Abstract
Bacterial symbionts have long been suspected to be the true producers of many drug candidates isolated from marine invertebrates. Sponges, the most important marine source of biologically active natural products, have been frequently hypothesized to contain compounds of bacterial origin. This symbiont hypothesis, however, remained unproven because of a general inability to cultivate the suspected producers. However, we have recently identified an uncultured Pseudomonas sp. symbiont as the most likely producer of the defensive antitumor polyketide pederin in Paederus fuscipes beetles by cloning the putative biosynthesis genes. Here we report closely related genes isolated from the highly complex metagenome of the marine sponge Theonella swinhoei, which is the source of the onnamides and theopederins, a group of polyketides that structurally resemble pederin. Sequence features of the isolated genes clearly indicate that it belongs to a prokaryotic genome and should be responsible for the biosynthesis of almost the entire portion of the polyketide structure that is correlated with antitumor activity. Besides providing further proof for the role of the related beetle symbiont-derived genes, these findings raise intriguing ecological and evolutionary questions and have important general implications for the sustainable production of otherwise inaccessible marine drugs by using biotechnological strategies.
For almost 30 years, symbiotic bacteria have been discussed as the likely true producers of numerous natural products isolated from marine invertebrates (1). Particularly suggestive of a bacterial origin is the similarity of many compounds from sponges, tunicates, or bryozoans to complex polyketides and nonribosomal peptides, two groups of metabolites that are otherwise known exclusively from microorganisms (2, 3). Into these structural families fall some of today's most promising drug candidates, such as discodermolide, bryostatin 1, aplidine, and ET-743 (4). The existence of producing symbionts could have far reaching consequences for the development of sustainable, fermentation-based sources of invertebrate-derived drug candidates, almost all of which are currently inaccessible in large amounts. Several studies have tried to pinpoint the producers by using cultivation (5), cell separation (6), immunolocalization (7), or in situ hybridization (8) approaches. However, as none of these methods have so far provided unambiguous results, the true origin of the compounds still remains an enigma. We have recently used a genetic strategy to provide insights into natural product symbiosis in terrestrial insects (9). Rove beetles of the genera Paederus and Paederidus are the source of the highly active antitumor polyketide pederin (Fig. 1), which they use as chemical defense (10). We isolated a set of putative pederin biosynthesis genes (9, 11) from the metagenome of Paederus fuscipes beetles and traced them to a bacterial symbiont with the closest relationship to Pseudomonas aeruginosa (12). The genes encode a mixed modular polyketide synthase (PKS)-nonribosomal peptide synthetase (NRPS) system and were found to be distributed among two distinct regions of the symbiont genome (pedIJK and pedABCDEF in Fig. 2 A), which is a rare exception for bacterial secondary metabolites. Bacterial modular PKSs are giant enzymes containing a large number of variable catalytic domains that are organized into repeated sets of modules (13). Because each module usually incorporates one building block into the growing polyketide chain, the domain architecture of PKSs in most cases closely mirrors the structure of the assembled metabolite. Such a colinearity at the genetic and metabolic level was also found in the cloned symbiont genes, which strongly indicated their involvement in pederin biosynthesis. However, the present unculturability of the symbiont has so far precluded a final proof of gene functions by knockout. Intriguingly, metabolites with high structural similarity to pederin are also known from several different marine sponges (14). Many of these compounds exhibit extremely potent antitumor activity, and psymberin (15) [also known as irciniastatin A (16)] has recently been demonstrated to exhibit remarkably selective activity against solid tumor cell lines, which makes it a promising drug lead. The biological complexity of sponges can be enormous, because many species contain hundreds of distinct bacteria that can occupy up to 40% of their biomass (17). We suspected that the close relationships among pederin-type polyketides could be exploited to target specific biosynthetic genes and thus provide insights into natural product symbiosis in this marine animal group.
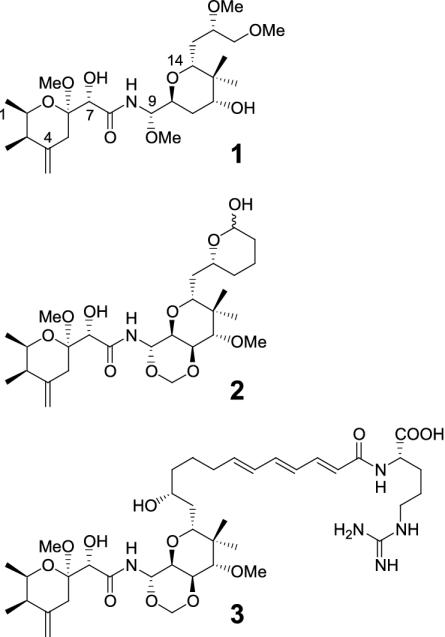
Fig. 1.
