Abstract
Coiled-coil motifs are ubiquitous mediators of specific protein-protein interactions through the formation of interlocking hydrophobic seams between α-helical chains. Residues that form these seams occur at the first (a) and fourth (d) positions of a characteristic 7-aa repeat and are primarily aliphatic. The potential of aromatic residues to promote helix association in a coiled coil was explored by engineering a “Trp-zipper” protein with Trp residues at all 14 a and d positions. The protein forms a discrete, stable, α-helical pentamer in water at physiological pH. Its 1.45-Å crystal structure reveals a parallel, five-stranded coiled coil, a previously uncharacterized type of “knobs-into-holes” packing interaction between interfacial Trp side chains, and an unusual ≈8-Å-diameter axial channel lined with indole rings that is filled with polyethylene glycol 400 and water and sulfate ion molecules. The engineered Trp-zipper pentamer enlarges current views of coiled-coil assembly, molecular recognition, and protein engineering, and may serve as a soluble model for membrane ion channels.
Keywords: coiled coils, protein design, protein structure
The coiled coil is among the most common motifs in protein structures, accounting for a significant fraction of all inter-domain interactions (1-3). Coiled-coil sequences, estimated to constitute 3-5% of the protein residues in known genomes, share a characteristic seven-residue repeat, the heptad repeat, denoted as a-b-c-d-e-f-g, with a predominance of the aliphatic side chains Leu, Ile, Val, and Ala at the a and d positions, and polar residues generally elsewhere (4-7). Despite their simple sequence pattern, coiled coils exhibit great diversity in the number and orientation of the chains involved: Proteins composed of 2 to 12 “strands” are known, and general principles for assembling dimers (8) as well as higher-order structures including hexamers and dodecamers have been deduced (9, 10). Coiled coils have served as an important test-bed for protein design efforts (11, 12). The Escherichia coli lipoprotein contains a three-stranded coiled-coil domain (Lpp-56) embedded between the outer membrane and the periplasmic peptidoglycan (13). The natural sequence of Lpp-56 includes three Ala side chains at adjacent a and d positions that lead to a significant narrowing in the diameter of the helices in this region. This unusual flexibility in the local helix geometry has not been seen in GCN4 leucinezipper models and led us to undertake a systematic analysis of the effect of the number and size of the side chains at a and d positions on the structure and stability of the protein. Replacement of all a and d residues by Ala, for example, completely unfolds the protein in solution, although the trimer is stable in crystals (14). To explore the limits of helix geometry on coiled-coil formation, we engineered an Lpp-56 variant that contains exclusively Trp residues at the a and d positions (Fig. 1). This Trp-zipper protein (denoted Trp-14) forms a stable pentameric coiled coil in solution. Its 1.45-Å crystal structure reveals an unprecedented interface between five parallel helices formed from interacting Trp residues. The arrangement of Trp side chains at the core a and d positions obeys an unusual pattern of knobs-into-holes packing. Indeed, in both a and d layers, we find evidence for a simple rule for parallel coiled coils relating to the number of strands to the Cα-Cα and Cα-Cβ bond vectors. The Trp-14 structure also contains an irregular hydrophobic cavity with a diameter of 8 Å running along the long axis that offers a soluble model for investigating the ligand properties of comparably sized membrane channels.
Fig. 1.
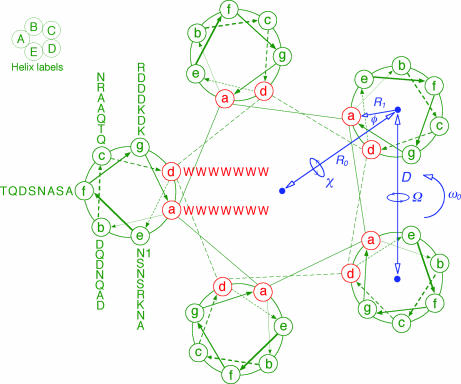
Coiled-coil schematic of Trp-14 as a pentamer. The view is from the N terminus looking toward the C terminus. Heptad-repeat positions are labeled a through g. The tryptophans at the first (a) and fourth (d) positions form the apolar interface of the pentamer. The Crick supercoil parameters are indicated as follows: R0 and R1, the radii of the supercoil and α-helix, respectively; ω0, the supercoil pitch; φ, the Cα phase angle, defined as the angle between vectors from the α-helix center to the supercoil center and to the Cα of the a-position residue; χ, superhelix crossing angle.
