S1-10, Supplemental Methods and Results), which determined the molecular structures shown in Fig. 5 (Figs. S2, 10). NMR analysis of the derivative D-I indicated a dehydrogenation reaction at the C3-OH position of apo-arabidiol. D-I was, therefore, named 3-keto-14-apo-arabidiol. In derivative D-R, the C3-OH group was acetylated, which was evident from the appearance of HMBC correlations and acetylation shifts (Figs. S1–3; Supplemental Results). In addition, the orientation of the C3-OH group is converted from β in apo-arabidiol to α in D-R indicating an epimerization reaction and suggesting a selective conversion of the carbonyl group in 3-keto-14-apo-arabidiol. We named this compound α-14-acetyl-apo-arabidiol. Unfortunately, we were unable to unambiguously identify the structure of D-M because of limited amount of the compound. From the mass spectral data (Fig. 3B), we assume that D-M is derived from D-I by re-hydrogenation at the C3 carbonyl group causing a β to α epimerization in this position prior to acetylation to form acetyl-apo-arabidiol (Fig. 5).
Figure 5.
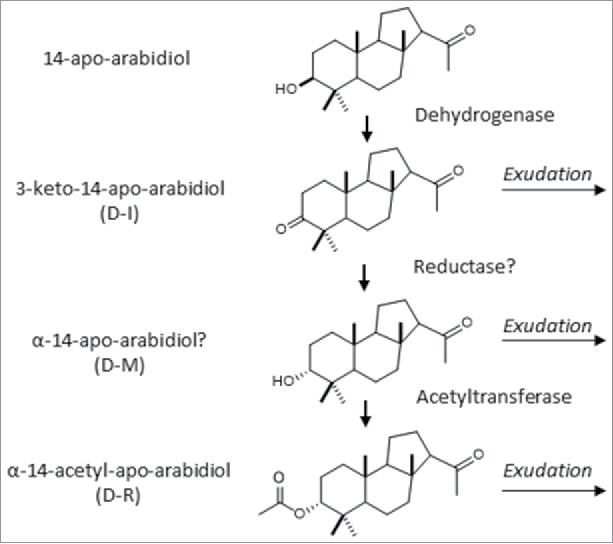
Predicted pathway for the formation of 14-apo-arabidiol derivatives.
To provide further proof for the sequence of the modification reactions, we performed feeding experiments with the purified individual compounds. Compounds were applied for 30 h to axenically grown abds-1 plants and products were extracted from the culture medium and analyzed by GC-MS. Upon feeding with apo-arabidiol, we detected 3-keto-14-apo-arabidiol, D-M (the putative α-14-apo-arabidiol), and small amounts of α-14-acetyl-apo-arabidiol (Fig. S11). Application of 3-keto-14-apo-arabidiol resulted in the substantial production of D-M suggesting an immediate conversion of this intermediate. Feeding with D-M only yielded α-14-acetyl-apo-arabidiol suggesting that the putative α-alcohol is a substrate of the subsequent acetyl transferase reaction. Collectively, these results suggested a sequential modification pathway of epimerization followed by an acylation of the C3 hydroxyl group to yield acetyl-apo-arabidiol as presented in Fig. 5.
The genes and enzymes responsible for the discovered enzymatic pathway are currently unknown and not readily predictable from gene positions in the extended baroul/arabidiol synthase gene cluster on chromosome 4 of the Arabidopsis genome.11 The baroul/arabidiol synthase cluster contains two putative acyltransferase genes (At4g15390; At4g15400), of which a full length cDNA could only be obtained for At4g15390 but not for At4g15400 from RNA of JA-treated roots. The amplified At4g15400 cDNA was missing a 58 bp fragment and thus encoded a truncated protein. The At4g15390 gene could not be associated with an acetyltransferase activity when we expressed the cDNA in yeast in the presence of the compounds apo-arabidiol, D-M, or D-I following methods described previously.11 Therefore, we concluded that other possible acyltransferase enzymes, which are not physically linked to the arabidiol synthase cluster, might be involved in this reaction. We further tested to what extent the modification steps of apo-arabidiol are conserved or vary among different Arabidopsis accessions. Since the accession Cvi-1 (ABRC stock center) is impaired in the formation of DMNT,11 we reasoned that this accession might also be unable to modify apo-arabidiol. However, when the roots of axenically grown Cvi-1 plants were treated with apo-arabidiol, we found a conversion of the compound into all downstream products that have been detected in the Col-0 accession (Fig. 6). By contrast, in the Ler-2 (ABRC stock center) accession, which breaks down arabidiol, apo-arabidiol was converted to 3-keto-14-apo-arabidiol and the putative