Some members of the pederin family of antitumor compounds. 1, Pederin from Paederus spp. rove beetles; 2, theopederin A from sponge T. swinhoei; 3, onnamide A from T. swinhoei.
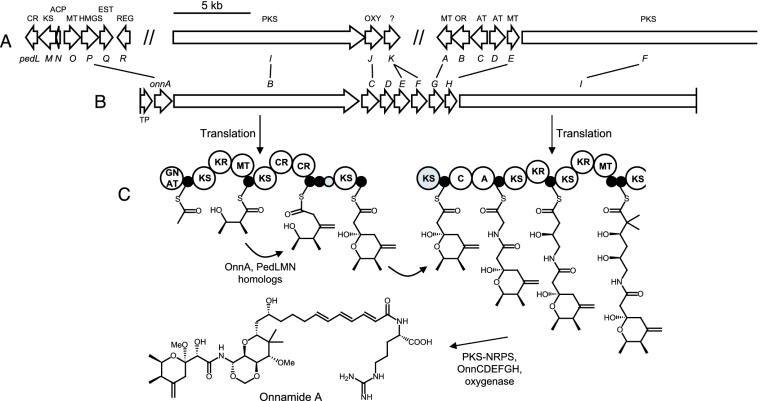
Fig. 2.
Comparison of the onn gene cluster with the ped system and proposed onnamide A biosynthesis. (A) Map of the ped cluster from the Pa. fuscipes symbiont. Double slashes separate the three genome regions. (B) Map of the onn cluster and correlation to ped homologs. (C) Protein products of the PKS-NRPS genes onnB and onnI and proposed biosynthetic pathway leading to onnamide/theopederin-type compounds. Each circle represents one domain. The intermediates are attached to ACP or, in the case of the second module of OnnI, peptidyl carrier protein (PCP) domains (shown as small, filled circles). Domains in gray lack active site motifs and are presumably nonfunctional. CR, CR superfamily; EST, esterase; REG, regulator; OXY, oxygenase; OR, oxidoreductase; TP, transposase; C, condensation domain; A, adenylation domain.
Materials and Methods
Sponge Collection and Workup. T. swinhoei Y and W specimens were collected by hand during scuba diving at Hachijo-jima Island, Japan, at a depth of 15 m. Immediately after collection they were either stored in 95% EtOH at 4°C (T. swinhoei Y1) or shock-frozen in liquid nitrogen, followed by storage at 80°C (T. swinhoei Y2 and W1). In addition, three freshly collected specimens (T. swinhoei Y3, Y4, and W2) were homogenized and subjected to cell separation by differential centrifugation as described previously (11), except that the mixture was not filtered and the pellets from four centrifugation steps at 5, 20, 400, and 1,700 × g were collected and stored in 95% EtOH at 4°C. DNA was isolated from these samples by a modified cetyltrimethylammonium bromide (CTAB) procedure. To each gram of separated cells or whole-sponge tissue ground in liquid nitrogen was added 5 ml of lysis buffer containing 100 mM Tris·HCl (pH 8), 1.4 M NaCl, 20 mM EDTA, 2 ml of CTAB solution at 55°C, 100 μl of 10% SDS, 350 μl of 100 mM diethyldithiocarbamate (DETC), 100 μl of mercaptoethanol, 6.3 g of polyvinylpyrrolidone, 10 mg of lysozyme, and 500 μg of proteinase K. The mixture was incubated at 30°C for 1.5 h and extracted once with 10 ml of chloroform, three times with equal volumes of phenol/chloroform, and twice with equal volumes of chloroform. DNA was precipitated from the aqueous phase by the addition of 1.2 volumes of isopropanol, washed with 10 ml of ice-cold EtOH, air-dried, and dissolved in water.
Analysis of Onnamide Content. The presence of onnamide A was checked from an EtOH extract of the sponges by liquid chromatography (LC)/electrospray ionization (ESI)/MS using a reversed-phase HPLC column and a gradient elution with aqueous methanol. The MH+ ion of onnamide A was used for identification.
Analysis of Ketosynthase (KS) Fragments from the Sponge Metagenome. KS fragments of PKS genes were amplified by PCR from the isolated DNA and sequenced as described previously (9). Sequences were placed into a phylogenetic PKS tree to assign them to functional types as described previously (18). Based on these data the primer pair sponge3f 5′-TGG AGC TCA GTT GGC AGG TA-3′ and sponge3r 5′-TGG TTT CAA CAG CAG CAT CAC-3′ was designed and used in diagnostic PCR to isolate the onn genes. For control PCRs, the primers sponge11f 5′-GCA TGA TGC TGG AGA CGA GCT G-3′ and sponge11r 5′-CGT CGA ACG CCT TGC ACT GC-3′ were used.