Materials and Methods
Protein Expression and Purification. Trp-14 was expressed in E. coli BL21(DE3)/pLysS by using a modified pET3a vector. Cells were lysed with glacial acetic acid and centrifuged to separate the soluble fraction from inclusion bodies. Trp-14 from the soluble fraction was purified to homogeneity by reverse-phase HPLC on a C18 preparative column. Selenomethionyl (SeMet) protein was produced in amino acid-supplemented minimal media.
Biophysical Experiments. CD spectra were acquired on an Aviv 62A/DS CD spectrometer (Aviv Associates, Lakewood, NJ) as described in ref. 15. Stability to chemical denaturation was determined by monitoring [θ]222 as a function of guanidinium chloride (GdmCl) concentration at 20°C in PBS. Data were recorded with an average time of 120 s per data point. Baselines were estimated by nonlinear fitting to the standard six-parameter, two-state transition curve using weighted averages. The ΔGunf value at 0 M GdmCl was calculated by using the linear extrapolation method (16). Sedimentation equilibrium measurements were performed with a ProteomeLab XL-A ultracentrifuge (Beckman Coulter) as described in ref. 15. Protein solutions were dialyzed overnight against PBS, loaded at initial concentrations of 10, 30, and 100 μM, and analyzed at rotor speeds of 21,000 and 24,000 rpm at 20°C. Data sets were fitted to a single-species model. The apparent molecular mass of Trp-14 is 33.8 kDa.
Crystallization and Structure Determination. Trp-14 was crystallized at room temperature by the vapor diffusion method against 2% polyethylene glycol (PEG) 400/0.1 M Tris·HCl (pH 8.0)/1.65 M (NH4)2SO4. Crystals belong to space group P1 (a = 30.55 Å, b = 30.67 Å, c = 72.41 Å, α = 97.4°, β = 91.9°, γ = 113.5°) with five molecules in the asymmetric unit. Data sets Native and SeMet were collected on beamline X4A at the National Synchrotron Light Source. Intensities were integrated and scaled by using denzo and scalepack (17). All five selenium sites in the asymmetric unit were located with solve (18), and ≈80% of the polypeptide chain was traced automatically into the electron-density maps. Iterative rounds of model building with O, refinement with refmac (19), and addition of ordered solvent clarified the trace, except for a few residues at the helix termini. Superhelical parameters for Trp-14 were obtained by fitting the Cα backbones to a supercoil parameterization suggested by Crick (8).
Results and Discussion
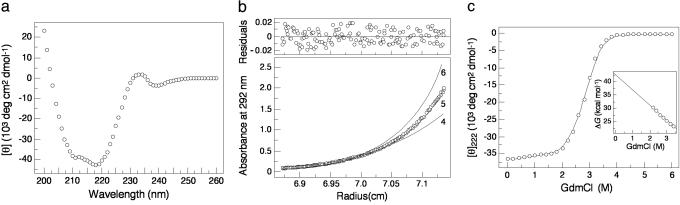
Solution Properties. The CD spectrum of Trp-14 has double minima at 211 and 218 nm, as well as a minor positive band around 233 nm (Fig. 2a). This CD signal differs from the characteristic signature of an α-helical conformation (with minima at 208 and 222 nm), which can be attributed to the presence of Trp residues because the indole chromophore is known to contribute significant ellipticity in the peptide region of the CD spectrum (20). Although not a quantitative measure, the mean residue ellipticity at either 208 nm or 222 nm would suggest >90% helical structure at 4°C in PBS. Sedimentation equilibrium measurements indicate that Trp-14 sediments as a discrete pentamer and exhibits no systematic dependence of molecular weight on concentration between 10 and 100 μM (Fig. 2b). The stability of Trp-14 toward denaturation by GdmCl was determined by monitoring ellipticity at 222 nm at 20°C (Fig. 2c). Analysis of the denaturation data using a two-state monomer-pentamer unfolding model indicates that the unfolding free energy of Trp-14 is 42.5 kcal·mol-1 with the m value of 5.7 kcal-1·mol-1. Thus, the Trp-14 protein forms a highly stable, five-stranded α-helical complex in solution.