α-14-apo-arabidiol without detection of the acetylated derivative (Fig. 6). These results suggest accession-dependent natural variation in the acylation of apo-arabidiol or its exudation from root tissue. Furthermore, as it is evident from the Cvi-1 accession, the arabidiol degradation pathway can be impaired at its first step without the loss of function of the subsequent enzymes, which indicates that the entire modification pathway was established prior to the divergence of the Cvi-1 accession. These accession-specific differences in the modification of arabidiol support the general notion of evolutionary plasticity and functional optimization in triterpene metabolic pathway assembly.9 Triterpene pathway plasticity is also apparent between the closely related species A. thaliana and A. lyrata because of the absence of functional arabidiol and marneral synthase clusters in A. lyrata.9, 11 To test whether any of the observed downstream enzymatic steps could be present in A. lyrata, we established axenic cultures by growing plants from seed (ABRC stock center) for 2 weeks on ½ x MS medium plates and then for 4 weeks in ½ x MS liquid culture. When we applied apo-arabidiol to these plants, we found a conversion to 3-keto-14-apo-arabidiol (Fig. 6). This finding suggests that structural modifications such as oxidation reactions might be adopted easily by the activity of enzymes with broader substrate specificity once an initial pathway has been established. Overall, our results support the extraordinary intra- and inter-specific modularity in the evolution of the terpenoid metabolic landscape. Questions remain to what extent such variation represents natural metabolic plasticity in interplay with evolutionary fine-tuning in response to different selective pressures. In this context, it will be of interest to further evaluate the possible functional roles and activities of the identified apo-arabidiol conversion products. Since these compounds are produced together with DMNT upon pathogen attack and partly released into the rhizopshere, it is possible that they contribute, as non-volatile metabolites, to the defensive activity that has been associated with the arabidiol-DMNT pathway (Sohrabi et al., 2015). Furthermore, the derivatives, may, even at low levels, function as signals or have synergistic effects in interactions with microbes or other target organisms in the endosphere and/or rhizosphere.
Figure 6.
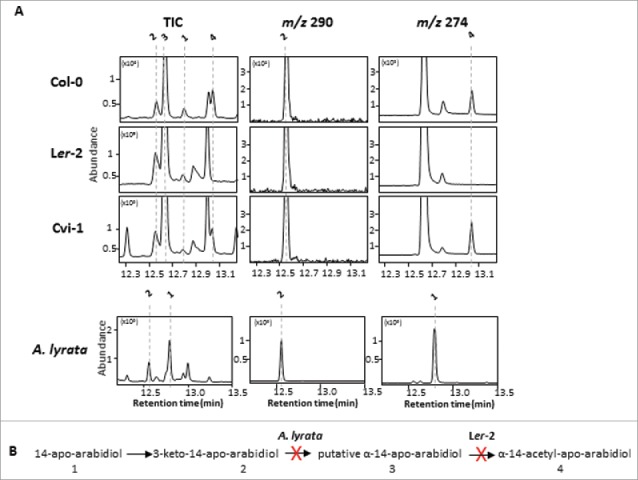
Naturally occurring variation in the modification steps of arabidiol-derived compounds between different accessions of A. thaliana and in A. lyrata. (A) Feeding experiments with 30 μM 14-apo-arabidiol were performed for 3 h with axenically grown roots of A. thaliana accessions Col-0, Ler-2, and Cvi-1–1 and A. lyrata. GC chromatograms of ethyl acetate extracts from the culture medium are shown. Col-0 and Cvi-1–1 converted 14-apo-arabidiol into all downstream products, whereas α-14-acetyl-apo-arabidiol was not detected for Ler-2. 14-Apo-arabidiol was oxidized in the presence of A. lyrata roots but no further conversion products were detected. TIC and SIM scan profiles are shown. Numbers indicate the different compounds as shown in (B). (B) The predicted pathway for 14-apo-arabidiol modification. The X represents steps that are not supported by A. lyrata or Ler-2. Note that 14-apo-arabidiol is not a natural product in Cvi-1 and A. lyrata.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgment
We thank the Ohio State University, College of Pharmacy, instrumentation facility and the Campus Chemical Instrument Center (CCIC) for the acquisition of the NMR and mass spectra.
Funding
This work was supported by the National Science Foundation under grant MCB-0950865.