Isolation of the Onnamide Genes. For library construction, equal portions of each cell type fraction of the sponge T. swinhoei Y4 were combined. A cosmid library of ≈60,000 clones was constructed from the total DNA and screened as described previously (9). For PCR screening, the primers sponge3f and sponge3r were used. The isolated cosmid pTS1E4 was completely sequenced and analyzed as described previously (11).
Results
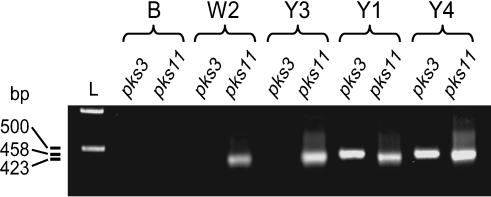
Identification of Putative Onnamide Biosynthesis Gene Fragments in the Metagenome of the Sponge T. swinhoei. To search for genes related to the pederin system, we selected the marine sponge T. swinhoei, a species with an exceptionally rich chemistry (14, 19). Japanese specimens occur in two distinct chemotypes that grow side by side in the same habitats at Hachijo-jima Island, Japan. The chemotype with a yellow interior (designated T. swinhoei Y) contains pederin-type metabolites among other biologically active polyketides and nonribosomal peptides, whereas specimens with a white interior (T. swinhoei W) harbor compounds unrelated to pederin. A survey of PKS genes in T. swinhoei Y1, an ethanol-preserved specimen, by PCR amplification of the KS domain regions of PKSs revealed a wide range of distinct sequences, which reflects the remarkable metabolic and microbial diversity of this sponge (18). The complexity of sponge metagenomes poses a serious technical problem in the search for specific biosynthesis genes. However, a phylogenetic analysis can provide useful information about PKS functions and thus greatly simplify the screening procedure. We have previously shown that the putative pederin PKS belongs to an evolutionarily distinct enzymatic group that we designated trans-AT PKSs (18). The PKS modules of these enzymes lack the acyltransferase (AT) domains that usually select the polyketide extension unit in each chain-elongation step (20). Instead, trans-AT systems contain discrete ATs encoded on one to three separate genes in the cluster. The AT-type of PKSs can be predicted by phylogenetic analysis of the corresponding KS sequences. Using this strategy, we isolated from the bacterial consortium of Pa. fuscipes beetles a set of putative pederin genes, pedIJK, that did not cluster with the remaining ped genes (Fig. 2) (11). The same approach revealed that within the large group of KS amplicons from the sponge metagenome only three sequences belonged to trans-AT PKSs (18). These sequences therefore represented good candidates for the onnamide/theopederin genes. Indeed, when the full sequence of pedI became available we discovered that the KS domain of the first extension module exhibited striking similarity (82%) to the translated protein of one of the trans-AT sponge sequences, TSY1-3. By inverse PCR we obtained an additional 1.5 kb adjacent to this sequence, which confirmed the absence of an AT domain and increased our confidence that we had selected the correct amplicon. Using specific primers for this sequence in diagnostic PCR experiments, we detected the gene in two of three T. swinhoei Y specimens, although it was absent in a T. swinhoei W sample (Fig. 3). A control PCR with primers based on the PKS sequence TSY1-11, which is present in both yellow and white sponge types, provided amplicons from all tested specimens. HPLC analysis revealed that the single TSY1-3-negative T. swinhoei Y3 was devoid of pederin-type compounds. This agreement at the genetic and metabolic levels strongly indicated that we had a PKS gene fragment from the onnamide producer in hand. To screen for the corresponding gene cluster, various DNA preservation and isolation protocols were evaluated. Among all procedures tried, only a cetyltrimethylammonium bromide (CTAB)-based protocol used with a sponge (T. swinhoei Y4) that had been dissociated into single cells and stored under ethanol yielded DNA of sufficient quality for cosmid library construction. Although the cell separation procedure had yielded fractions significantly enriched in specific cell types as judged by microscopy, PCR studies suggested that the DNA isolated from each cell type contained the TSY1-3 target sequence in similar concentrations (data not shown). This result may indicate that a part of the symbiont population could remain attached to sponge cells during the dissociation process or could even be located intracellularly. We therefore recombined equal portions of each cell fraction and used the isolated DNA for the construction of a metagenomic library in the vector pWEB. We expected this library to be extremely complex, because it contained genomes of several hundred different bacteria in addition to a large sponge genome (21). For the screening of such demanding libraries we have found that an approach based on the PCR analysis of clone pools produces reliable results (9). By this rapid procedure we were able to isolate a positive cosmid of ≈60,000 clones. This cosmid, designated pTS1E4, was completely sequenced.
Fig. 3.
PCR analysis of PKS genes in different specimens of T. swinhoei. B, blind control without template DNA; W2, T. swinhoei W2 (onnamide-free); Y3, T. swinhoei Y3 (onnamide-free); Y1 and Y4, two different onnamide-containing specimens of T. swinhoei Y; pks3, TSY1-3-specific PKS primers sponge3f/sponge3r; pks11, TSY1-11-specific PKS primers sponge11f/sponge11r.