Fig. 2.
Folding of Trp-14 as a stable, α-helical pentamer. (a) CD spectrum at 4°C in PBS and 5 μM protein concentration. (b) Sedimentation equilibrium of a 10 μM sample at 20°C and 21 krpm in PBS. The data fit closely to a pentameric complex. Curves expected for tetrameric and hexameric models are indicated for comparison. (Upper) The deviation in the data from the linear fit for a pentameric model is plotted. (c) GdmCl-induced unfolding at 20°C in PBS and 20 μM protein concentration. Inset shows the linear extrapolation of the ΔG value determined in the transition region to the limit of zero concentration of GdmCl.
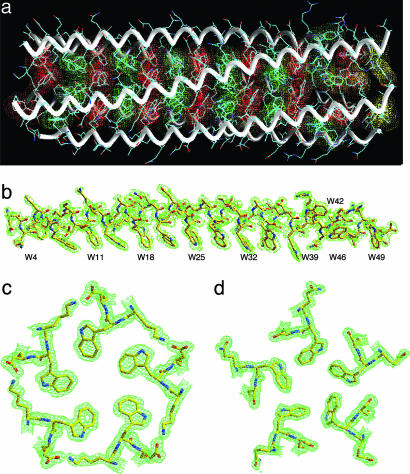
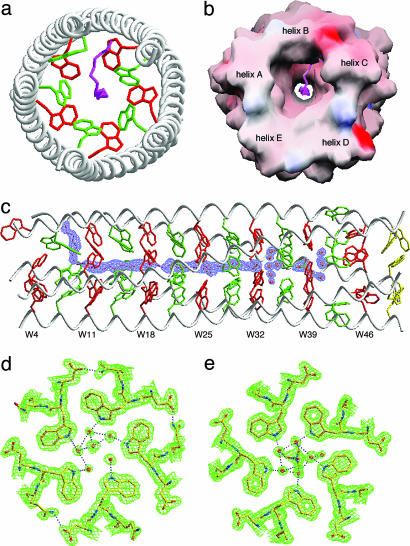
Overview of the Structure. To evaluate the Trp core packing in atomic detail, we determined the x-ray crystal structure of Trp-14 at 1.45 Å by the method of multiwavelength anomalous diffraction (21), using a selenomethionine derivative (Table 1). The Trp-14 structure shows that five parallel α-helices intertwine in a gradual left-handed supercoil (Fig. 3a). The superhelix forms a cylinder with an overall diameter of ≈29 Å and length of ≈75 Å. An approximate 5-fold axis of symmetry coincides with the superhelical axis. The individual helices in the pentamer can be superimposed on each other with a rms deviation for α carbon atoms of 0.29-0.69 Å and with the largest deviations occurring at their ends. With the exception of solvent-exposed W42 residues, the Trp side chains at the a and d positions point into the interior of the pentamer, forming an indole ring core at the interface between the five α-helices (Fig. 3 a and b). Cross-sectional layers containing one Trp structural pattern at the a positions alternate with layers containing a different Trp pattern at the d positions (Fig. 3 c and d). An unexpected feature of the Trp-14 structure is the loss of core packing of the W42 side chains (although they are well defined in the electron-density map; Fig. 3 a and b). It is possible that the a and d Trp residues (because of their flat rigid shape) shift gradually across the helix surface over five heptads, producing a cumulative drift in heptad frame at the W42 position. The dihedral angles χ1 and χ2 of the Trp side chains (excluding six that are at the helix termini) are approximately -177°, -105° or -177°, 90°, corresponding to their most commonly occurring rotamers in α-helices (22). The Trp residue in the a layer closest to the C terminus of the A helix assumes dihedral angles near -80°, 165°, thereby directing its indole ring to bend away from the supercoil axis toward solvent (Fig. 3a). The Trp side chains in the B4, A46, C46, D46, and E49 positions all show modest bond-angle strains but still bend toward the hydrophobic interface. Imperfect packing of Trp at these positions causes the superhelix to be locally underwound at its ends (Fig. 3a).