References
- 1.Tholl D. Biosynthesis and biological functions of terpenoids in plants In: Schrader J, Bohlmann J, eds. Biotechnology of Isoprenoids, Advances in biochemical engineering-biotechnology. Cham (Switzerland): Springer; 2015:63-106; PMID:25583224; http://dx.doi.org/24588567 10.1007/10_2014_295 [DOI] [PubMed] [Google Scholar]
- 2.Mithöfer A, Boland W. Plant defense against herbivores: Chemical aspects In: Merchant SS, ed. Annual Review of Plant Biology, Vol 63. Palo Alto (USA): Annual Reviews; 2012: 431-50; PMID:22404468; http://dx.doi.org/24588567 10.1146/annurev-arplant-042110-103854 [DOI] [PubMed] [Google Scholar]
- 3.Muhlemann JK, Klempien A, Dudareva N. Floral volatiles: from biosynthesis to function. Plant Cell Environ 2014; 37:1936-49; PMID:24588567; http://dx.doi.org/ 10.1111/pce.12314 [DOI] [PubMed] [Google Scholar]
- 4.Maag D, Erb M, Köllner TG, Gershenzon J. Defensive weapons and defense signals in plants: Some metabolites serve both roles. Bioessays 2015; 37:167-74; PMID:25389065; http://dx.doi.org/ 10.1002/bies.201400124 [DOI] [PubMed] [Google Scholar]
- 5.Niemeyer HM. Hydroxamic acids derived from 2-hydroxy-2h-1,4-benzoxazin-3(4h)-one: Key defense chemicals of cereals. J Agric Food Chem 2009; 57:1677-96; PMID:19199602; http://dx.doi.org/ 10.1021/jf8034034 [DOI] [PubMed] [Google Scholar]
- 6.Vaughan MM, Wang Q, Webster FX, Kiemle D, Hong YJ, Tantillo DJ, Coates RM, Wray AT, Askew W, O'Donnell C, et al.. Formation of the unusual semivolatile diterpene rhizathalene by the Arabidopsis class I terpene synthase TPS08 in the root stele is involved in defense against belowground herbivory. Plant Cell 2013; 25:1108-25; PMID:23512856; http://dx.doi.org/ 10.1105/tpc.112.100057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu MM, Galhano R, Wiemann P, Bueno E, Tiernan M, Wu W, Chung IM, Gershenzon J, Tudzynski B, Sesma A, et al.. Genetic evidence for natural product-mediated plant-plant allelopathy in rice (Oryza sativa). New Phytol 2012; 193:570-5; PMID:22150231; http://dx.doi.org/ 10.1111/j.1469-8137.2011.04005.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Osbourn AE, Qi XQ, Townsend B, Qin B. Dissecting plant secondary metabolism - constitutive chemical defences in cereals. New Phytol 2003; 159:101-8; http://dx.doi.org/ 10.1046/j.1469-8137.2003.00759.x [DOI] [PubMed] [Google Scholar]
- 9.Field B, Fiston-Lavier AS, Kemen A, Geisler K, Quesneville H, Osbourn AE. Formation of plant metabolic gene clusters within dynamic chromosomal regions. Proc Natl Acad Sci USA 2011; 108:16116-21; PMID:21876149; http://dx.doi.org/ 10.1073/pnas.1109273108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Field B, Osbourn AE. Metabolic diversification - Independent assembly of operon-like gene clusters in different plants. Science 2008; 320:543-7; PMID:18356490; http://dx.doi.org/ 10.1126/science.1154990 [DOI] [PubMed] [Google Scholar]
- 11.Sohrabi R, Huh JH, Badieyan S, Rakotondraibe LH, Kliebenstein DJ, Sobrado P, Tholl D. In planta variation of volatile biosynthesis: An alternative biosynthetic route to the formation of the pathogen-induced volatile homoterpene DMNT via triterpene degradation in Arabidopsis roots. Plant Cell 2015; 27:874-90; PMID:25724638; http://dx.doi.org/ 10.1105/tpc.114.132209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tholl D, Sohrabi R, Huh J-H, Lee S. The biochemistry of homoterpenes – Common constituents of floral and herbivore-induced plant volatile bouquets. Phytochemistry 2011; 72:1635-46; PMID:21334702; http://dx.doi.org/ 10.1016/j.phytochem.2011.01.019 [DOI] [PubMed] [Google Scholar]
- 13.Kappers IF, Aharoni A, van Herpen TWJM, Luckerhoff LLP, Dicke M, Bouwmeester HJ. Genetic engineering of terpenoid metabolism attracts bodyguards to Arabidopsis. Science 2005; 309:2070-2; PMID:16179482; http://dx.doi.org/ 10.1126/science.1116232 [DOI] [PubMed] [Google Scholar]
- 14.Wang Q, Sohrabi R, Tholl D. Analysis of diterpenes and triterpenes from plant foliage and roots In: Rodriguez Concepcion M, ed. Plant Isoprenoids: Methods and Protocols, Methods in Molecular Biology, Totowa (USA): Humana Press Inc.; 2014:149-59; PMID:24777795; http://dx.doi.org/ 10.1007/978-1-4939-0606-2_10 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