Onnamide/Theopederin Biosynthesis Genes Belong to a Symbiotic Bacterium. By similarity searches and GeneMark (http://opal.biology.gatech.edu/GeneMark) prediction, 10 putative genes were identified in a 36.5-kb region with an overall GC content of 53.4%; these are shown in Fig. 2B and Table 1. All of the genes were devoid of internal stop codons and frameshifts and therefore appeared to be intact. As in the case of the pederin cluster, the genes clearly exhibited a bacterial architecture, because they lacked introns and polyadenylation sites and were preceded by putative Shine-Dalgarno sequences. An important indicator of the prokaryotic origin was the small intergenic distances between several genes, which would leave no room for promoters in the monocistronic mRNA of eukaryotes. For example, we measured 16 bp between onnB and onnC and 18 bp between onnE and onnF. A putative rho-independent transcriptional terminator with a perfect 18-bp inverted repeat was found directly downstream of onnF, which suggested that the corresponding mRNA transcript carried at least six genes. Moreover, we identified one ORF with high protein homology to bacterial transposases. Homology searches further revealed that 9 of the 10 ORFs (designated onn genes) strikingly resembled genes of the previously isolated ped system. Close homologs to PedA, PedE, PedG, PedI, PedJ, PedK, and a large portion of PedF were found, with protein similarity values ranging between 64% and 81%. The architectures of PedI and PedF are mirrored in OnnB and OnnI to an astonishing degree, which clearly shows that they encode the biosynthesis of closely related compounds. The domain structure of PedI is of especially high diagnostic value for pathway prediction, because it contains a GCN5-related N-acyltransferase (GNAT) and two crotonase (CR) superfamily domains, which were previously unique among PKSs (18). All three domains were also identified in OnnB at identical locations. One of the few architectural differences between the PKS genes isolated from both organisms is that onnB lacks a ketoreductase (KR) and the upstream low-homology region in module 4. However, the function of these elements in pedI has been unclear, because they do not correspond to any structural counterpart of pederin (18). They are therefore most likely an inactive evolutionary remnant, a feature that has been reported in several other PKS systems (13). Another presumably nonfunctional domain is the additional acyl carrier protein (ACP)-like region in module 3 of onnB, which lacks the strictly conserved serine residue necessary for polyketide biosynthesis (22). An important finding is that, in contrast to the disjointed structure of the ped genes, the onn homologs are clustered together. This confirms our previous prediction that the two ped regions belong to the same pathway and have been separated by genetic recombination (11). Several tailoring genes that should act at the post-PKS stage were also identified. These include the pedJ homolog onnC, which is likely responsible for hydroxylation at either C7 or C9, and the three methyltransferase (MT) genes onnD, onnG, and onnH, which should catalyze all three O-methylations of the sponge polyketides. Other homologs are onnE and onnF, which both resemble pedK with unknown function.
Table 1. Deduced functions of the ORFS identified in this work.
| Protein | Size, aa | Proposed function | Closest homolog, protein (origin) | S/I, %/% |
|---|---|---|---|---|
| TP | 140* | Transposase | MM1044 (M. mazei) | 72/52 |
| OnnA | 420 | HMGS-like activity | PksG (B. subtilis) | 85/71 |
| PedP (Pa. fuscipes symbiont) | 76/63 | |||
| OnnB | 4,359† | PKS (domains: GNAT, ACP, KS, KR, MT, ACP, KS, CR, CR, ACP, ACP, ACP‡, KS, ACP) | Pedl (domains: GNAT, ACP, KS, KR, MT, ACP, KS, CR, CR, ACP, ACP, KS, ?, KR, ACP) | 63/46 |
| OnnC | 386 | Oxygenase | PedJ | 81/65 |
| OnnD | 317 | MT | PedA | 68/50 |
| OnnE | 356 | Unknown | PedK | 66/50 |
| OnnF | 340 | Unknown | PedK | 71/55 |
| OnnG | 321 | MT | PedA | 70/53 |
| OnnH | 268 | MT | PedE | 71/52 |
| Onnl | 5,027* | PKS (domains: KS‡, ACP, C, A, PCP, KS, KR, ACP, KS, KR, MT, ACP, ACP, KS) | PedF (domains: KS‡, ACP, C, A, PCP, KS, KR, ACP, KS, KR, MT, ACP, KS, DH, DH, KR, ACP, KS, KR, ACP, KS, DH‡ | 64/49 |
| PedL | 251 | CR superfamily | PksH (B. subtilis) | 69/53 |
| PedM | 443 | KS | PksF (B. subtilis) | 56/35 |
| PedN | 80 | ACP | AcpK (B. subtilis) | 57/41 |
| PedO | 403 | MT | PedA | 68/54 |
| PedP | 430 | HMGS-like activity | PksG (B. subtilis) | 79/65 |
| OnnA (T. swinhoei symbiont) | 76/63 | |||
| PedQ | 314 | Esterase | PA2949 (Ps. aeruginosa) | 47/30 |
| PedR | 297 | LysR-type regulator | PA0784 (Ps. aeruginosa) | 64/49 |
onn genes were from T. swinhoei, and ped genes were from Pa. fuscipes. S/I, protein similarity/identity; M. mazei, Methanosarcina mazei; B. subtilis, Bacillus subtilis; GNAT, GCN5-related N-acetyltransferase; C, condensation; A, adenylation; PCP, peptidyl carrier protein; DH, dehydratase.