Table 1. Crystallographic data and refinement statistics.
| Diffraction data | ||||
| Data set | Native | SeMet λ1 | SeMet λ2 | SeMet λ3 |
| Wavelength, Å | 0.9793 | 0.9795 | 0.9800 | 0.9757 |
| Resolution, Å | 70.7-1.45 | 50-2.3 | 50-2.3 | 50-2.3 |
| Rsym, % | 5.8 | 3.8 | 3.8 | 4.0 |
| Completeness | 93.7 | 99.7 | 99.7 | 99.6 |
| I/σ(I) | 13.7 | 17.0 | 16.4 | 16.0 |
| Phasing | ||||
| Resolution, Å | 20-2.9 | |||
| No. of sites | 5 | |||
| Phasing power (ano/iso) | 0.9/0.2 | 0.6/0.7 | 0.8/— | |
| Overall figure of merit | 0.50 | |||
| Refinement | ||||
| Resolution range, Å | 70.7-1.45 | |||
| No. of reflections | 38,990 | |||
| Total no. of atoms | 2,572 | |||
| Rcryst/Rfree, % | 21.6/27.6 | |||
| rmsd bond lengths, Å | 0.021 | |||
| rmsd bond angles, ° | 1.7 | |||
| Average B factor, Å2 | 29.8 | |||
| rmsd B values, Å2 | 2.6 | |||
—, not applicable; rmsd, rms deviation.
Fig. 3.
Crystal structure of Trp-14. (a) Side view of the pentamer. Red van der Waals surfaces identify tryptophans at the a positions, green surfaces identify tryptophans at the d positions, and yellow surfaces identify the C-terminal-most tryptophan d layer. (b) The 2Fo - Fc electron-density map at 1.0σ contour superposed onto the final model of the C helix. Electron densities are shown in broken lines to clarify the drawing. (c) The 2Fo - Fc electron-density map (contoured at 1.0σ) showing the cross-section of the pentamer in the W18 (a) layer. (d) The cross-section of the pentamer in the W21 (d) layer.
The indole ring planes in both the a and d layers incline by ≈45° toward the C terminus of each helix with respect to the plane perpendicular to the pentamer axis (Fig. 3b). This geometry results in a greater degree of surface overlap between the side chains. Residues at positions e and g pack against tryptophans at d and a, respectively, to complete the bulky hydrophobic core. Compared with the side chains of isolated helices, residues at the a, d, e, and g positions of the pentamer are substantially buried (≈78%); residues at the b and c positions are partly buried (≈13%); and the f positions remain completely exposed. Approximately 2,170 Å2 of accessible surface area (43% of the total accessible surface area of the five helical monomers) is buried in the pentamer. As a result, the net hydrophobic stabilization energy estimated by the method of Eisenberg and McLachlan (23) would be -84.8 kcal·mol-1 for the pentamer, greater than the value derived from isothermal GdmCl-denaturation experiments but consistent with the lower hydrophobicity of Trp itself (Fig. 2c). Solvent-exposed electrostatic interactions may also contribute to the stabilization of the pentamer conformation. On the basis of distance criteria, charged or polar residues on the surface of the pentamer form eighteen interhelical g to b and c to e salt bridges and charge-stabilized hydrogen bonds (for example, K17-D19; Fig. 3c). A comparable number of electrostatic interactions can be discerned between adjacent pentamers in the crystal as well.
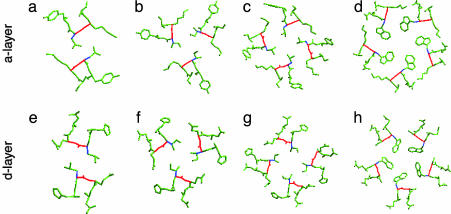
Knobs-into-Holes Packing. The Trp-14 pentamer interface shows the classical knobs-into-holes type of packing interactions between helices (8, 24), whereby each Trp indole ring at the a and d positions fits into a cavity formed by the spaces between side chains on a partnering helix. In the a layers, the Cα-Cβ bond of a Trp knob forms an ≈72° angle with the Cα-Cα vector at the base of the recipient hole (Fig. 4d). Similarly, the Trp side chains in the d layers pack with the Cα-Cβ bond of each Trp knob at position d pointing out of the interface at a 198° angle (Fig. 4h) (24, 25). Interestingly, the pentamer core in COMP, composed of 12 a and d layers of aliphatic residues (26), also shows similar packing geometry in the a and d layers (data not shown). Thus, this pattern may be independent of buried Trp residues and characteristic of parallel five-stranded coiled coils.
Fig. 4.
Four types of knobs-into-holes packing in parallel coiled coils (8, 24, 25). (a) Parallel packing at position a (180°) in the p1 dimer (32). The view is from the N terminus looking down the superhelical axis. The Cα-Cβ bond of each knob (blue) is oriented parallel to the Cα-Cα vector (red) at the base of the recipient hole on the neighboring helix. (b) Acute packing at position a (120°) in the pII trimer. (c) Perpendicular packing at position a (90°) in the pLI tetramer. (d) Packing at position a (72°) in the Trp-14 pentamer. (e) Perpendicular packing at position d (90°) in the dimer. (f) Acute packing at position d (150°) in the trimer. (g) Parallel packing at position d (180°) in the tetramer. (h) Packing at position d (198°) in the pentamer.
The packing geometry in the pentamer differs from that seen in dimeric, trimeric, and tetrameric leucine-zipper core variants. In the GCN4-p1 dimer, parallel and perpendicular knobs-into-holes packing occurs in the a and d layers, respectively (Fig. 4 a and e) (24, 25). This packing mode is reversed in the GCN4-pIL tetramer, which adopts perpendicular geometry in the a layers and parallel geometry in the d layers (Fig. 4 c and g) (24). On the other hand, the GCN4-pII trimer conformation places all of the a and d residues in acute geometry (Fig. 3 b and f) (25). There is an interesting geometrical relationship in the angles between the Cα-Cα and Cα-Cβ vectors at the a and d positions as the number of strands increases. In a layers, these angles are 180° × 2 = 360° for the dimer, 120° × 3 = 360° for the trimer, 90° × 4 = 360° for the tetramer, and 72° × 5 = 360° for the pentamer. In the d layers, these relationships switch: for the dimer, 270° - 180° (observed for a) = 90° (observed for d); for the trimer, 270° - 120° (observed for a) = 150° (observed for d); for the tetramer, 270° - 90° (observed for a) = 180° (observed for d); and for the pentamer, 270° - 72° (observed for a) = 198° (observed for d).
Helix-Helix Interactions. The linkage between Trp side chain packing and superhelix parameters is clearly evident in the Trp-14 structure despite the fact that the superhelical radius, pitch, and residues per turn and the radius of helix curvature all increase with the number of helices (8, 24, 25, 27). In contrast to the COMP pentamer that has a pitch value of 204 Å, the pitch of the Trp-14 supercoil is 277 Å (Table 2). This unusually large pitch allows Trp side chains to face toward the axis of supercoil rotation and mesh when the five α-helical chains interlock; recognition sites for intermolecular interactions must be positioned in relation to the exact pitch of the helix (28). Accordingly, the a-position orientation angle for the Trp-14 supercoil, ≈2°, is small as compared to its value of ≈20° in other coiled-coil proteins, producing an effective seam of interlocking Trp residues with their indole rings stably anchored in the hydrophobic core. As a consequence, the backbones of the five helices flatten out and wrap less tightly around the superhelical axis, resulting in a large spacing between adjacent helices (11.2 Å). These stereochemical details of the Trp residues that affect local packing are thus mirrored in the overall pentameric coiled-coil assembly and geometry.