Fragment.
Size of 4,376 aa when measured from alternate start codon.
Domain lacking active site motifs.
Isolation of a Third Set of Pederin Genes from Pa. fuscipes Beetles. The only onn gene on pTS1E4 for which we did not find a ped counterpart was onnA, the protein product of which resembles 3-hydroxy-3-methylglutaryl-CoA synthases (HMGSs). In polyketide biosyntheses, these enzymes are discussed as being responsible for the generation of unusual one- or two-carbon branches at β-positions of former acyl-enzyme intermediates (”β-branches”) (23), such as the exomethylene group at C4 in pederin and the onnamides. Because an HMGS gene had not been among the previously isolated ped genes and we could not amplify such a gene with degenerate PCR primers from the total DNA, we had initially suspected that the unprecedented CR domains in pedI are solely responsible for exomethylene group formation in pederin (11). However, we recently sequenced the Pa. fuscipes symbiont genome to a coverage of ≈95% (unpublished data), which allowed us to search for possible HMGS homologs that might have been missed. Indeed, such an analysis revealed an ORF encoding an HMGS-like protein that is most similar to OnnA and, directly upstream, a methyl transferase gene with highest homology to pedA (Fig. 2 A). In addition, stand-alone KS, ACP, and CR genes, as well as an esterase gene and an ORF encoding a putative LysR-type regulator, were found in this region. These homologies suggest that we had identified an additional set of pederin biosynthesis genes presumably involved in exomethylene group formation and O-methylation. The fragmentation of the Pseudomonas sp. ped system into three distinct genome regions is most unusual for bacterial PKS pathways but correlates with the large number of transposase pseudogenes that frame the two previously isolated ped regions (9, 11, 12).
Discussion
The aim of this study was to trace the biosynthetic origin of antitumor polyketides of the onnamide series in the sponge T. swinhoei. Among sponges, this animal contains an unusually complex microbial community (21) and exceptionally rich chemistry (24) and is therefore a challenging but appropriate model organism to study natural product symbiosis. A phylogeny-guided approach has allowed us to isolate a set of natural product biosynthesis genes from the sponge metagenome. Several architectural as well as functional features clearly show that the host of these genes is indeed a bacterium, including gene clustering, lack of promoters, polyadenylation sites and introns, and the presence of putative Shine-Dalgarno sequences and a bacterial transposase gene. A recent study has demonstrated the horizontal transfer of a gene cluster from a symbiotic Wolbachia sp. bacterium to the X chromosome of its beetle host (25). Although this is the only known case of such a gene transfer, we cannot exclude the possibility that similar mechanisms exist in sponges with a dense bacterial community, which would lead to bacterial-like gene architectures on the sponge chromosome. However, in the Wolbachia case many genes were degraded, and it is unclear whether any of the ORFs were functional. In contrast, the presence of the polyketide products in the sponge requires that polycistronic (in the onnA to onnF section, at least hexacistronic), bacterial-type mRNA is transcribed from the sequenced region and recognized by the translational machinery of the animal. Polycistronic transcription in eukaryotes is very rare, and involvement of more than two genes is, to our knowledge, restricted to nematodes and trypanosomes, but the architecture and processing mechanism significantly differ from bacteria (26). In particular, neither cleavage and polyadenylation-specificity-factor binding sites nor U-rich elements required for trans splicing, which would be expected for eukaryotic premRNA processing (27), were found on the sequenced cosmid. These different lines of evidence therefore all point to a bacterium as the carrier of the PKS genes. Further cloning and sequencing of phylogenetically relevant genes outside the PKS system will be necessary to determine whether the symbiont is a member of the genus Pseudomonas as shown for the pederin producer. Because our data indicate that pederin-type genes have been subject to horizontal gene transfer (12), production of the compounds may well be independent from bacterial taxonomy. So far, data about the microbiology of T. swinhoei exist only for a specimen collected at Palau (21). By 16S rRNA analysis, taxonomically diverse but highly sponge-specific bacteria were found that are largely unrelated to species from other habitats, including an entire candidate phylum described as “Poribacteria” (28). It will be interesting to see whether the different chemistry of the Palauan specimen and our Japanese T. swinhoei is also reflected in the composition of a part of the symbiotic consortium.