Table 2. Helix-helix interactions in parallel coiled coils.
| GCN4 leucine-zipper variant* | |||||
|---|---|---|---|---|---|
| Parameter | Dimer | Trimer | Tetramer | COMP* pentamer | Trp-14 pentamer |
| Superhelix | |||||
| Supercoil radius, Ro, Å | 4.9 | 6.7 | 7.6 | 8.6 | 9.7 |
| Residues per supercoil turn, ωo | 100 | 118 | 139 | 140 | 190 |
| Supercoil pitch, Å | 148 | 175 | 205 | 204 | 277 |
| Radius of curvature, Å | 118 | 124 | 149 | — | 211 |
| Superhelix crossing angle, χ ° | −11.7 | −13.4 | −13.0 | — | −12.4 |
| Position a orientation angle, φ, ° | 21.6 | 20.4 | 19.8 | 19.5 | 2.0 |
| α-Helix | |||||
| Residues per α-helix turn, n | 3.62 | 3.60 | 3.59 | 3.58 | 3.60 |
| Rise per residue, d, Å | 1.51 | 1.53 | 1.52 | 1.52 | 1.49 |
| α-Helix radius (Cα), R1, Å | 2.28 | 2.24 | 2.26 | 2.20 | 2.06 |
| Pairwise helix-crossing angle, Ω, ° | 23.4 | 23.2 | 18.3 | 18.5 | 14.0 |
| Pairwise interhelix distance, D, Å | 9.8 | 11.5 | 10.6 | 10.2 | 11.2 |
Hydrophobic Channel. An axial view of the Trp-14 pentamer reveals a continuous central channel formed by spaces at the middle of each Trp layer and lined exclusively with indole rings (Fig. 5 a and b). The diameter of this channel varies from 7.2 to 9.0 Å, a size range of interest for a number of solutes in membrane channels such as maltoporin (29). During refinement, a strong string of electron density appeared on the coiled-coil pentamer axis between W7 and W28. This string is modeled as a PEG 400 molecule (a nonionic precipitant present in the crystallization buffer) because the central channel provides a snug fit to its alternating polar and nonpolar pattern of bonds. Indeed, the distances between the carbon and oxygen atoms of the modeled PEG 400 and the interacting Trp atoms seem optimal for van der Waals contacts (3.0-4.0 Å), although the higher B factor of the PEG 400 molecule (46.6 Å2 as compared with an average B factor for the protein of 28.9 Å2) implies that the polymer chains (which are heterogeneous in length) are flexible. Beyond the enclosed PEG 400 chain, five coordinated water molecules and a single water molecule are observed to be anchored to the W32 layer through hydrogen bonding to the five indole nitrogens (Fig. 5d). Another six sulfate ion-coordinated water molecules are positioned in the W39 layer such that favorable hydrogen-bonding interactions can occur between the indole nitrogens and the water molecules (Fig. 5e). The average B factor value of the structured water molecules, 35.4 Å2, is only slightly higher than the 20.8-Å2 B factor of the nearest protein groups. The crystal structure thus reveals that the pentameric channel formed from the parallel Trp-14 α-helices can interact effectively with both included PEG 400 and structured water molecules. Determining the structural and energetic consequences of varying solvents should further define the detailed properties of the channel.
Fig. 5.
Hydrophobic channel in Trp-14. (a) Axial view of the pentamer. The view is from the N terminus looking down the superhelical axis. The side chains of W7 (d) and W11 (a) are shown in green and red, respectively. A single PEG 400 molecule (pink) is located in the axial channel. (b) Molecular surface representation of the PEG 400 binding site. The solvent-accessible surface is colored according to the local electrostatic potential, ranging from +24 V in dark blue (most positive) to -32 V in deep red (most negative). (c) Side view showing the position of the buried PEG 400 molecule. A simulated annealing omit map covering the bound PEG 400 in a 2Fo - Fc difference Fourier synthesis was calculated with the ligand removed from the model and contoured at 1.0σ. (d) The 2Fo - Fc electron-density map at 1.0σ contour showing the hydrogen bonding network of structured waters in the W32 (a) layer. Water molecules are shown as red spheres, and hydrogen bonds are denoted by pink dotted lines. (e) The 2Fo - Fc electron-density map at 1.0σ contour showing a sulfate ion-mediated hydrogen bonding network of structured waters in the W39 (a) layer. The indole NH group in ≈35% of the Trp residues is hydrogen-bonded in the structure.