The striking architectural similarity and remarkable homology between the onn and ped systems provide clear evidence that the cloned region is involved in the biosynthesis of one or more members of the onnamide/theopederin series. Several mixed polyketides/nonribosomal peptides in addition to the pederin-type compounds have been reported from T. swinhoei Y, including members of the aurantoside series (29), the pseudotheonamides, and orbiculamide A (24). None of these natural products features structural elements that match the modular architecture of the cloned genes. The aurantosides are highly repetitive chlorinated polyenes with a single terminal asparagine unit, whereas the remaining compounds contain almost exclusively amino acid moieties and should be assembled by multi-modular NRPS systems. In contrast, each of the >20 identified domains can be reasonably correlated to structural counterparts of onnamide and the theopederins. The domain architecture perfectly matches the region between C1 and C14, which corresponds to almost the entire portion in common with the pederin molecule. The sequenced system shares almost every feature with the ped PKS and contains the unusual GCN5-related N-acyltransferase (GNAT) and CR domains, both MT domains, and an NRPS module that, according to a sequence motif in the adenylation domain (30), should incorporate a glycine unit into the polyketide chain (31). Of the genes that likely act at the tailoring stage we found all three MT genes expected for the O-methylations and at least one of the three oxygenase genes that should be required for the introduction of hydroxyl groups at C7, C9, and C11. The sequenced region lacks AT genes, homologs of the suspected exomethylene genes pedL, pedM, and pedN identified in this work, and the part of the PKS responsible for the final polyketide extension steps. Because the isolated cosmid contains only the 5′ part of onnI, we expect that the downstream region harbors the remaining genes. However, attempts to isolate this region by chromosome walking have not yet met with success because of the high complexity of the library. Cloning of these genes should also provide an answer to the interesting question of whether the broad range of onnamides and theopederins are generated by the same PKS system via promiscuous downstream processing mechanisms, as known from the pikromycin group of polyketides (32), or whether the onn cluster exists in various polymorphic versions in the sponge metagenome that belong to different bacteria. The isolation of downstream regions would also help to answer questions regarding pederin biosynthesis. Pederin differs from the sponge polyketides in its shortened terminus, which is presumably generated by oxidative cleavage of an elongated precursor (9). The ped cluster indeed contains an oxygenase gene inserted into a PKS region that precisely corresponds to the position of the cleaved terminus, followed by a larger number of seemingly superfluous modules. Cloning of the remaining onn genes would provide insights on whether the sponge symbiont lacks a homolog of this oxygenase, as would be expected for the biosynthesis of uncleaved polyketides, and what the biosynthetic function and evolutionary origin of the additional pederin modules might be.
Surprisingly, our data on the onn cluster have also revealed the presence of a third set of pederin genes. In addition to the HMGS-type gene pedP, several other genes were identified that putatively participate in exomethylene bond biosynthesis. Evidence for the role of PedL, PedM, and PedN in this pathway comes from the comparison of gene clusters involved in the biosynthesis of polyketides with β-branches, including mupirocin (23), the antibiotic TA (33), leinamycin (34), and the jamaicamides (35). All gene clusters contain a conserved, clustered set of genes encoding an HMGS, a variable number of CR genes (usually described as enoyl-CoA hydratases but most likely catalyzing different reactions), and a monofunctional KS and ACP. The pederin PKS differs from these systems only in that two CR domains are integrated into a PKS module (11). The MT found in addition to these proteins is the only enzyme that has no close homolog to onnamide MTs and could be responsible for the attachment of the two terminal O-methyl groups at C16 and C17 that are unique for pederin. With the entire set of ped genes available, sustainable sources for the production of pederin can now be created: 25 million beetles alone were killed to elucidate the structure of this compound (36).
This work, which to our knowledge is the first isolation of natural product biosynthesis genes from a marine invertebrate, confirms the long-suspected role of symbiotic bacteria in the production of marine polyketides and peptides. The isolated genes correspond to almost the entire region of the onnamide molecule that would be needed to obtain an antitumor-active marine compound according to structure-activity relationship studies (37). The clustered architecture of these pharmacologically relevant genes has important consequences not only for the production of known and novel pederin-type anticancer agents but also for marine biotechnology and ecology in general. It suggests that, by similar gene cloning and expression strategies, renewable and environmentally sound sources of other marine polyketides and nonribosomal peptides could be created. Even highly valuable animal specimens that have been collected only a single time should provide sufficient amounts of DNA for library construction. However, to provide a generally useful alternative to mass harvesting, several technical limitations have yet to be overcome, such as the current lack of efficient cloning strategies from complex libraries. As our knowledge about the molecular genetics of the bacterial secondary metabolism increases, many other biosynthetic pathways will likely become targetable, for example, by focusing on pathway-specific-tailoring enzyme families or on additional phylogenetic subgroups of PKSs and NRPSs. Our data also raise several fundamental biological questions. So far nothing is known about the evolution and mechanisms of natural product symbiosis. The close relationship of the pederin-producing beetle symbiont with the insect pathogen Ps. aeruginosa suggests that pederin symbiosis evolved from an infection of a Paederus spp. ancestor. For sponges, several scenarios can be envisioned. These and many other marine invertebrate groups are filter feeders that take up large amounts of food bacteria. Some of these microorganisms may be resistant to lysis, which would permit them to recolonize the tissue of each new sponge generation. It is unlikely that the onnamides impart a direct competitive advantage to the producer against other members of the bacterial consortium, because they exhibit only weak antibacterial activity. However, because these cytotoxic agents might chemically defend the host, selective sequestration mechanisms could have evolved that would also explain why onnamide producers are present in only one subgroup of a single sponge species, whereas the large majority of sponge symbionts seem to be ubiquitous according to 16S rRNA studies (21). As an alternative to a continuous sequestration of symbionts, onnamide-producing bacteria might have been acquired from the environment by an ancestral sponge and passed on vertically to the following generations. Because sequence data of the onn system have become available, mechanisms underlying marine natural product symbiosis can now be studied. The design of in situ hybridization probes and PCR primers will allow us to conduct comparative phylogenetic studies of hosts and symbionts and to trace bacteria in artificial feeding experiments.
What is the origin of the pederin-type biosynthetic pathways in these diverse animals? The close similarity and complexity of the ped and onn systems suggest that they are derived from a common ancestral gene cluster. As a direct symbiont or gene transfer between sponges and beetles is very unlikely, this cluster should have once belonged to a free-living bacterium. This idea raises the intriguing question of why metabolites of the pederin group, which are highly conspicuous in biological assays because of their potent activity, have never been found in a nonsymbiotic strain. One possible explanation is that if these compounds confer an advantage to animal hosts rather than to the producers themselves, the biosynthetic genes would be preferentially maintained in symbionts. Ecological studies on the effect of the polyketides on sponge epibionts or predators would help to clarify this issue.
Acknowledgments
This paper is dedicated to Professor Heinz G. Floss on the occasion of his 70th birthday. We thank K. Takada and E. Meier for technical assistance; U. Hentschel, W. E. G. Müller, B. S. Moore and E. W. Schmidt for discussion; C. R. Hutchinson and G. Pohnert for valuable comments on the manuscript; and W. Boland for support. This work was funded by the Deutsche Forschungsgemeinschaft, the Society of Chemical Engineering and Biotechnology (DECHEMA), the Japan Society for the Promotion of Science, and the Max Planck Society (to J.P.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PKS, polyketide synthase; NRPS, nonribosomal peptide synthetase; KS, ketosynthase; AT, acyltransferase; CR, crotonase; KR, ketoreductase; ACP, acyl carrier protein; MT, methyltransferase; HMGS, 3-hydroxy-3-methylglutaryl-CoA synthase.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY688304).