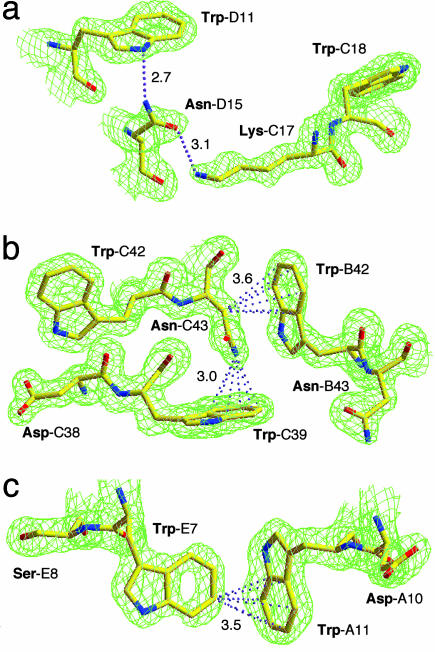
Role of Trp in Protein Structure. Analysis of the chemical groups that interact with the aromatic ring of Trp residues in the Trp-14 structure reveals a set of weak but specific interactions that have been implicated in stabilizing protein structure (30). First, conventional hydrogen bonding between the indole NH group of Trp and an acceptor (nitrogen, oxygen, or sulfur) atom likely contributes to specific packing of interfacial Trp side chains and organization of residues around them (Fig. 5 d and e and Fig. 6a). Second, C-H and N-H···π interactions involving the sixmembered ring of indole can be identified in the Trp-14 structure (e.g., Fig. 6b), consistent with the notion that the benzene ring of indole is the preferred cation-π binding site over the five-membered pyrrole-type ring (31). Third, interactions between aromatic C-H donor groups and aromatic π-acceptors may help define Trp interaction geometries in the pentamer interface (Fig. 6c). Thus, the engineered Trp-14 pentamer should provide a test ground for both experimental studies and computational simulations of the role of these interactions in protein structure, binding, and stability.
Fig. 6.
Stabilizing interactions of Trp side chains. (a) The 2Fo - Fc electron-density map (contoured at 1.2σ) showing imino hydrogen bonding. Hydrogen bonds are denoted by pink dotted lines. Hydrogen bond distances are given in Å. (b) The 2Fo - Fc electron-density map at 1.2σ contour showing presumptive C-H···π and N-H···π interactions. The distances between the carbon or nitrogen atom and the center of the Trp-π are given in Å. (c) The 2Fo - Fc electron-density map at 1.2σ contour showing π···π interactions.
Acknowledgments
We thank X. Yang and R. Abramowitz for support at beamline X4A. This work was supported by National Institutes of Health Grant AI511151 and the Irma T. Hirschl Trust.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PEG, polyethylene glycol; GdmCl, guanidinium chloride.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1T8Z).