References
- 1.Piel, J. (2004) Nat. Prod. Rep. 21, 519-538. [DOI] [PubMed] [Google Scholar]
- 2.Faulkner, D. J. (2000) Nat. Prod. Rep. 17, 1-6. [DOI] [PubMed] [Google Scholar]
- 3.Schwarzer, D. & Marahiel, M. A. (2001) Naturwissenschaften 88, 93-101. [DOI] [PubMed] [Google Scholar]
- 4.Schwartsmann, G., da Rocha, A. B., Mattei, J. & Lopes, R. M. (2003) Expert Opin. Investig. Drugs 12, 1367-1383. [DOI] [PubMed] [Google Scholar]
- 5.Schmidt, E. W., Obraztsova, A. Y., Davidson, S. K., Faulkner, D. J. & Haygood, M. G. (2000) Mar. Biol. (Berlin) 136, 969-977. [Google Scholar]
- 6.Faulkner, J., Unson, M. D. & Bewley, C. A. (1994) Pure Appl. Chem. 66, 1983-1990. [Google Scholar]
- 7.Gillor, O., Carmeli, S., Rahamim, Y., Fishelson, Z. & Ilan, M. (2000) Mar. Biotechnol. 2, 213-223. [DOI] [PubMed] [Google Scholar]
- 8.Davidson, S. K., Allen, S. W., Lim, G. E., Anderson, C. M. & Haygood, M. G. (2001) Appl. Environ. Microbiol. 67, 4531-4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piel, J. (2002) Proc. Natl. Acad. Sci. USA 99, 14002-14007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kellner, R. L. L. & Dettner, K. (1996) Oecologia 107, 293-300. [DOI] [PubMed] [Google Scholar]
- 11.Piel, J., Wen, G., Platzer, M. & Hui, D. (2004) Chembiochem 5, 93-98. [DOI] [PubMed] [Google Scholar]
- 12.Piel, J., Höfer, I. & Hui, D. (2004) J. Bacteriol. 186, 1280-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rawlings, B. J. (2001) Nat. Prod. Rep. 18, 231-281. [DOI] [PubMed] [Google Scholar]
- 14.Bewley, C. A. & Faulkner, D. J. (1998) Angew. Chem. Int. Ed. 37, 2163-2178. [DOI] [PubMed] [Google Scholar]
- 15.Cichewicz, R. H., Valeriote, F. A. & Crews, P. (2004) Org. Lett. 6, 1951-1954. [DOI] [PubMed] [Google Scholar]
- 16.Pettit, G. R., Xu, J.-X., Chapuis, J.-C., Pettit, R. K., Tackett, L. P., Doubek, D. L., Hooper, J. N. A. & Schmidt, J. M. (2004) J. Med. Chem. 47, 1149-1152. [DOI] [PubMed] [Google Scholar]
- 17.Brantley, S. E., Molinski, T. F., Preston, C. M. & Delong, E. F. (1995) Tetrahedron 51, 7667-7672. [Google Scholar]
- 18.Piel, J., Hui, D., Fusetani, N. & Matsunaga, S. (2004) Environ. Microbiol. 6, 921-927. [DOI] [PubMed] [Google Scholar]
- 19.Fusetani, N. & Matsunaga, S. (1993) Chem. Rev. 93, 1793-1806. [Google Scholar]
- 20.Liou, G. F. & Khosla, C. (2003) Curr. Opin. Chem. Biol. 7, 279-284. [DOI] [PubMed] [Google Scholar]
- 21.Hentschel, U., Hopke, J., Horn, M., Friedrich, A. B., Wagner, M., Hacker, J. & Moore, B. S. (2002) Appl. Environ. Microbiol. 68, 4431-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaworski, J. G., Post-Beittenmiller, M. A. & Ohlrogge, J. B. (1989) Eur. J. Biochem. 184, 603-609. [DOI] [PubMed] [Google Scholar]
- 23.El-Sayed, A. K., Hothersall, J., Cooper, S. M., Stephens, E., Simpson, T. J. & Thomas, C. M. (2003) Chem. Biol. 10, 419-430. [DOI] [PubMed] [Google Scholar]
- 24.Matsunaga, S. & Fusetani, N. (2003) Curr. Org. Chem. 7, 945-966. [Google Scholar]
- 25.Kondo, N., Nikoh, N., Ijichi, N., Shimada, M. & Fukatsu, T. (2002) Proc. Natl. Acad. Sci. USA 99, 14280-14285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blumenthal, T. (1998) BioEssays 20, 480-487. [DOI] [PubMed] [Google Scholar]
- 27.Huang, T., Kuersten, S., Deshpande, A. M., Spieth, J., MacMorris, M. & Blumenthal, T. (2001) Mol. Cell. Biol. 21, 1111-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fieseler, L., Horn, M., Wagner, M. & Hentschel, U. (2004) Appl. Environ. Microbiol. 70, 3724-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsunaga, S., Fusetani, N., Kato, Y. & Hirota, H. (1991) J. Am. Chem. Soc. 113, 9690-9692. [Google Scholar]
- 30.Stachelhaus, T., Mootz, H. D. & Marahiel, M. A. (1999) Chem. Biol. 6, 493-505. [DOI] [PubMed] [Google Scholar]
- 31.Du, L. C., Sanchez, C., Shen, M., Edwards, D. J. & Shen, B. (2000) Chem. Biol. 7, 623-642. [DOI] [PubMed] [Google Scholar]
- 32.Xue, Y. Q. & Sherman, D. H. (2000) Nature 403, 571-575. [DOI] [PubMed] [Google Scholar]
- 33.Paitan, Y., Orr, E., Ron, E. Z. & Rosenberg, E. (1999) Microbiology 145, 3059-3067. [DOI] [PubMed] [Google Scholar]
- 34.Cheng, Y. Q., Tang, G. L. & Shen, B. (2003) Proc. Natl. Acad. Sci. USA 100, 3149-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edwards, D. J., Marquez, B. L., Nogle, L. M., McPhail, K., Goeger, D. E., Roberts, M. A. & Gerwick, W. H. (2004) Chem. Biol. 11, 817-833. [DOI] [PubMed] [Google Scholar]
- 36.Pavan, M. & Bo, G. (1953) Physiol. Comp. Oecol. 3, 307-312. [Google Scholar]
- 37.Narquizian, R. & Kocienski, P. J. (2000) in The Role of Natural Products in Drug Discovery, eds. Mulzer, R. & Bohlmann, R. (Springer, Heidelberg), Vol. 32, pp. 25-56. [Google Scholar]