References
- 1.Cohen, C. & Parry, D. A. (1994) Science 263, 488-489. [DOI] [PubMed] [Google Scholar]
- 2.Wolf, E., Kim, P. S. & Berger, B. (1997) Protein Sci. 6, 1179-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lupas, A. (1997) Curr. Opin. Struct. Biol. 7, 388-393. [DOI] [PubMed] [Google Scholar]
- 4.McLachlan, A. D. & Stewart, M. (1975) J. Mol. Biol. 98, 293-304. [DOI] [PubMed] [Google Scholar]
- 5.Parry, D. A. (1982) Biosci. Rep. 2, 1017-1024. [DOI] [PubMed] [Google Scholar]
- 6.Lupas, A., Van Dyke, M. & Stock, J. (1991) Science 252, 1162-1164. [DOI] [PubMed] [Google Scholar]
- 7.Hodges, R. S., Sodek, J., Smillie, L. B. & Jurasek, L. (1972) Cold Spring Harbor Symp. Quant. Biol. 37, 299-310. [Google Scholar]
- 8.Crick, F. H. C. (1953) Acta Crystallogr. 6, 689-697. [Google Scholar]
- 9.North, B., Summa, C. M., Ghirlanda, G. & DeGrado, W. F. (2001) J. Mol. Biol. 311, 1081-1090. [DOI] [PubMed] [Google Scholar]
- 10.Calladine, C. R., Sharff, A. & Luisi, B. (2001) J. Mol. Biol. 305, 603-618. [DOI] [PubMed] [Google Scholar]
- 11.Oakley, M. G. & Hollenbeck, J. J. (2001) Curr. Opin. Struct. Biol. 11, 450-457. [DOI] [PubMed] [Google Scholar]
- 12.Betz, S. F., Bryson, J. W. & DeGrado, W. F. (1995) Curr. Opin. Struct. Biol. 5, 457-463. [DOI] [PubMed] [Google Scholar]
- 13.Shu, W., Liu, J., Ji, H. & Lu, M. (2000) J. Mol. Biol. 299, 1101-1112. [DOI] [PubMed] [Google Scholar]
- 14.Liu, J. & Lu, M. (2002) J. Biol. Chem. 277, 48708-48713. [DOI] [PubMed] [Google Scholar]
- 15.Shu, W., Ji, H. & Lu, M. (1999) Biochemistry 38, 5378-5385. [DOI] [PubMed] [Google Scholar]
- 16.Pace, C. N. (1986) Methods Enzymol. 131, 266-280. [DOI] [PubMed] [Google Scholar]
- 17.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307-326. [DOI] [PubMed] [Google Scholar]
- 18.Terwilliger, T. C. & Berendzen, J. (1999) Acta Crystallogr. D 55, 849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997) Acta Crystallogr. D 53, 240-255. [DOI] [PubMed] [Google Scholar]
- 20.Adler, A. J., Greenfield, N. J. & Fasman, G. D. (1973) Methods Enzymol. 27, 675-735. [DOI] [PubMed] [Google Scholar]
- 21.Hendrickson, W. A. (1991) Science 254, 51-58. [DOI] [PubMed] [Google Scholar]
- 22.Lovell, S. C., Word, J. M., Richardson, J. S. & Richardson, D. C. (2000) Proteins 40, 389-408. [PubMed] [Google Scholar]
- 23.Eisenberg, D. & McLachlan, A. D. (1986) Nature 319, 199-203. [DOI] [PubMed] [Google Scholar]
- 24.Harbury, P. B., Zhang, T., Kim, P. S. & Alber, T. (1993) Science 262, 1401-1407. [DOI] [PubMed] [Google Scholar]
- 25.Harbury, P. B., Kim, P. S. & Alber, T. (1994) Nature 371, 80-83. [DOI] [PubMed] [Google Scholar]
- 26.Malashkevich, V. N., Kammerer, R. A., Efimov, V. P., Schulthess, T. & Engel, J. (1996) Science 274, 761-765. [DOI] [PubMed] [Google Scholar]
- 27.Seo, J. & Cohen, C. (1993) Proteins 15, 223-234. [DOI] [PubMed] [Google Scholar]
- 28.Pauling, L. & Corey, R. B. (1953) Nature 171, 59-61. [DOI] [PubMed] [Google Scholar]
- 29.Schirmer, T., Keller, T. A., Wang, Y. F. & Rosenbusch, J. P. (1995) Science 267, 512-514. [DOI] [PubMed] [Google Scholar]
- 30.Burley, S. K. & Petsko, G. A. (1985) Science 229, 23-28. [DOI] [PubMed] [Google Scholar]
- 31.Ma, J. C. & Dougherty, D. A. (1997) Chem. Rev. 97, 1303-1324. [DOI] [PubMed] [Google Scholar]
- 32.O'Shea, E. K., Klemm, J. D., Kim, P. S. & Alber, T. (1991) Science 254, 539-544. [DOI] [PubMed] [Google Scholar]